Abstract
The interaction of xeroderma pigmentosum group A protein (XPA) and replication protein A (RPA) with damaged DNA in nucleotide excision repair (NER) was studied using model dsDNA and bubble-DNA structure with 5-{3-[6-(carboxyamido-fluoresceinyl)amidocapromoyl]allyl}-dUMP lesions in one strand and containing photoreactive 5-iodo-dUMP residues in defined positions. Interactions of XPA and RPA with damaged and undamaged DNA strands were investigated by DNA–protein photocrosslinking and gel shift analysis. XPA showed two maximums of crosslinking intensities located on the 5′-side from a lesion. RPA mainly localized on undamaged strand of damaged DNA duplex and damaged bubble-DNA structure. These results presented for the first time the direct evidence for the localization of XPA in the 5′-side of the lesion and suggested the key role of XPA orientation in conjunction with RPA binding to undamaged strand for the positioning of the NER preincision complex. The findings supported the mechanism of loading of the heterodimer consisting of excision repair cross-complementing group 1 and xeroderma pigmentosum group F proteins by XPA on the 5′-side from the lesion before damaged strand incision. Importantly, the proper orientation of XPA and RPA in the stage of preincision was achieved in the absence of TFIIH and XPG.
INTRODUCTION
The nucleotide excision repair (NER) is one of the major repair systems to remove a wide range of helix distorting lesions from DNA, including those formed by UV light, various environmental mutagens and certain chemotherapeutic agents (1–3). Defects in NER are associated with several human autosomal hereditary diseases (4,5). NER can be dissected into two partly overlapping subpathways: global genome NER (GG-NER), operating wide genome, and transcription-coupled repair (TC-NER), focusing on lesions in the transcribed strand of active genes (6). The only difference between TC-NER and GG-NER is their mode of damage sensing. Thereafter the two sub-pathways merge into a common multi-step reaction mechanism. Reconstitution of GG-NER with purified proteins on artificial DNA templates has revealed sequential damage detection, helix opening, dual incision of the damaged strand 5′ and 3′ to the lesion, release of the 24–32 nt oligonucleotide, gap filling DNA synthesis, and ligation. The recognition of damaged sites is crucial for successful repair. The complex consisting of xeroderma pigmentosum group C protein (XPC), human Rad23B protein (HR23B) and centrin 2 (CEN2) has been considered the damage sensing structure of GG-NER (7–9). XPC bound to the lesion then recruits the transcription factor II (TFIIH) to the site of the lesion (10), and XPD, XPB helicases of TFIIH partially open the DNA helix (11). The structure is fully opened upon recruitment of excision repair cross-complementing group 1 and xeroderma pigmentosum group F proteins (ERCC1–XPF), XPG, replication protein A (RPA) and xeroderma pigmentosum group A protein (XPA). The DNA incision 3′ to the damage is carried out by XPG, the one 5′ to the damage by ERCC1–XPF (12,13). The coordination of the assembly of the NER preincision complexes and the sequential individual reactions is achieved through multiple protein interactions (14). Following the removal of the damaged oligonucleotide, the gap is filled by the replication machinery, and DNA ligase I or DNA ligase III-XRCC1 seals the remaining nick (15). Although the overall NER mechanism is fairly well understood, details of the damage recognition and of the spatial orientation of proteins in preincision complex have not been resolved.
Here we analyzed the interaction of the two inherent participants of the NER process, XPA and RPA, with DNA structures that mimic DNA intermediates arising in the NER process. Some studies initially suggested XPA and RPA as primary damage sensors (16–18). Subsequently it became clear that XPA and RPA work at a later stage, after the action of XPC–RAD23B and before cleavage by ERCC1–XPF and XPG (11,19). To shed light on the unsolved question of the RPA and XPA roles in damaged DNA recognition and preincision complex assemblage, we investigated the strand specificity of these proteins to bind damaged and/or undamaged DNA strands to resolve the ‘topography’ of the preincision complex. We used photoaffinity labeling to reveal the binding loci of RPA and XPA on damaged DNA. By using lesion-mimicking photoreactive groups we have previously shown that the method could identify contacts of NER proteins with such photoreactive groups attached to nucleotide bases at the site of damage (20,21). However, it remained unclear of how protein factors of the NER system contacted with DNA regions around the lesion in the damaged and undamaged strands. To this end, we developed the method to indicate the spatial assembly of proteins in the preincision complex. We have constructed DNA duplexes and bubbled DNA structures bearing 5I-dUMP residue in different positions of damaged or undamaged strands and fluorescein group linked to uridine residue (Flu-dUMP) as the lesion (Figure 1). The choice of 5I-dUMP was motivated by its minimal effect on the structure of DNA double helix (22). The size of the studied DNA bubble was similar to the partially open region of DNA duplex under TFIIH action (23). We have combined in this study photocrosslinking and footprinting techniques to analyze protein–nucleic acid interaction of XPA and RPA with damaged DNA. We find that XPA and RPA bind cooperatively to the 5′-side of the damaged DNA duplex. Both RPA and XPA interact with the ss/dsDNA junction in the bubbled DNA structure on the 5′-side of the damaged strand. This positioning of XPA recruits ERCC1–XPF for the incision of the damaged strand (24).
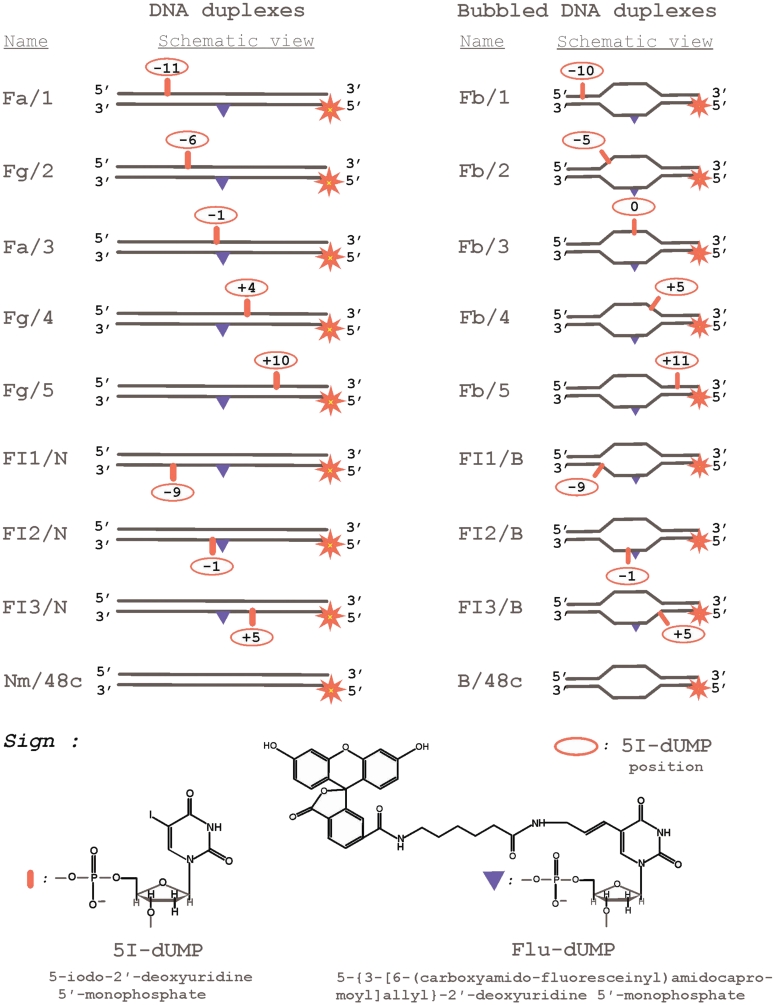
Figure 1.
