Abstract
Programmed ribosomal frameshifting is a translational recoding mechanism commonly used by RNA viruses to express two or more proteins from a single mRNA at a fixed ratio. An essential element in this process is the presence of an RNA secondary structure, such as a pseudoknot or a hairpin, located downstream of the slippery sequence. Here, we have tested the efficiency of RNA oligonucleotides annealing downstream of the slippery sequence to induce frameshifting in vitro. Maximal frameshifting was observed with oligonucleotides of 12–18 nt. Antisense oligonucleotides bearing locked nucleid acid (LNA) modifications also proved to be efficient frameshift-stimulators in contrast to DNA oligonucleotides. The number, sequence and location of LNA bases in an otherwise DNA oligonucleotide have to be carefully manipulated to obtain optimal levels of frameshifting. Our data favor a model in which RNA stability at the entrance of the ribosomal tunnel is the major determinant of stimulating slippage rather than a specific three-dimensional structure of the stimulating RNA element.
INTRODUCTION
Programmed ribosomal frameshifting is a translational recoding event that increases the versatility of gene expression. It is mainly utilized by eukaryotic RNA viruses (1–3), though some prokaryotic (4) and mammalian genes (5–7) are also controlled by ribosomal frameshifting. The requirements for −1 ribosomal frameshifting are the presence of a slippery heptanucleotide sequence X XXY YYZ (where X can be A, U, G or C; Y can be A or U; and Z does not equal Y; the spaces indicate the original reading frame) (8) followed by a downstream structural element, such as a pseudoknot, a hairpin or an antisense oligonucleotide duplex [for reviews, see (9)]. Although the mechanism of frameshifting is still elusive, a promising model has been proposed by Brierley and co-workers using cryo-electron microscopy to image mammalian 80S ribosomes (10). In their model, the ribosome is paused by its inability to unwind a pseudoknot structure resulting in a blockage of the A-site by eEF-2. During translocation, the P-site tRNA is bent in the 3′-direction by opposing forces. To release the tension, the P-site tRNA may un-pair and subsequently re-pair in the −1 frame with a certain frequency, followed by A-site tRNA delivery into the new −1 reading frame. These and other recent data obtained by mechanical unfolding of frameshifter pseudoknots suggest that mRNA secondary structures with certain conformational features that resist ribosomal helicase-mediated unwinding and eEF-2 catalyzed translocation are key players in ribosomal frameshifting.
Small oligonucleotides have been used for several years to regulate gene expression by RNaseH-dependent RNA degradation (11), blocking translation (12), or re-directing splicing (13). More recently, microRNAs (miRNAs) (14) and small interfering RNAs (siRNAs) have appeared on the scene of post-transcriptional gene regulation (15). siRNAs may be effective in treatment of chronic hepatitis-B virus infection (16), HIV infection (17), cancer (18) and age-related macular degeneration (19). Very few antisense oligonucleotides, for example against the bcl-2 oncogene have reached the stage of clinical trials (20) or have actually been approved by the FDA, for instance for the treatment of human cytomegalovirus retinitis (21).
Enhancing the stability of small oligonucleotides to prolong circulation and meanwhile increasing target specificity are major concerns for therapeutic applications. Various kinds of modifications in backbones, sugars or even analogs have already been studied extensively [for reviews, see (22,23)] to meet these requirements. Locked nucleic acid (LNA) is a rather novel nucleic acid analog comprising a class of bicyclic high-affinity RNA analogs in which the furanose ring of LNA monomers is conformationally locked in an RNA-mimicking C3′-endo/N-type conformation (24). The LNA modification also resists degradation by cellular nucleases. Furthermore, introducing LNA into DNA or RNA oligonucleotides improves the affinity for complementary sequences and increases the melting temperature by several degrees (25). A recent study showed that LNA/DNA mix-mers against miRNA-122 can be acutely administered at high dosage with long lasting effects without any evidence of LNA-associated toxicities or histopathological changes in the studied animals (26). These data suggests that LNA is a promising candidate for small oligonucleotide applications.
We and others have demonstrated that small RNA oligonucleotides are able to mimic the function of frameshifter pseudoknots or hairpins by redirecting ribosomes into new reading frames (27,28). In this article, we have investigated the length and concentration of RNA oligonucleotides for optimal frameshifting, as well as the effects of introducing LNA-type sugars in DNA oligonucleotides.
MATERIALS AND METHODS
Frameshift reporter construct and oligonucleotides
The −1 ribosomal frameshifting events were monitored by the SF reporter construct described earlier (27). Complementary oligonucleotides (Eurogentec, Liege, Belgium) SF462 (CTAGTTGACCTCAACCCTTGGAA) and SF463 (CATGTTCCAAGGGTTGAGGTCAA) and SF468 (CTAGTTGAGCGCGCTGGAGGCCATGG) and SF469 (CATGCCATGGCCTCCAGCGCGCTCA) were annealed and ligated into SpeI/NcoI digested SF reporter to construct the SF462 and SF468 templates, respectively. All constructs were verified by DNA sequencing on an ABI PRISM® 3730xl analyzer (LGTC, Leiden, The Netherlands). RNA oligonucleotides (except for RNA13 which was obtained from Invitrogen) were purchased from Dharmacon (Lafayette, USA). The RNAs from Dharmacon carried a 2′-O-ACE protection group, which was removed by incubation with 100 mM acetic acid pH 3.8 and TEMED at 60°C for 30 min. The sequences of RNA oligos were as follows:
RNA6: GCGCGC, RNA9: CCAGCGCGC, RNA12: CCUCCAGCGCGC, RNA15: UGGCCUCCAGCGCGC, RNA18: CCAUGGCCUCCAGCGCGC, 18RNA: GCGCGCUGGAGGCCAUGG, and RNA13: CCAAGGGGUUGAGG.
DNA and LNA/DNA mix-mers were synthesized by Eurogentec. Custom oligonucleotides were extracted by phenol/chloroform followed by ethanol precipitated before use. The sequences of DNA and LNA/DNA mix-mers were as follows (lower case represents the LNA modification and capital represents DNA): DNA18: CCATGGCCTCCAGCGCGC. DNA13: CCAAGGGTTGAGG, LNA2: CCATGGCCTCCAGCGCgc, LNA4: CCATGGCCTCCAGCgcgc, LNA6: CCATGGCCTCCAgcgcgc, LNA2-1: CCATGGCCTCCAGCgcGC, LNA2-2: CCATGGCCTCCAgcGCGC, LNA2-3: ccATGGCCTCCAGCGCGC, LNA2-4: CCATGGCCTCCAGCgCGc, LD1: CCAAGGGTTGAGg, LD2: CCAAGGGTTGAgg, LD4: CCAAGGGTTgagg, LD6: CCAAGGGttgagg.
In vitro transcription
Plasmids were linearized by BamHI and purified by phenol/chloroform extraction followed by ethanol precipitation. In vitro transcription was conducted by SP6 RNA polymerase and carried out in the 30 μl reaction mixture of: 1 μg of linearized template, 5 mM of rNTPs, 20 units of RNase inhibitor and 15 units of SP6 RNA polymerase with buffer (all from Promega, Benelux). After 2 h incubation at 37°C, the integrity and quantity of transcripts were checked by agarose gel and appropriate amount of the RNA were diluted in nuclease free water for in vitro translation.
In vitro translation
In vitro translations were carried out in nuclease treated rabbit reticulocyte lysate (RRL) (Promega). The amount of mRNA was 0.025 pmol and different amounts of oligonucleotides (0.025–15.625 pmol) were mixed with template for 20 min at room temperature. After incubation, 4 μl of RRL, 0.01 mM amino acids mixture except methionine, 2 μCi of 35S methionine (10 mCi/ml, MP Biomedicals, in vitro translational grade) were added in total volume of 10 μl and incubated at 28°C for 1 h. After translation, samples were mixed with 2× Laemmli buffer, boiled at 90°C for 5 min and resolved by 13% SDS polyacrylamide gels. Gels were fixed in 10% acetic acid and 30% methanol for 20 min, dried under vacuum, and exposed to phosphoimager screens (Biorad). The screen was scanned and the 0 frame and −1 frameshift protein products were quantified by Quantity One software (Biorad). Frameshift percentages were calculated by dividing the amount of −1 frameshift product by the amount of 0-frame and −1 frameshift products after correction for the number of methionines in the protein sequence, multiplied by 100.
Determination of the melting temperature of oligonucleotide duplexes
RNA oligonucleotide 18RNA (5′GCGCGCUGGAGGCCAUGG3′, Dharmacon, USA) was mixed in a 1:1 molar ratio with RNA18, DNA18 or one of the various DNA/LNA mix-mers, in UV-melting buffer (100 mM NaCl, 10 mM Cacodylate acid, pH 6.8). The analysis was performed on a Varian Cary 300 spectrophotometer using temperature ramps of 0.25°C /min during heating and cooling. The absorbance at 260 nm was recorded and normalized to the blank control.