DNA structures and nucleotide analogues used.
MATERIALS AND METHODS
Materials
[γ-32P]ATP (3000 Ci/mmol) was produced in the Laboratory of Radiochemistry of the Institute of Chemical Biology and Fundamental Medicine, Siberian Branch of the Russian Academy of Sciences; phage T4 polynucleotide kinase was purchased from Biosan (Russia); Escherichia coli exonuclease III (exoIII) and Mung Bean nuclease were products from SibEnzyme (Russia); DNase I and glycogen were obtained from Sigma (USA); stained molecular mass markers were from BioRad (USA), reagents for electrophoresis and buffer components from Sigma (USA) or made in Russia (extra pure grade). Oligonucleotides bearing 5I-dUMP or fluorescein dUMP derivative were synthesized by Dr V. Silnikov (Nanotech-C, Russia). Structures of oligonucleotides and nucleotide analogs are presented in Table 1 and Figure 1, respectively.
Table 1.
Sequences of the 48-mer oligonucleotides
| Name | Sequences |
|---|---|
| 1 | 5′-ctatggcgaggcgaItaagttgggcaacgtcagggtcttccgaacgac-3′ |
| 2 | 5′-ctatggcgaggcgattaagItgggcaacgtcagggtcttccgaacgac-3′ |
| 3 | 5′-ctatggcgaggcgattaagttgggIaacgtcagggtcttccgaacgac-3′ |
| 4 | 5′-ctatggcgaggcgattaagttgggcaacgIcagggtcttccgaacgac-3′ |
| 5 | 5′-ctatggcgaggcgattaagttgggcaacgtcagggIcttccgaacgac-3′ |
| FI1 | 5′-gtcgttcggaagaccctgacgFtacccaacItaatcgcctcgccatag-3′ |
| FI2 | 5′-gtcgttcggaagaccctgacgFIacccaacttaatcgcctcgccatag-3′ |
| FI3 | 5′-gtcgttcggaagacccIgacgFtacccaacttaatcgcctcgccatag-3′ |
| Fa | 5′-gtcgttcggaagaccctgacgtFacccaacttaatcgcctcgccatag-3′ |
| Fg | 5′-gtcgttcggaagaccctgacgtFgcccaacttaatcgcctcgccatag-3′ |
| Fb | 5′-gtcgttcggaagacccactgcaaFgggttgataatcgcctcgccatag-3′ |
| N | 5′-ctatggcgaggcgattaagttgggtaacgtcagggtcttccgaacgac-3′ |
| Nm | 5′-ctatggcgaggcgattaagttgggсaacgtcagggtcttccgaacgac-3′ |
| 48c | 5′-gtcgttcggaagaccctgacgttgcccaacttaatcgcctcgccatag-3′ |
| B | 5′-ctatggcgaggcgattatcaacccattgcagtgggtcttccgaacgac-3′ |
The modifications are indicated as follows: I – 5I-dUMP, F – Flu-dUMP
Protein purification
Recombinant hRPA was isolated from E. coli according to (25). The plasmid containing cDNA of hRPA was a kind gift of Dr K. Weisshart (Leibniz Institute for Age Research—Fritz Lipmann Institute, Jena, Germany). Recombinant hXPA bearing N-terminal polyhistidine fragment was expressed in E. coli BL21(DE3)LysS strain, using pETI5b-XPA recombinant plasmid kindly provided by Dr O. Schärer (SUNY Stony Brook, USA). Protein isolation was performed according to (26) with one modification: EDTA was not added during purification.
Preparation of 5′-32P-labeled DNA duplexes
Radioactive label was inserted into the 5′-end of oligonucleotides using phage T4 polynucleotide kinase as described in ref. (27). Labeled oligonucleotides were purified by electrophoresis under denaturing conditions with subsequent elution. To obtain DNA duplexes or bubble-DNA structures, 5′-32P-labeled oligonucleotides were annealed with overall or partial complementary oligonucleotides in the ratio 1:1, incubated for 5 min at 95°C, then slowly cooled to 70°C, kept for 15 min at this temperature, and slowly cooled to room temperature. The hybridization degree was monitored by electrophoresis in 10% polyacrylamide gel (acrylamide/bisacrylamide = 40:1). TBE buffer (50 mM Tris–HCl, 50 mM H3BO3, 1 mM EDTA, pH 8.3) was used as the electrode buffer.
Protein binding to DNA
Protein–DNA complexes were analyzed by gel retardation. The reaction mixture (10 µl) contained 50 mM Tris–HCl, pH 7.5, 100 mM KCl, 1 mM dithiothreitol, 0.6 mg/ml BSA, 10 nM 5′-32P-labeled DNA and RPA, XPA or both proteins at various concentrations. A 50-nM unlabeled DNA duplex was used as a competitor in some experiments. Protein complexes with DNA were formed for 20 min at 37°C. Then loading buffer (1:5 v/v) containing 20% glycerol and 0.015% Bromophenol Blue was added to the sample. Protein–nucleic acid complexes were electrophoresed under non-denaturing conditions. To separate the products of complex formation of RPA or XPA, 5% polyacrylamide gel (acrylamide/bis-acrylamide = 60:1) was used. TBE was the electrode buffer. Electrophoresis was performed with voltage decrease 17 V/cm and at 4°C. Positions of radioactively labeled oligonucleotide and protein–nucleic acid complexes were determined autoradiographically using a Molecular Imager FX Pro+ from BioRad.