RESULTS
Length-dependent RNA oligonucleotide-induced ribosomal frameshifting
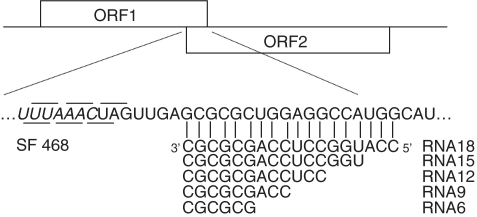
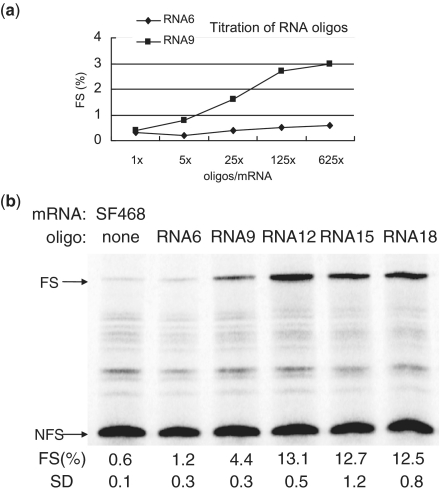
Although antisense oligonucleotides were found to induce ribosomal frameshifting (27,28), the optimal number of base pairs has not been addressed yet. To investigate this we designed antisense RNA oligonucleotides that are 6, 9, 12, 15 and 18 bases complementary to the region downstream of an UUUAAAC slippery sequence in our reporter plasmid SF468 (Figure 1). First, titration with RNA6 and RNA9 oligonucleotides revealed that a 625-fold molar excess of oligonucleotides over mRNA resulted in the highest level of frameshifting (Figure 2a); this ratio was used in the following experiments. The shortest oligonucleotide, RNA6, was not capable of inducing significant levels of frameshifting (Figure 2b), whereas RNA9 induced ∼3.5% of frameshifting. Maximum levels were obtained with RNA12, RNA15 and RNA18; all three induced ∼12% of frameshifting. In the following experiments oligonucleotides between 12 and 18 nt in length were used.
Figure 1.
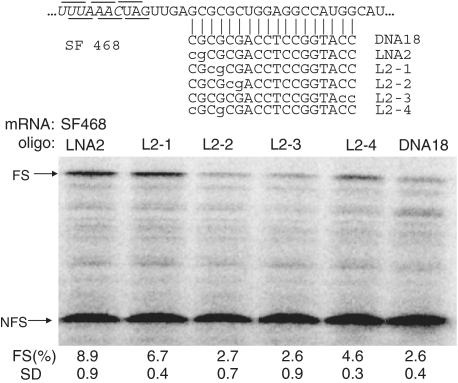
Schematic representation of frameshift reporter constructs. ORF1 (19 kD) is in the 0 frame and ORF2 (46 kD) is in the −1 translational frame with respect to ORF1. The appearance of the 65 kD fusion protein represents the occurrence of −1 frameshifting. The UUUAAAC slippery sequence is indicated in italics. The 0-reading frame and −1 reading frames codons are indicated above and below the sequences, respectively. RNA18, 12, 15, 9 and 6 are antisense RNA oligonucleotides complementary to the indicated region downstream of the slippery sequence in SF468 mRNA.
Figure 2.
Optimizing the ratio and lengths of frameshift-inducing RNA oligonucleotides. (a) An amount of 0.05 pmol of SF468 mRNA were mixed with 0.05, 0.25, 1.25, 6.25 and 31.25 pmol of RNA6 and RNA9 oligonucleotides, respectively. Mixtures were subsequently translated in the presence of 35S-methionine and the labeled proteins were examined by 13% SDS–PAGE. Frameshift efficiencies [FS (%)] were calculated after quantification and correction of in-frame and frameshifted product. (b) A total of 625 M excess of different lengths of RNA oligos (RNA6, 9, 12, 15, 18, respectively) and control without added oligonucleotide were mixed with 0.05 pmol of SF468 mRNA. Mixtures were translated and examined by 13% SDS–PAGE. In-frame and frameshifted protein products are indicated by NFS and FS, respectively. Frameshifting efficiency [FS (%)] and standard deviation (SD) of three independent duplicate assays are indicated below each lane.
LNA/DNA mix-mers induced-ribosomal frameshifting
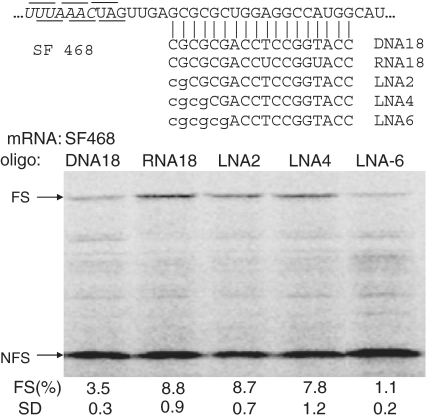
Since we have absent knowledge about the efficacy of LNA-induced ribosomal frameshifting, LNA/DNA mix-mers of 18 nt in length were designed to investigate this (Figure 3). A DNA oligonucleotide, as expected, was less capable (3.5%) of inducing frameshift due to the lower thermodynamic stability of RNA–DNA duplexes, see also below. Surprisingly, substituting the 3′-cytosine and guanosine in this DNA oligonucleotide by their LNA analogs enhanced its frameshift inducing capacity to 8.7%, i.e. as high as an RNA oligonucleotide (8.8%). Increasing the LNA content of this oligonucleotide further did not lead to higher frameshifting. On the contrary, the efficiency of LNA4 was with 7.7% lower than that of LNA2 and that of LNA6 was a mere 1.1%. Since the overall translation efficiency seemed not affected by LNA6 we suspected an effect of the oligonucleotide itself (see below).
Figure 3.
Effect of LNA substitutions on oligonucleotide-induced frameshifting. SF468 mRNA was translated in the presence of a 625-fold molar excess of DNA, RNA or LNA substituted DNA oligonucleotides in rabbit reticulocyte lysate. DNA and RNA oligonucleotides are indicated in capital and LNA substitutions are denoted by lowercase characters. See legend to Figure 2 for more details.
The effectiveness of LNA/DNA mix-mers is universal
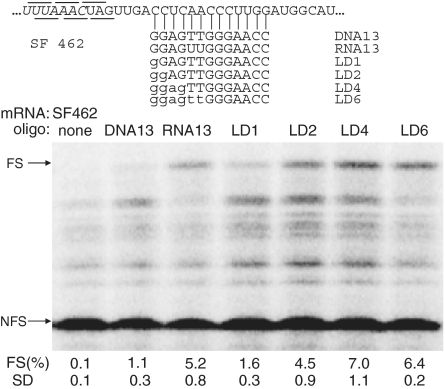
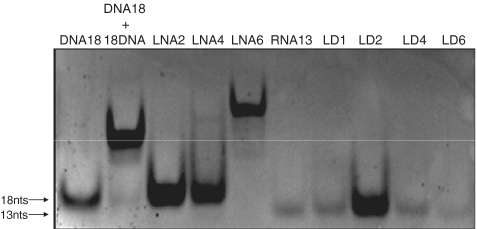
To demonstrate that the enhanced effect of LNA oligonucleotides is a general feature we designed another construct (SF462) in which the target sequence was replaced by an unrelated sequence (Figure 4). LNA/DNA mix-mers were designed in which nucleotides starting from the 3′-end were gradually replaced by LNA (Figure 4). Increasing the number of LNAs from one to two and four in these DNA oligonucleotides improved their frameshift inducing ability, reaching an apparent optimum of 7.0% with four LNA substitutions. Further increase of the LNA content to 6 nt (LD6) did not improve frameshift efficiency, but, on the other hand, LD6 also did not lead to the dramatic decrease as observed above for the LNA6 oligonucleotide applied in the SF468 construct. We suspected that (partial) self-complementarity may be limiting the effective concentration of free LNA/DNA oligonucleotides. To check this possibility, we ran all the oligonucleotides on a non-denaturing polyacrylamide gel. Figure 5 showes that the LNA6 oligonucleotide indeed migrated more slowly indicative of partial dimer formation, presumably by intermolecular base pairing of the palindromic GCGCGC sequences in each oligonucleotide (compare the migration to that of the full dimer formed by annealing of oligonucleotides DNA18 and 18DNA). The LD series, as predicted, migrated as monomers. We noted that LD2, though loaded in equal amount, based on its UV absorbance, showed a higher affinity to ethidium bromide than its counterparts. At present we have no explanation for this unexpected behavior of LD2, since its migration and therefore its conformation was identical to the other LNA/DNA mix-mers.
Figure 4.