Photoaffinity labeling
Protein modification by photoreactive DNA structures was performed in reaction mixture (10 µl) containing 50 mM Tris–HCl, pH 7.5, 100 mM KCl, 2 mM MgCl2 (if the reaction was performed in the presence of Mg2+), 1 mM dithiothreitol, 0.6 mg/ml BSA, 10 nM 5′-32P-labeled photoreactive DNA duplex and corresponding protein or protein mixture at the studied concentrations. Mixtures were incubated for 20 min at 37°C and then UV-irradiated for 1 h in an ice bath using a Bio-Link BLX-312 cross-linker from Vilber Lourmant (France), wavelength 312 nm, light intensity 5 mJ/cm2s. The reaction was terminated by 1:5 (v/v) dilution of the sample with stop buffer (5% SDS, 5% 2-mercaptoethanol, 0.3 M Tris–HCl, pH 7.8, 50% glycerol and 0.005% Bromophenol Blue). Modification products were separated by electrophoresis according to Laemmli (28) with subsequent autoradiography using the Molecular Imager FX Pro+.
Footprinting analyses
Reaction mixtures (10 µl) were prepared as in the gel retardation assay. After incubation at 37°C for 20 min, samples were mixed with one of the following nucleases. Samples with Mung Bean nuclease (20 U) were incubated at 37°C for 20 min. Samples containing MgCl2 (final concentration 2 mM) and exoIII (0.02 U) or DNase I (0.01 U) were incubated at 37°C for 2 min or for 4 min. The digestion reaction was terminated by addition of SDS and EDTA to give final concentrations of 0.05% and 0.05 M followed by rapid cooling to 0°C (in the case of Mung Bean nuclease) or heating to 95°C (for exoIII and DNaseI). Samples were adjusted to 50 µl by water, and proteins were extracted from water phase with two portions of phenol:chloroform (1:1). Then the aqueous phases were combined, and pH 4.0 was adjusted by the addition of 0.5 M NaAc buffer. DNA was precipitated with ethanol in the presence of 20 µg glycogen at −40°C. The DNA samples were then dissolved in loading solution (90% formamide, 50 mM EDTA, 0.1% xylene cyanole and 0.1% Bromophenol Blue), heated for 5 min at 95°C and subjected to 10% denaturing polyacrylamide gel (acrylamide/bis-acrylamide = 19:1) electrophoresis and autoradiography.
RESULTS
Proteins XPA and RPA bind to both damaged and undamaged strands of damaged DNA duplex
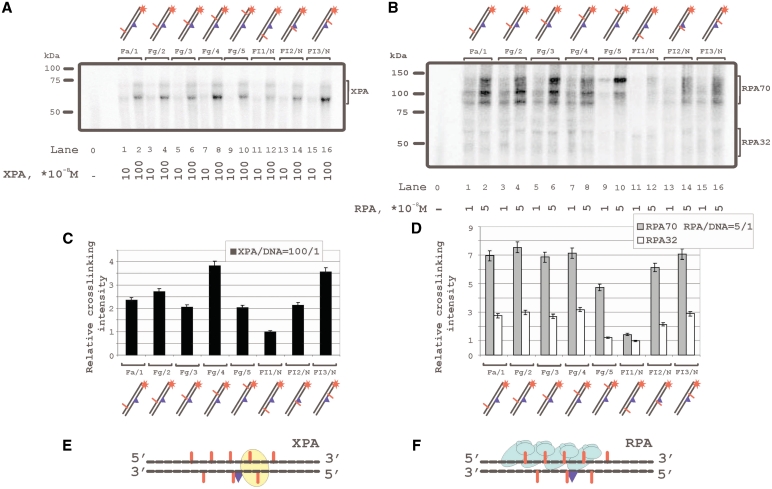
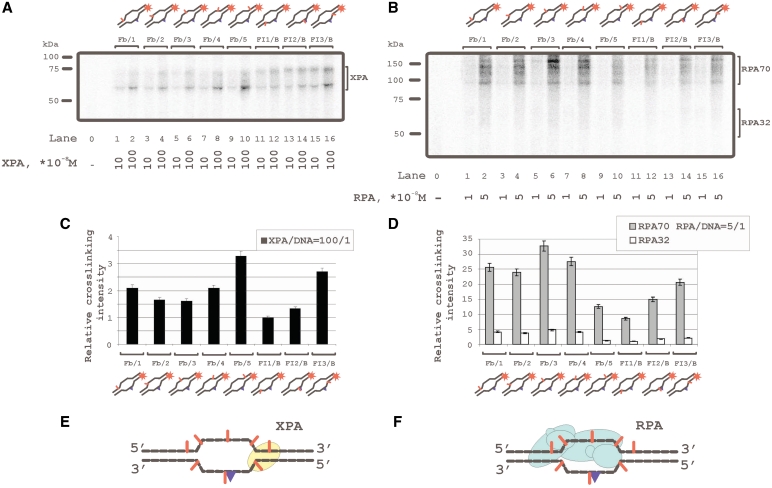
Despite numerous data on the interaction of XPA and RPA with damaged DNA, their localization on damaged and/or undamaged DNA strands remained unclear. We have used 48-mer DNA duplexes bearing fluorescein residue as the damage recognized by the NER system and 5I-dUMP residue as the photoreactive group for protein crosslinking. The 5I-dUMP can be regarded as non-damaging due to the similarity in dimension of iodine and the thymidine methyl group (17). Thus thymidine has been substituted in defined positions of the DNA strands by 5I-dUMP (Figure 1) that allowed footprinting of XPA and RPA by photocrosslinking and application to the recognition of damage and processing of damaged DNA. XPA was found crosslinked with 5I-dUMP in both damaged and undamaged strands (Figure 2A, C and E). The results show two positions with highest yield of DNA–protein adducts: one on the undamaged strand in the 5I-dUMP position of +4 (Figure 2A, lanes 7, 8; structure Fg/4 in Figure 1), and one on the damaged strand in the 5I-dUMP position of +5 (Figure 2A, lanes 15 and 16; structure FI3/N in Figure 1). Figure 2C shows relative efficiencies of XPA crosslinking versus 5I-dUMP position and Figure 2E shows the position of XPA. The RPA70 subunit crosslinked efficiently with undamaged strand but had also at least two contacts with damaged strand (Figure 2B, lanes 13–16). One of these contact positions coincided with the one for XPA, another one locates in the position 3′ adjacent to damage (Figure 2D). The lowest yield of crosslinking was observed for the photoreactive group in the damaged strand closest to the 3′ end (Figure 2B, lanes 11 and 12). The RPA32 subunit was photocrosslinked in the same manner as RPA70 although significantly less efficiently. The results in Figure 2B, D and F indicated photocrosslinking of RPA to the undamaged strand in various positions spaced over several nucleotide residues. The phenomenon could be attributed to different RPA molecules binding cooperatively to the damaged DNA duplexes. This was confirmed by the results in previous studies (26,29) and also in following experiments shown in Figure 6 and 7.
Figure 2.