Frameshift enhancing activity of LNA-substituted deoxy-oligonucleotides. mRNA SF462 was translated in the absence or presence of 625-fold molar excess of DNA, RNA or LNA substituted DNA oligonucleotides, in rabbit reticulocyte lysate. LNA modifications are indicated by lowercase characters. See legend to Figure 2 for more details.
Figure 5.
Self-dimerization of frameshift-inducing oligonucleotides. An amount of 31.25 pmol of the indicated oligonucleotides were left at room temperature for 10 min. and then loaded on a 15% non-denaturing polyacrylamide gel. After electrophoresis, the gel was stained by EtBr and photographed under UV-light.
These results demonstrate that LNA modifications indeed enhance the antisense-induced frameshifting efficiency probably due to higher thermodynamic stability and RNA-like structural properties. This phenomenon appears to be general, at least in our experiments.
Position effect of LNA substitutions
To investigate which positions in a DNA oligonucleotide would exert the largest effect when substituted by an LNA analog, we designed LNA/DNA mix-mer mutants based on LNA2, which is the most efficient LNA/DNA mix-mer in our experiments and would give a good read-out. When the two LNA substitutions were moved two positions more inward (L2-1), compared to LNA2, frameshift efficiency decreased to 6.7% (Figure 6). However, when the LNA modifications were moved another two positions more inward (L2-2), activity dropped to 2.7% (Figure 6) which is comparable to an unmodified DNA oligonucleotide. Similarly, when the LNA groups were introduced at the other end of the oligonucleotide, activity was as low as DNA18 (Figure 6). Finally, L2-4, in which the first and fourth position were LNA, was only half as efficient as LNA2. These results indicate that the choice of the location of the LNA modifications is crucial for the frameshift-inducing efficiency of an oligonucleotide.
Figure 6.
Position effect of LNA substitutions on their frameshift-inducing activity. LNA substitutions are indicated by lowercase characters. See legend to Figure 2 for more details.
Thermodynamic stability of frameshift-inducing oligonucleotides
Theoretically the position effect of the LNA substitutions could simply be explained by differences in thermodynamic stability of the resulting mRNA/oligonucleotide duplexes. To investigate this possibility we carried out UV-melting studies of the 18-nt LNA oligonucleotides in a 1:1 complex with an 18-nt RNA (18RNA) representing the mRNA. The melting temperatures (Tm) are shown in Table 1. The Tm of the 18RNA/RNA18 duplex was the highest with 82°C in agreement with its high frameshifting efficiency. The 18RNA/DNA18 duplex had a much lower Tm of 72°C, which is expected for an RNA/DNA hybrid, and also agreed with the lower frameshifting efficiency. The LNA substituted oligonucleotides, all had higher Tms (+4 to +9°C) than DNA18. The Tm of L2-2 was with 81°C almost as high as that of RNA18. Remarkably there was no correlation between the Tm of the LNA oligonucleotides and their frameshifting inducing capacity. For example, the Tm of LNA2 was rather low with 76°C but it had the highest frameshifting activity, and L2-2, which had the highest Tm, actually had the lowest frameshifting activity. Tms of L2-3 and L2-4 were identical but their frameshifting activities were 2.3 and 4.6%, respectively. We also noted that both L2-2 and L2-3 were comparable to DNA18 in frameshifting activity but formed far more stable duplexes. These data suggest that the position effect of the LNA substitutions is related to the mechanism of frameshifting and not per se to their thermodynamic stability.
Table 1.
Tm measurements of frameshift-inducing oligos hybridized to complementary RNA
| Frameshift-inducing oligo | Tm (°C) |
|---|---|
| DNA18 | 72 |
| RNA18 | 82 |
| LNA2 | 76 |
| LNA2-1 | 80 |
| LNA2-2 | 81 |
| LNA2-2 | 78 |
| LNA2-4 | 78 |
DISCUSSION
Previously, we have demonstrated that antisense oligonucleotides can induce high levels of −1 frameshifting (27). The optimal length of small antisense oligonucleotides, however, was not investigated. Understanding the optimal length of trans-acting oligonucleotides that can induce the most efficient frameshifting and, at the same time, escape RNAi interference will be an important issue for future in vivo applications. Here we found that maximum levels of frameshifting were obtained with oligonucleotides of 12 nt and more. This is comparable to the stem lengths (S1+S2) of known examples of highly frameshift inducing H-type pseudoknots, such as the 6+6 bp of the Simian retrovirus type−1 pseudoknot (29), the 11+6 bp of the minimal Infectious Bronchitis virus (IBV) pseudoknot (30), and the 6+6 bp chimeric Mouse Mammary Tumor virus (MMTV)-IBV pseudoknot (31). In addition, in known examples of hairpin-induced frameshifts, the stem length of hairpins is around 12 bp (32–34). This may imply that a full helical turn of an RNA helix either in one single stem or in two stacking stems of a pseudoknot (S1+S2) is selected by viruses to induce efficient ribosomal frameshifting.