Topography of the XPA and RPA protein location on damaged DNA duplex by photoaffinity labeling. The damaged DNA model substrates carry photoreactive 5I-dUMP-substitutions at the indicated positions of radioactively 5′-end labeled damaged or undamaged strands. The reaction mixtures (10 μl) contained 50 mM Tris–HCl 7.5, 100 mM KCl, 1 mM DTT, 0.6 mg/ml BSA, 10 nM 5′-32P-labeled photoreactive DNA and protein factors at analyzed concentrations: (A) 10 or 100 × 10−8 M XPA; (B) 1 or 5 × 10−8 M RPA. The photocrosslinking products were separated by SDS–PAGE and visualized by autoradiography. The (C) and (D) panels shows quantitative analysis of the data from the (A) and (B) photocrosslinking experiments. (C) Diagram of the relative intensities of the XPA photocrosslinked products. (D) diagram of the relative intensities of the RPA photocrosslinked products. Averages and experimental errors were taken from three experiments. (E) and (F) localization of XPA and RPA, respectively, on damaged DNA duplex (in accordance with photocrosslinking intensity maximums).
Figure 6.
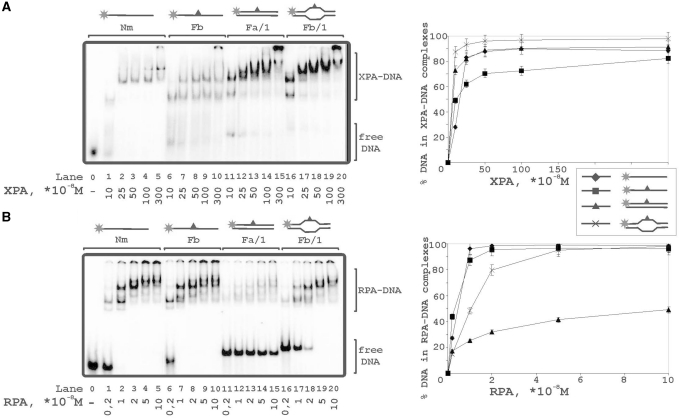
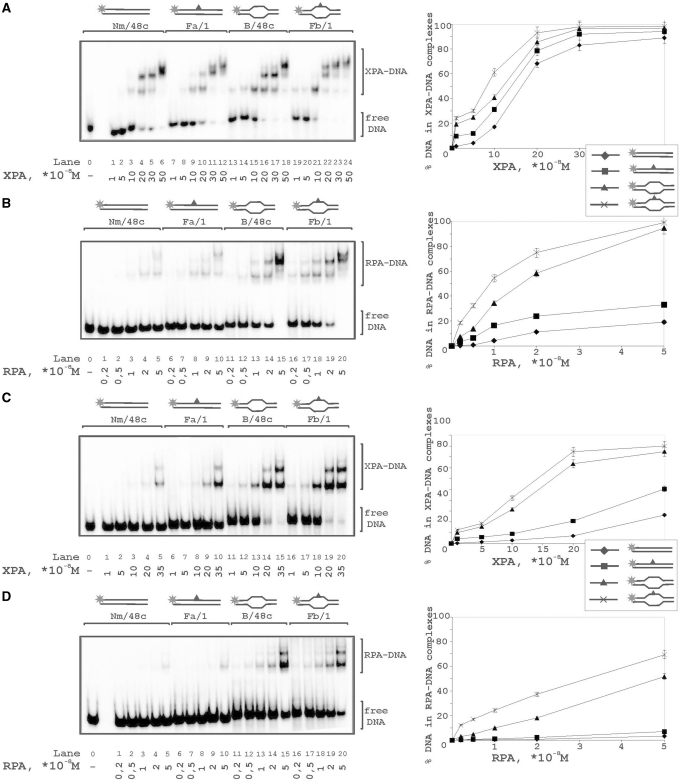
Binding of XPA and RPA to various types of DNA structures. The reaction mixtures (10 μl) contained 50 mM Tris–HCl 7.5, 100 mM KCl, 1 mM DTT, 0.6 mg/ml BSA, 10 nM 5′-32P-labeled DNA structure (lanes 0–5: undamaged ssDNA; lanes 6–10: damaged ssDNA; lanes 11–15: damaged DNA duplex; lanes 16–20: damaged DNA duplex with bubble) and protein factors at analyzed concentrations: (A) 10, 25, 50, 100 or 300 × 10−8 M XPA; (B) 0.2, 1, 2, 5 or 10 × 10−8 M RPA. The right panels show quantitative analysis from the A and B experiments. Bars indicate error of five independent sets of experiments.
Figure 7.
Comparative analysis of XPA and RPA binding to damaged and undamaged DNA. Increasing amounts of XPA (A) or RPA (B) were added to undamaged DNA duplex (lanes 1–5), damaged DNA duplex (lanes 6–10), undamaged DNA duplex with 15 nt bubble (lanes 11–15), or damaged bubble (lanes 16–20). A 5-fold excess of non-radioactive native 48-mer duplex was added in the reaction mixtures to compete binding of the studied DNA structures with XPA (C) or RPA (D). The right panels show quantitative analysis from the A, B,C and D experiments. Bars indicate error of five independent sets of experiments.
Similar crosslinking profiles for both RPA and XPA were obtained with bubbled DNA structures (Figure 3). These imitate DNA intermediates at the preincision step of NER. Data on the topography of these complexes have not been reported before. We have selected the size of bubble equal to 15 nt in agreement with the size of bubble opened by TFIIH in the damaged DNA duplex (23). In the case of bubbled DNA the maximum of XPA crosslinking with undamaged strand shifted to the ultimate 5I-dUMP on the 3′-side (compare corresponding lanes in Figures 2A and 3A) whereas that in the damaged strand remained in the site seen before in Figure 2. Both positions with maximum XPA crosslinking located near the ss/dsDNA junction as indicated in Figure 3E. This position of XPA protein was in agreement with the one found previously for XPA binding with branched DNA (30). The pattern of RPA crosslinking with duplex and bubbled DNA did not differ; however, levels of RPA crosslinking to DNA were higher with bubbled DNA. Thus, both XPA and RPA crosslinks located to the ss/dsDNA junction on the 5′-side from a lesion.
Figure 3.
Topography of the XPA and RPA protein location on damaged DNA duplexes containing 15-nt bubble by photoaffinity labeling. The damaged DNA model substrates carry photoreactive 5I-dUMP-substitution at the indicated positions of radioacetively labeled damaged or undamaged strands. The reaction mixtures (10 μl) contained 50 mM Tris–HCl 7.5, 100 mM KCl, 1 mM DTT, 0.6 mg/ml BSA, 10 nM 5′-32P-labeled photoreactive DNA and protein factors at analyzed concentrations: (A) 10 or 100 × 10−8 M XPA; (B) 1 or 5 × 10−8 M RPA. The photocrosslinking products were separated by SDS–PAGE and visualized by autoradiography. The (C) and (D) panels shows quantitative analysis of the data from the (A) and (B) photocrosslinking experiments. (C) diagram of the relative intensities of the XPA photocrosslinked products. (D) Diagram of the relative intensities of the RPA photocrosslinked products. Averages and experimental errors were taken from three experiments. (E) and (F) localization of XPA and RPA, respectively, on damaged DNA bubble (in accordance with photocrosslinking intensity maximums).