In addition to RNA oligonucleotides, we demonstrated that LNA/DNA mix-mers are also capable of stimulating efficient −1 ribosomal frameshifting in contrast to DNA oligonucleotides. Replacing 2 nt in a DNA oligonucleotide by LNA was already sufficient to reach the same level of frameshifting as with a comparable RNA oligonucleotide. However, the excellent affinity of LNA oligonucleotides could be a double-edged sword in certain cases. In our experimental system, the oligonucleotides are partly self-complementary and this resulted in the formation of dimers (Figure 5), which were apparently unable to induce frameshifting (LNA6, Figure 3). Hence, LNA substitutions should be optimized in a sequence that is prone to form dimers. In our SF462 construct (Figure 4), the optimal number of LNA substitutions to induce the most significant amount of frameshifting is four. LD6 with two additional LNA substitutions did not improve the efficiency. Thus, our results suggest that the first 4 bp are critical for antisense-induced frameshifting. A likely explanation is that when a ribosome that is translating the slippery sequence, the helicase active site is around position +11, with respect to the first nucleotide of the P-site, which is close to the first base pair of the mRNA/oligonucleotide duplex (35). Increasing the local thermodynamic stability in this region may prevent ribosomes to unwind RNA structures, causing ribosomal pausing at the slippery sequence, and finally results in a higher frequency of ribosomal frameshifting.
Our data also showed that a single LNA modification is not sufficient to turn a DNA oligonucleotide into an efficient frameshift inducer but that a second LNA is needed. The best position for the second modification appeared to be also close to the 3′-end of the oligonucleotide. Although one could expect that spacing of two LNA groups by two non-modified sugars as applied in probes for miRNAs, results in the optimal induction of the 3′-endo conformation in the neighboring sugars (36), this was not the case in our frameshift assays. Here such a spacing was less efficient (see data for L2-4, Figure 6). However, we have not investigated if possible differences of self-dimerization behavior of these oligonucleotides accounts for the different stimulating activities, since such effects were only observed when six LNA modifications were introduced in an oligonucleotide of this sequence.
The observation that different positions of LNA substitutions induced different levels of ribosomal frameshifting is interesting. Even though the overall thermodynamic stability of these oligonucleotides is roughly the same, they still create different degrees of barriers for ribosomes to unwind and these differences could be the reason for different level of induced frameshifting.
The finding that local stability at the 3′-end of the LNA/DNA mix-mers is important for frameshifting is in agreement with the observation that in natural examples of frameshift stimulators, most of them have high GC content in the first few nucleotides (1). Hence, our data support the notion that the stability of the 3′-end of the oligonucleotide, which may reside in the active site of the ribosomal helicase, is critical for frameshift-inducing structural elements. In pseudoknots this stability is probably attained by triple interactions, since nature has no other way to increase the stability of a GC-rich A-type helix. Triplex structures have been documented for a number of frameshifter pseudoknots, e.g. BWYV (37), SRV−1 (38) and in a telomerase pseudoknot (39).
Several models of ribosomal frameshifting have been proposed (1,40,41). The consistency from these studies is that ribosomal pausing at shifty sites by downstream structural elements is important but that pausing caused by RNA secondary structure, does not always result in frameshifting. In addition, a lack of correlation between the extent of pausing and the efficiency of frameshifting by IBV pseudoknots has been observed (42). A recent study also showed that pseudoknots with a similar global structure can still induce very different levels of frameshifting although their thermodynamic stabilities were different (43). These data complicate the view on the role of the downstream structure. Experiments involving simple oligonucleotides such as shown here may be better alternatives to elucidate the role of the downstream element.
Several groups have correlated the mechanical force of unfolding of a pseudoknot with its frameshifting efficiency by using optical tweezers (39,44–46) and suggest that frameshift efficiency is dependent on the unfolding force rather than on differences of thermodynamic stability between folded and unfolded states. Since we showed here that antisense oligonucleotides can induce frameshifting presumably by serving as a physical barrier for the elongating ribosome, it will be interesting to measure the strength of these linear oligonucleotides in complex with (a piece of) mRNA by optical tweezers and see if there is a correlation with their frameshifting efficiency.