XPA and RPA protect DNA against nuclease digestion cooperatively
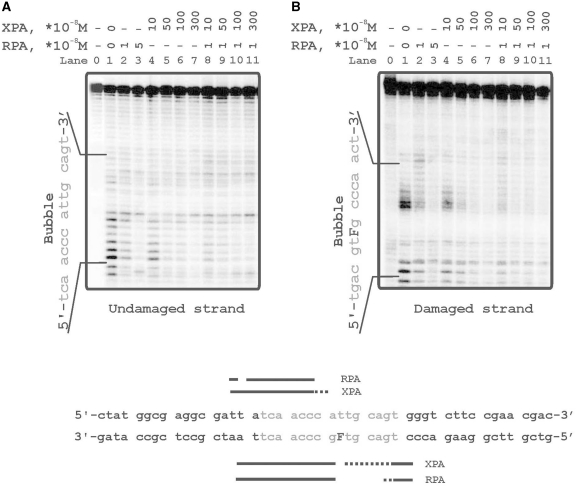
To confirm the contact of XPA and RPA with particular sites of damaged DNA structures we mapped their binding sites using footprinting assays. When DNAse I was used for footprinting, it did not reveal any XPA footprints in digestion profiles of DNA duplexes (data not shown), in agreement with previous data (31). In contrast, digestion with ExoIII revealed footprints on both undamaged (left panel) and damaged (right panel) strands of bubbled DNA in the presence of various concentrations of XPA, RPA either alone or in combination (Figure 4). ExoIII 3′→5′ exonuclease activity for DNA duplex, but significantly less activity toward ssDNA, explained the digestion patterns of the downstream regions flanking the bubble on both damaged and undamaged strand in Figure 4. RPA protected these sites more effectively than XPA, which showed low protection ability even at high excess of XPA over DNA (Figure 4, lanes 2 and 3 in comparison with lanes 6 and 7). XPA in high concentration slightly stimulated the DNA protection by RPA both on undamaged and damaged strands (Figure 4, lanes 10 and 11 in comparison with lane 2). Despite both XPA and RPA crosslinked preferentially with undamaged strand, footprinting experiments demonstrated no difference in protection of either strand. This discrepancy may be explained by the ability of crosslinking technique to fix unstable and transient Protein–DNA interactions, whereas the tight interactions were required to shield DNA against nuclease degradation.
Figure 4.
ExoIII footprinting analysis of RPA and XPA binding to the damaged DNA bubble. The reaction mixtures (10 μl) contained 50 mM Tris–HCl 7.5, 100 mM KCl, 1 mM DTT, 0.6 mg/ml BSA, protein factors at analyzed concentrations and 10 nM 5′-32P-labeled damaged DNA bubble substrates. Bases in the bubble are marked in grey. The left panel 5′-terminally labeled undamaged strand of DNA duplex. The right panel 5′-terminally labeled damaged strand of DNA duplex. The bottom panel shows schematic representation of the protection pattern for the bubbled substrate. Strongly and weakly protected regions are designated by solid and shaded bars, respectively.
To clarify further topography of the interaction of XPA and RPA with bubbled DNA structure we have used Mung Bean nuclease footprinting (Figure 5). One can see from these data that both XPA and RPA protect single stranded (ss) region more effectively as compared to duplex part (Figure 4). This result was expected for RPA but not for XPA because of lack of clear data on its affinity to ssDNA as well branched DNA (30,32,33). When both proteins were added simultaneously, a cooperative effect on DNA protection was observed in particular on damaged strand (Figure 5, right panel, lane 9 in comparison with lanes 1 and 5). The data indicated that each XPA and RPA showed low protection alone and cooperative protection when together. We also observed mutual effects by XPA and RPA on their interaction with damaged DNA by using DNA-crosslinking and gel shift assays (Supplementary Figure S1).
Figure 5.
Mung Bean footprinting analysis of RPA and XPA binding to the damaged DNA bubble. The reaction mixtures (10 μl) contained 50 mM Tris–HCl 7.5, 100 mM KCl, 1 mM DTT, 0.6 mg/ml BSA, protein factors at analyzed concentrations and 10 nM 5′-32P-labeled damaged DNA duplex. The left panel 5′-[32P] labeled undamaged strand of DNA duplex. The right panel terminally labeled damaged strand of DNA duplex. After the Mung Bean DNA digestion reaction mixtures was separated on 10% denaturing polyacrylamide gel. The bottom panel shows schematic representation of the protection pattern for the DNA substrate. Strongly and weakly protected regions are designated by solid and shaded bars, respectively.
Complex formation of RPA and XPA with damaged DNA substrates
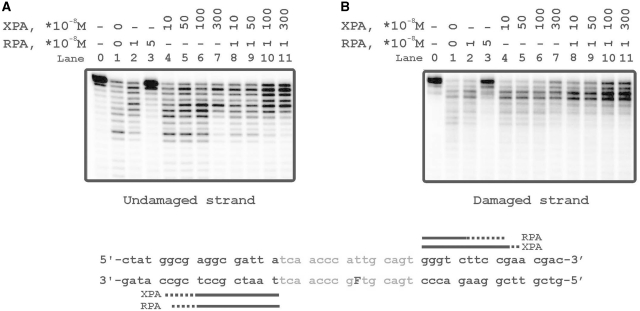
The photocrosslinking experiments demonstrated contacts of RPA and XPA with both strands of damaged DNA duplex with some preference in contacts with undamaged strand (Figures 2 and 3). Binding of these proteins was analyzed with (i) undamaged 48-mer ss oligonucleotide, (ii) its damaged version bearing fluorescein group as the lesion, (iii) damaged DNA duplex and (iv) damaged bubble-DNA (Figure 6). XPA showed the highest affinity to bubbled DNA (Figure 6A, lanes 16–20) and bound undamaged ssDNA in comparison with damaged DNA duplex with similar affinities (Figure 6A, lanes 1–5 and 11–15, respectively). RPA preferred ssDNA (Figure 6B, lanes 1–5 and 6–10) and bubbled DNA over damaged duplex DNA (Figure 6B, lanes 16–20 in comparison with lanes 11–15). While XPA bound more efficiently undamaged ssDNA than damaged ssDNA, RPA bound equally well and showed no selectivity for damaged ssDNA. Moreover, RPA located to damaged DNA duplex in a cooperative manner displaying numerous DNA–protein complexes of different mobility in the presence of free, uncomplexed DNA (Figure 6, lanes 11–15).
To compare the sensitivities of XPA and RPA to a lesion within a duplex or bubble we have analyzed binding of these proteins with both damaged and undamaged DNA structures. Both proteins demonstrated light preference in the binding of damaged DNA in comparison with undamaged structures (Figure7A and B). Although XPA demonstrated the highest affinity to damaged bubble, its preference for this structure was not substantial (protein concentrations for 50% binding level increase by factor of 1.5–2.0). To exclude non-specific interactions we performed binding experiments in the presence of 5-fold excess of undamaged non-radioactively labeled 48-mer duplex as a competitor (Figure7C and D). Differences in XPA binding to the various structures became more distinct in the presence of competitor, particularly the difference between DNA duplex and bubbled DNA (Figure7C, lanes 1–10 in comparison with 11–20). Surprisingly, RPA binding to bubble was inhibited by competitor in the same range as to duplex DNA (Figure7D). Using circular ds plasmid DNA as another competitor, no inhibition effect on RPA binding to bubble was observed (data not shown). Probably this result was due to the contribution of the duplex blunt ends in the RPA binding.
DISCUSSION
Both XPA and RPA belong to obligate protein factors required for NER activity in vivo as well as in reconstituted in vitro system (34,35). XPA, a zinc-finger containing protein, recognizes and binds to damaged DNA (36,37), the zinc-finger domain is also involved in a protein–protein interaction with RPA (38). XPA interacts with many of the core repair factors in NER and, without XPA, the stable preincision complex and NER are not assembled (11,39). Reduction of XPA to low levels in cells has been demonstrated to reduce NER activity and increase sensitivity to UV irradiation (40). Using reconstituted NER, XPA catalyzed the assembly of TFIIH and downstream NER factors towards the incision/excision of the damaged oligonucleotide (41). Taken together, XPA functions at several stages during NER performance. In addition, recent results suggest that XPA functions in DNA damage checkpoints and in cellular DNA damage responses, depending on its state of protein phosphorylation (42,43).
RPA is an evolutionarily conserved, heterotrimeric protein complex that binds and stabilizes ssDNA regions (25). The RPA heterotrimer binds ssDNA in several modes, with occlusion lengths of 8–10, 13–14 and 30 nt corresponding to global, transitional and elongated conformations of protein (44,45). In NER, RPA plays an integral role in damage recognition preceding the incision of the damage, and then again in post-excision DNA repair synthesis. RPA polarity appears crucial for positioning of the excision repair nucleases XPG and ERCC1–XPF on the DNA and RPA seems to bind in preincision complex in elongated form (46). However RPA-binding analysis and size of the open region of 13 nt generated by TFIIH (23) allow to assume a contribution of transitional conformation of RPA in NER preincision complex. Not only XPA but also RPA binds to damaged DNA with some degree of specificity (47–49), which is significantly enhanced by a cooperative interaction between these proteins (50). The XPA–RPA complex was the originally proposed factor responsible for primary damage recognition, but in vivo and in vitro studies later on led to the suggestion that recruitment of XPA to lesion sites occurred sometimes after TFIIH recruitment (51,52).
Based on our results, XPA and RPA will be essential for the assembly of the preincision complex by the following arguments: It is known that purified XPA protein exhibits specific binding affinities for certain kinked DNA substrates, namely three-way or four-way junctions (33,53) and ss/dsDNA junctions (30). This suggests that XPA would recognize a certain intermediate conformation of DNA that emerged during unwinding action by TFIIH helicases or during further events in the DNA damage processing. The open DNA duplex (bubble), which is formed as DNA is unwound around the damaged site, has both DNA bends and ssDNA regions and thus would be efficiently recognized by the XPA–RPA complex. In the present study we have analyzed XPA and RPA interaction with various types of DNA structures attributed to NER intermediates. Indeed, RPA and XPA bound bubbled DNA with affinities that were higher than those for binding DNA duplex. Footprinting analysis suggested time-limited contacts of both proteins, XPA and RPA, with ssDNA in the bubbled DNA structures, especially of RPA with intact ssDNA. On this basis we suggest only transient binding of XPA and RPA to ss regions of DNA intermediate that would be sufficient to protect ssDNA intermediates from the action of non-specific nucleases.
Our crosslinking experiments display higher yields of DNA–protein adducts of XPA and RPA with undamaged strand using both DNA duplex (Figure 2) and bubbled DNA (Figure 3). No data exist on XPA and (or) RPA crosslinking with bubble DNA structures. Our data on DNA duplex modification disagree with earlier published results (54) claiming RPA and XPA crosslinks mainly to 5I-dUMP residues in the damaged strand of cisplatin-modified 24-mer duplex. This difference is likely due to the use of different lengths and/or sequences of DNA duplexes and/or different types of lesions. Each can influence DNA secondary structure, resulting in changes of the DNA–protein binding surface. It has been thus recently shown for DNA bearing benzo[a]pyrene adduct that the structural distortions provoked on dsDNA by this adduct depended on sequence contexts (55). The important influence of the type of lesion on modulating duplex geometry has been reported (56). It should be noted that RPA crosslinking to undamaged strand is also demonstrated using DNA duplex with cholesterol lesion (22).
XPA interacted with damaged bubble asymmetrically, i.e. mainly with the ds/ssDNA junction toward the 5′-end of damaged strand (Figure 3). And XPA was located to the NER intermediate in the absence of protein partners. Surprisingly, XPA demonstrated asymmetric localization also on damaged DNA duplex although it binds more specifically with bubbled DNA (Figure 7C). The difference in XPA-binding affinity between duplex and bubbled DNA was much stronger than that between damaged and undamaged DNA structures supporting the view that XPA would be involved in NER process at a time following damaged DNA recognition and partial duplex opening. XPA asymmetric localization as well as RPA-binding polarity would play an important role in the ultimate asymmetry of preincision complex preceding the loading of ERCC1–XPF and XPG endonucleases.