Finally, several properties of LNA, including its good aqueous solubility, low toxicity, highly efficient binding to complementary nucleic acids, high biostability, and, improved mismatch discrimination relative to natural nucleic acid (47) make LNA a promising candidate for in vivo applications of antisense-induced frameshifting.
FUNDING
Funding for open access charge: Leiden Institute of Chemistry, Leiden university.
Conflict of interest statement. None declared.
REFERENCES
- 1.Giedroc DP, Theimer CA, Nixon PL. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J. Mol. Biol. 2000;298:167–85. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.BrierleyI, Dos Ramos FJ. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 2006;119:29–42. doi: 10.1016/j.virusres.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinman JD, Icho T, Wickner RB. A -1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc. Natl Acad. Sci. USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baranov PV, Fayet O, Hendrix RW, Atkins JF. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 2006;22:174–181. doi: 10.1016/j.tig.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Wills NM, Moore B, Hammer A, Gesteland RF, Atkins JF. A functional -1 ribosomal frameshift signal in the human paraneoplastic Ma3 gene. J. Biol. Chem. 2006;281:7082–7088. doi: 10.1074/jbc.M511629200. [DOI] [PubMed] [Google Scholar]
- 6.Manktelow E, Shigemoto K, Brierley I. Characterization of the frameshift signal of Edr, a mammalian example of programmed -1 ribosomal frameshifting. Nucleic Acids Res. 2005;33:1553–563. doi: 10.1093/nar/gki299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark MB, Jänicke M, Gottesbühren U, Kleffmann T, Legge M, Poole ES, Tate WP. Mammalian gene PEG10 expresses two reading frames by high efficiency −1 frameshifting in embryonic-associated tissues. J. Biol. Chem. 2007;282:37359–37369. doi: 10.1074/jbc.M705676200. [DOI] [PubMed] [Google Scholar]
- 8.Brierley I, Jenner AJ, Inglis SC. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crooke ST. Progress in antisense technology. Annu. Rev. Med. 2004;55:61–95. doi: 10.1146/annurev.med.55.091902.104408. [DOI] [PubMed] [Google Scholar]
- 12.Heasman J. Morpholino oligos: making sense of antisense? Dev. Biol. 2002;243:209–214. doi: 10.1006/dbio.2001.0565. [DOI] [PubMed] [Google Scholar]
- 13.Lu QL, Mann CJ, Lou F, Bou-Gharios G, Morris GE, Xue SA, Fletcher S, Partridge TA, Wilton SD. Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse. Nat. Med. 2003;9:1009–1014. doi: 10.1038/nm897. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 17.Berkhout B. RNA interference as an antiviral approach: targeting HIV–1. Curr. Opin. Mol. Ther. 2004;6:141–145. [PubMed] [Google Scholar]
- 18.Huang C, Li M, Chen C, Yao Q. Small interfering RNA therapy in cancer: mechanism, potential targets, and clinical applications. Expert Opin. Ther. Targets. 2008;12:637–645. doi: 10.1517/14728222.12.5.637. [DOI] [PubMed] [Google Scholar]
- 19.Fattal E, Bochot A. Ocular delivery of nucleic acids: antisense oligonucleotides, aptamers and siRNA. Adv. Drug Deliv. Rev. 2006;58:1203–1223. doi: 10.1016/j.addr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 20.Moreira JN, Santos A, Simões S. Bcl-2-targeted antisense therapy (Oblimersen sodium): towards clinical reality. Rev Recent Clin Trials. 2006;1:217–235. doi: 10.2174/157488706778250050. [DOI] [PubMed] [Google Scholar]
- 21.Perry CM, Balfour JA. Fomivirsen. Drugs. 1999;57:375–380. doi: 10.2165/00003495-199957030-00010. [DOI] [PubMed] [Google Scholar]
- 22.Sahu NK, Shilakari G, Nayak A, Kohli DV. Antisense technology: a selective tool for gene expression regulation and gene targeting. Curr. Pharm. Biotechnol. 2007;8:291–304. doi: 10.2174/138920107782109985. [DOI] [PubMed] [Google Scholar]