It has been recently shown that protein–protein interactions between XPA and ERCC1 were specifically required for NER but not in the context of other DNA repair pathways (24). The endonuclease ERCC1–XPF incises the damaged strand of DNA in 5′-position to a lesion. XPA interacts with ERCC1 and recruits it to sites of damage. ERCC1–XPF binds and cleaves at junctions between ss and double-stranded DNA incising the 3′ssDNA overhang. Our data demonstrate for the first time that XPA localizes nearby ss/dsDNA junction that is positioned 5′ to a lesion. In this position XPA will initiate the assembly of the DNA preincision complex by recruiting ERCC1. The assembly will be followed by the incision in the damaged ssDNA towards the 3′ direction. Thus, binding of XPA is necessary for specific cleavage of the damaged strand by NER. This scenario is in agreement with reported 5′ ERCC1–XPF dependent incision of the DNA strand prior to the 3′ DNA strand incision by XPG (57). Despite numerous studies on XPA interactions with various DNA structures, there has been no direct evidence for its positioning 5′ to a lesion. It is important that the proper orientation of XPA and RPA in the stage of preincision is achieved in the absence of TFIIH. Thus, XPA binding mainly depends on position of the DNA damage relatively to ss/ds transition in the bubbled DNA. It is proposed that RPA contributes to the positioning of XPA at ss/dsDNA junction because RPA and XPA demonstrate cooperativity in their binding to such DNA structures. Although RPA locates almost at the same position as does XPA to the damaged DNA strand (Figure 3), it will bind to the undamaged DNA strand. Similar such a positioning activity by RPA is seen for the DNA polymerase dependent DNA synthesis after the cleavage of the damaged DNA strand. Together XPA and RPA play a structural role and ensure a proper 3D arrangement of the DNA intermediate for excision in addition of being involved in the DNA damage strand recognition. The proposed model of interaction of proteins within the preincision NER complex is shown in Supplementary Figure S2.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Foundation for Basic Research (grant numbers 08-04-91202, 10-04-00837); Russian Ministry of Education and Science [contract number 02.740.11.0079]; Program of Presidium of RAS ‘Molecular and Cellular Biology’; SB RAS grant for Young Scientists. Funding for open access charge: INTAS-SB RAS (The International Association for the Promotion of Co-operation with Scientists from the New Independent States of the Former Soviet Union – Siberian Branch of the Russian Academy of Science) grant, RFBR (Russian Foundation of Basic Research) grant.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are thankful to Prof. Dr Eggehard Holler, University of Regensburg, Germany, and the lab colleague Dr Pavel Pestryakov, for careful reading of the manuscript and useful comments.
REFERENCES
- 1.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 2.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 3.Schärer OD. Chemistry and biology of DNA repair. Angew. Chem. Int. Ed. Engl. 2003;42:2946–2974. doi: 10.1002/anie.200200523. [DOI] [PubMed] [Google Scholar]
- 4.Bootsma D, Kraemer KH, Cleaver JE, Hoeijmakers JH. Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. In: Vogelstein B, Kinzler KW, editors. The Genetic Basis of Human Cancer. NY: McGraw-Hill Book Co; 1998. pp. 245–274. [Google Scholar]
- 5.Berneburg M, Lehman AR. Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv. Genet. 2001;43:71–102. doi: 10.1016/s0065-2660(01)43004-5. [DOI] [PubMed] [Google Scholar]
- 6.Sweder KS, Hanawalt PC. Transcription-coupled DNA repair. Science. 1993;262:439–440. doi: 10.1126/science.8211165. [DOI] [PubMed] [Google Scholar]
- 7.Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 8.Volker M, Moné MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 9.Araki M, Masutani C, Takemura M, Uchida A, Sugasawa K, Kondoh J, Ohkuma Y, Hanaoka F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 2001;276:18665–18672. doi: 10.1074/jbc.M100855200. [DOI] [PubMed] [Google Scholar]
- 10.Yokoi M, Masutani C, Maekawa T, Sugasawa K, Ohkuma Y, Hanaoka F. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J. Biol. Chem. 2000;275:9870–9875. doi: 10.1074/jbc.275.13.9870. [DOI] [PubMed] [Google Scholar]
- 11.Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Donovan A, Davies AA, Moggs JG, West SC, Wood RD. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature. 1994;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- 13.Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei YF, Moggs JG, Carter KC, Shell BK, Evans E, de Jong MC, et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 14.Araújo SJ, Nigg EA, Wood RD. Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol. Cell. Biol. 2001;21:2281–2291. doi: 10.1128/MCB.21.7.2281-2291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Araújo SJ, Tirode F, Coin F, Pospiech H, Syväoja JE, Stucki M, Hübscher U, Egly JM, Wood RD. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 2000;14:349–359. [PMC free article] [PubMed] [Google Scholar]
- 16.Jones CJ, Wood RD. Preferential binding of the xeroderma pigmentosum group A complementing protein to damaged DNA. Biochemistry. 1993;32:12096–12104. doi: 10.1021/bi00096a021. [DOI] [PubMed] [Google Scholar]
- 17.Hey T, Lipps G, Krauss G. Binding of XPA and RPA to damaged DNA investigated by fluorescence anisotropy. Biochemistry. 2001;40:2901–2910. doi: 10.1021/bi002166i. [DOI] [PubMed] [Google Scholar]
- 18.Wakasugi M, Sancar A. Order of assembly of human DNA repair excision nuclease. J. Biol. Chem. 1999;274:18759–18768. doi: 10.1074/jbc.274.26.18759. [DOI] [PubMed] [Google Scholar]
- 19.Rademakers S, Volker M, Hoogstraten D, Nigg AL, Moné MJ, Van Zeeland AA, Hoeijmakers JH, Houtsmuller AB, Vermeulen W. Xeroderma pigmentosum group A protein loads as a separate factor onto DNA lesions. Mol. Cell. Biol. 2003;16:5755–5767. doi: 10.1128/MCB.23.16.5755-5767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maltseva EA, Rechkunova NI, Gillet LC, Petruseva IO, Schärer OD, Lavrik OI. Crosslinking of the NER damage recognition proteins XPC-HR23B, XPA and RPA to photoreactive probes that mimic DNA damages. Biochim. Biophys. Acta. 2007;1770:781–789. doi: 10.1016/j.bbagen.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Maltseva EA, Rechkunova NI, Petruseva IO, Vermeulen W, Schärer OD, Lavrik OI. Crosslinking of nucleotide excision repair proteins with DNA containing photoreactive damages. Bioorg. Chem. 2008;36:77–84. doi: 10.1016/j.bioorg.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Hermanson-Miller IL, Turchi JJ. Strand-specific binding of RPA and XPA to damaged duplex DNA. Biochemistry. 2002;41:2402–2408. doi: 10.1021/bi0112863. [DOI] [PubMed] [Google Scholar]