- 23.Corey DR. Chemical modification: the key to clinical application of RNA interference? J. Clin. Invest. 2007;117:3615–3622. doi: 10.1172/JCI33483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 25.Braasch DA, Corey DR. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 26.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 27.Olsthoorn RC, Laurs M, Sohet F, Hilbers CW, Heus HA, Pleij CW. Novel application of sRNA: stimulation of ribosomal frameshifting. RNA. 2004;10:1702–1703. doi: 10.1261/rna.7139704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howard MT, Gesteland RF, Atkins JF. Efficient stimulation of site-specific ribosome frameshifting by antisense oligonucleotides. RNA. 2004;10:1653–1661. doi: 10.1261/rna.7810204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ten Dam E, Brierley I, Inglis S, Pleij C. Identification and analysis of the pseudoknot-containing gag-pro ribosomal frameshift signal of simian retrovirus–1. Nucleic Acids Res. 1994;22:2304–2310. doi: 10.1093/nar/22.12.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liphardt J, Napthine S, Kontos H, Brierley I. Evidence for an RNA pseudoknot loop-helix interaction essential for efficient −1 ribosomal frameshifting. J. Mol. Biol. 1999;288:321–335. doi: 10.1006/jmbi.1999.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Napthine S, Liphardt J, Bloys A, Routledge S, Brierley I. The role of RNA pseudoknot stem 1 length in the promotion of efficient −1 ribosomal frameshifting. J. Mol. Biol. 1999;288:305–320. doi: 10.1006/jmbi.1999.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dulude D, Baril M, Brakier-Gingras L. Characterization of the frameshift stimulatory signal controlling a programmed −1 ribosomal frameshift in the human immunodeficiency virus type 1. Nucleic Acids Res. 2002;30:5094–5102. doi: 10.1093/nar/gkf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucchesi J, Mäkeläinen K, Merits A, Tamm T, Mäkinen K. Regulation of −1 ribosomal frameshifting directed by cocksfoot mottle sobemovirus genome. Eur. J. Biochem. 2000;267:3523–3529. doi: 10.1046/j.1432-1327.2000.01379.x. [DOI] [PubMed] [Google Scholar]
- 34.Larsen B, Gesteland RF, Atkins JF. Structural probing and mutagenic analysis of the stem-loop required for Escherichia coli dnaX ribosomal frameshifting: programmed efficiency of 50% J. Mol. Biol. 1997;271:47–60. doi: 10.1006/jmbi.1997.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 36.Válóczi A, Hornyik C, Varga N, Burgyán J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su L, Chen L, Egli M, Berger JM, Rich A. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat. Struct. Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michiels PJ, Versleijen AA, Verlaan PW, Pleij CW, Hilbers CW, Heus HA. Solution structure of the pseudoknot of SRV–1 RNA, involved in ribosomal frameshifting. J. Mol. Biol. 2001;310:1109–1123. doi: 10.1006/jmbi.2001.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen G, Chang KY, Chou MY, Bustamante C, Tinoco I., Jr Triplex structures in an RNA pseudoknot enhance mechanical stability and increase efficiency of −1 ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 2009;106:12706–12711. doi: 10.1073/pnas.0905046106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brierley I, Pennell S. Structure and function of the stimulatory RNAs involved in programmed eukaryotic–1 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 2001;66:233–248. doi: 10.1101/sqb.2001.66.233. [DOI] [PubMed] [Google Scholar]
- 41.Plant EP, Jacobs KL, Harger JW, Meskauskas A, Jacobs JL, Baxter JL, Petrov AN, Dinman JD. The 9-A solution: how mRNA pseudoknots promote efficient programmed −1 ribosomal frameshifting. RNA. 2003;9:168–174. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kontos H, Napthine S, Brierley I. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell Biol. 2001;21:8657–8670. doi: 10.1128/MCB.21.24.8657-8670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cornish PV, Stammler SN, Giedroc DP. The global structures of a wild-type and poorly functional plant luteoviral mRNA pseudoknot are essentially identical. RNA. 2006;12:1959–1969. doi: 10.1261/rna.199006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansen TM, Reihani SN, Oddershede LB, Sørensen MA. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 2007;104:5830–5835. doi: 10.1073/pnas.0608668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazauric MH, Seol Y, Yoshizawa S, Visscher K, Fourmy D. Interaction of the HIV–1 frameshift signal with the ribosome. Nucleic Acids Res. 2009;37:7654–7664. doi: 10.1093/nar/gkp779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green L, Kim CH, Bustamante C, Tinoco I., Jr Characterization of the mechanical unfolding of RNA pseudoknots. J. Mol. Biol. 2008;375:511–528. doi: 10.1016/j.jmb.2007.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grünweller A, Hartmann RK. Locked nucleic acid oligonucleotides: the next generation of antisense agents? BioDrugs. 2007;21:235–243. doi: 10.2165/00063030-200721040-00004. [DOI] [PubMed] [Google Scholar]