- 23.Tapias A, Auriol J, Forget D, Enzlin JH, Schärer OD, Coin F, Coulombe B, Egly JM. Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors. J. Biol. Chem. 2004;279:19074–19083. doi: 10.1074/jbc.M312611200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orelli B, McClendon TB, Tsodikov OV, Ellenberger T, Niedernhofer LJ, Schärer OD. The XPA-binding domain of ERCC1 is required for nucleotide excision repair but not other DNA repair pathways. J. Biol. Chem. 2010;285:3705–3712. doi: 10.1074/jbc.M109.067538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 26.Krasikova YS, Rechkunova NI, Maltseva EA, Petruseva IO, Silnikov VN, Zatsepin TS, Oretskaya TS, Schärer OD, Lavrik OI. Interaction of nucleotide excision repair factors XPC-HR23B, XPA, and RPA with damaged DNA. Biochemistry. 2008;73:886–896. doi: 10.1134/s0006297908080063. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Kim C, Wold MS. Recombinant human replication protein A binds to polynucleotides with low cooperativity. Biochemistry. 1995;34:2058–2064. doi: 10.1021/bi00006a028. [DOI] [PubMed] [Google Scholar]
- 30.Yang Z, Roginskaya M, Colis LC, Basu AK, Shell SM, Liu Y, Musich PR, Harris CM, Harris TM, Zou Y. Specific and efficient binding of xeroderma pigmentosum complementation group A to double-strand/single-strand DNA junctions with 3′- and/or 5′-ssDNA branches. Biochemistry. 2006;45:15921–15930. doi: 10.1021/bi061626q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maillard O, Camenisch U, Blagoev KB, Naegeli H. Versatile protection from mutagenic DNA lesions conferred by bipartite recognition in nucleotide excision repair. Mutat. Res. 2008;658:271–286. doi: 10.1016/j.mrrev.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Asahina H, Kuraoka I, Shirakawa M, Morita EH, Miura N, Miyamoto I, Ohtsuka E, Okada Y, Tanaka K. The XPA protein is a zinc metalloprotein with an ability to recognize various kinds of DNA damage. Mutat. Res. 1994;315:229–237. doi: 10.1016/0921-8777(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 33.Camenisch U, Dip R, Schumacher SB, Schuler B, Naegeli H. Recognition of helical kinks by xeroderma pigmentosum group A protein triggers DNA excision repair. Nat Struct. Mol. Biol. 2006;13:278–284. doi: 10.1038/nsmb1061. [DOI] [PubMed] [Google Scholar]
- 34.Mu D, Park CH, Matsunaga T, Hsu DS, Reardon JT, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 35.Cleaver JE, States JC. The DNA damage-recognition problem in human and other eukaryotic cells: the XPA damage binding protein. Biochem. J. 1997;328:1–12. doi: 10.1042/bj3280001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuraoka I, Morita EH, Saijo M, Matsuda T, Morikawa K, Shirakawa M, Tanaka K. Identification of a damaged-DNA binding domain of the XPA protein. Mutat. Res. 1996;362:87–95. doi: 10.1016/0921-8777(95)00038-0. [DOI] [PubMed] [Google Scholar]
- 37.Robins P, Jones CJ, Biggerstaff M, Lindahl T, Wood RD. Complementation of DNA repair in xeroderma pigmentosum group A cell extracts by a protein with affinity for damaged DNA. EMBO J. 1991;10:3913–3921. doi: 10.1002/j.1460-2075.1991.tb04961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikegami T, Kuraoka I, Saijo M, Kodo N, Kyogoku Y, Morikawa K, Tanaka K, Shirakawa M. Solution structure of the DNA- and RPA-binding domain of the human repair factor XPA. Nat. Struct. Biol. 1998;5:701–706. doi: 10.1038/1400. [DOI] [PubMed] [Google Scholar]
- 39.Mu D, Wakasugi M, Hsu D, Sancar A. Characterization of reaction intermediates of human excision repair nuclease. J. Biol. Chem. 1997;272:28971–28979. doi: 10.1074/jbc.272.46.28971. [DOI] [PubMed] [Google Scholar]
- 40.Koberle B, Roginskaya V, Wood RD. XPA protein as a limiting factor for nucleotide excision repair and UV sensitivity in human cells. DNA Repair. 2006;5:641–648. doi: 10.1016/j.dnarep.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Coin F, Oksenych V, Mocquet V, Groh S, Blattner C, Egly JM. Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol. Cell. 2008;31:9–20. doi: 10.1016/j.molcel.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 42.Bomgarden RD, Lupardus PJ, Soni DV, Yee MC, Ford JM, Cimprich KA. Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase eta on ATR checkpoint signaling. EMBO J. 2006;25:2605–2614. doi: 10.1038/sj.emboj.7601123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu X, Shell SM, Yang Z, Zou Y. Phosphorylation of nucleotide excision repair factor xeroderma pigmentosum group A by ataxia telangiectasia mutated and Rad3-related-dependent checkpoint pathway promotes cell survival in response to UV irradiation. Cancer Res. 2006;66:2997–3005. doi: 10.1158/0008-5472.CAN-05-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lavrik OI, Kolpashchikov DM, Weisshart K, Nasheuer HP, Khodyreva SN, Favre A. RPA subunit arrangement near the 3′-end of the primer is modulated by the length of the template strand and cooperative protein interactions. Nucleic Acids Res. 1999;27:4235–4240. doi: 10.1093/nar/27.21.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21:1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Laat WL, Appeldoorn E, Sugasawa K, Weterings E, Jaspers NG, Hoeijmakers JH. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev. 1998;12:2598–2609. doi: 10.1101/gad.12.16.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burns JL, Guzder SN, Sung P, Prakash S, Prakash L. An affinity of human replication protein A for ultraviolet-damaged DNA. J. Biol. Chem. 1996;271:11607–11610. doi: 10.1074/jbc.271.20.11607. [DOI] [PubMed] [Google Scholar]
- 48.Clugston CK, McLaughlin K, Kenny MK, Brown R. Binding of human single-stranded DNA binding protein to DNA damaged by the anticancer drug cis-diamminedichloroplatinum (II) Cancer Res. 1992;52:6375–6379. [PubMed] [Google Scholar]
- 49.Lao Y, Gomes XV, Ren Y, Taylor JS, Wold MS. Replication protein A interactions with DNA. III. Molecular basis of recognition of damaged DNA. Biochemistry. 2000;39:850–859. doi: 10.1021/bi991704s. [DOI] [PubMed] [Google Scholar]
- 50.He Z, Henricksen LA, Wold MS, Ingles CJ. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 51.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 52.Riedl T, Hanaoka F, Egly JM. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003;22:5293–5303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Missura M, Buterin T, Hindges R, Hubscher U, Kasparkova J, Brabec V, Naegeli H. Double-check probing of DNA bending and unwinding by XPA-RPA: an architectural function in DNA repair. EMBO J. 2001;20:3554–3564. doi: 10.1093/emboj/20.13.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schweizer U, Hey T, Lipps G, Krauss G. Photocrosslinking locates a binding site for the large subunit of human replication protein A to the damaged strand of cisplatin-modified DNA. Nucleic Acids Res. 1999;27:3183–3189. doi: 10.1093/nar/27.15.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kropachev K, Kolbanovskii M, Cai Y, Rodríguez F, Kolbanovskii A, Liu Y, Zhang L, Amin S, Patel D, Broyde S. The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived DNA lesions: insights into the structural factors that favor dual incisions. J. Mol. Biol. 2009;386:1193–1203. doi: 10.1016/j.jmb.2008.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hess MT, Schwitter U, Petretta M, Giese B, Naegeli H. Bipartite substrate discrimination by human nucleotide excision repair. Proc. Natl Acad. Sci. USA. 1997;94:6664–6669. doi: 10.1073/pnas.94.13.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, Giglia-Mari G, Clarkson SG, Vermeulen W, Schärer OD. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.