Abstract
Agarose gel electrophoresis, circular dichroism and differential scanning calorimetry showed that single-stranded RNA from satellite tobacco mosaic virus transforms from a conformationally ‘closed state’ at 4°C to a more conformationally ‘open state’ at 65°C. The transition is reversible and shows no hysteresis. Atomic force microscopy (AFM) allowed visualization of the two states and indicated that the conformationally ‘closed state’ probably corresponds to the native encapsidated conformation, and that the ‘open state’ represents a conformation, characterized as short, thick chains of domains, as a consequence of the loss of tertiary interactions. Heating from 75°C to 85°C in the presence of EDTA was necessary to further unravel the ‘open’ conformation RNA into extended chains of lengths >280 nm. Virus exposed to low concentrations of phenol at 65°C, extruded RNA as distinctive ‘pigtails’ in a synchronous fashion, and these ‘pigtails’ then elongated, as the RNA was further discharged by the particles. Moderate concentrations of phenol at 65°C produced complete disruption of virions and only remains of decomposed particles and disordered RNA were evident. AFM images of RNA emerging from disrupted virions appear most consistent with linear arrangements of structural domains.
INTRODUCTION
Satellite viruses (1–3) provide particularly useful models for the assembly of icosahedral viruses because of their small size, capsid simplicity, small nucleic acid complement and minimum coding capacity. Four encapsidated, T = 1 icosahedral satellite viruses are known to infect plants in the presence of their helper viruses, and the crystallographic structures have been determined at high resolution for three of them (4). Each virus codes only for its coat protein, using single-stranded RNA genomes ranging from 728 nt for satellite panicum mosaic virus to 1457 nt for satellite tobacco necrosis virus.
Satellite tobacco mosaic virus (STMV) is the most thoroughly studied of the satellite viruses (4–7). Its virion is composed of 60 identical copies of a 17.5 kD coat protein and a single-stranded RNA molecule of 1058 nt. Its structure has been refined to 1.45 Å resolution by X-ray crystallography (S. Larson et al., 2010, submitted for publication), the highest for any virus and ∼45% of the encapsidated RNA is present in electron density maps (8–10). This implies that nearly half of the RNA is structured in a manner consistent with icosahedral symmetry. Portions of the remainder of the encapsidated RNA structure can be inferred. The distribution of RNA density has been interpreted as a linear series of local stem–loop and pseudo-knot substructures that are bound firmly by coat protein dimers at the interior surface of the capsid (11). The 2-fold axes at the centers of double helical stems coincide with protein dimer dyads, and hence with icosahedral 2-fold axes.
An assembly pathway has been proposed for STMV, which is based on cooperative interactions between coat protein dimers and stem–loop elements of the RNA, proceeding in an orderly pathway as RNA is synthesized on replication complexes (11). In this model, RNA spontaneously forms a sequence of local stem–loops as it is synthesized, essentially as predicted by earlier researchers (12–15). The stem–loop elements are then immediately bound and maintained by protein dimers. The RNA is driven into a more compact, encapsidated conformation by the condensation of the coat protein into an icosahedral shell, a consequence of interactions and interfaces inherent in the protein structure. Thus, both the protein and the RNA are complicit in the assembly process.
An important feature of the current model for the structure of the encapsidated STMV RNA (11,16) is that it is not assumed to be in its minimum energy conformation in which 60–70% of its nucleotides would be predicted to form base pairs (9,17). It is, rather, assumed to be in a more fluid, metastable conformation, one more readily unfolded upon decapsidation and one more conducive to dynamic, physiological processes such as replication, translation and intercellular transport. On the other hand, some physiologically important substructures may be present throughout the life of the RNA. These might include the histidine accepting, tRNA-like structure at the 3′ end of the molecule (18), and the 5′ structural element forming the recognition site for the tobacco mosaic virus (TMV) helper virus replicase (19–20).
Although the secondary structure of the RNA contains helical stem–loops, pseudo knots and perhaps more complex substructures, the overall tertiary structure of the encapsidated RNA must be consistent with an approximately spherical shape of diameter ∼10 nm in order to be contained within the capsid. Presumably, this is maintained by protein–nucleic acid interactions, but also, at least in part, by RNA tertiary interactions that subsequently form. Earlier experiments on STMV that had been exhaustively digested with proteinase K showed, by mass spectrometry, that RNA cores resulted which still had bound within them peptides that included the basic amino terminal tails of the coat protein (21). These cores, by quasi-elastic light scattering (QELS), had uniform diameters of ∼10 nm, the inside diameter of the STMV protein shell. This was confirmed by atomic force microscopy (AFM) (16). Somewhat surprisingly, the cores did not readily unfold into more extended structures. The RNA—amino terminal peptide cores had to be heated >75°C before they began to lose their integrity.
In another paper, we described an AFM study of genomic single-stranded RNA, extracted by the conventional phenol procedure, from a number of small icosahedral viruses (22). These included poliovirus and STMV among others. We also included in that study, for comparison, the RNA extracted from the rod shaped, helical virus, TMV. We showed that RNA, freshly extracted from icosahedral viruses, existed initially as roughly spherical masses of highly condensed nucleic acid having diameters corresponding to the insides of their respective viral capsids. This suggested, consistent with the QELS results, that the RNA, upon extraction, maintained the same, or a very similar conformation to that which it had when encapsidated.
With time, at 25°C, the conformation of the RNA from poliovirus transformed into linear arrays of secondary structural domains, helical stem–loop structures and possibly pseudo knots. The substructure was occasionally interrupted by stretches of extended single-stranded RNA. The RNA from STMV showed similar behavior, but only under more rigorous conditions.
To further characterize the structure of the encapsidated STMV RNA, and the structures that it assumed upon transformation from the condensed, encapsidated conformation to the extended array of domains, we undertook a further study of the RNA, and that is described in the present article. We combined AFM observations with a series of biophysical experiments. We also imaged the release of RNA from STMV that was intentionally disrupted by various means, both physical and chemical.
MATERIALS AND METHODS
Preparation of STMV
STMV was prepared as described previously (6–7,23), principally using fractionation by PEG followed by crystallization from salt solutions. For these studies, the STMV was crystallized, washed, dissolved and re-crystallized at least once. This was necessary to minimize the presence of RNA from TMV in the samples used for AFM, biophysical and biochemical characterizations. A significant modification was introduced into our procedures for crystallization. Crystals in the past were obtained by dialysis of the crude virus solutions against 10–20% w/v ammonium sulfate. Crystals grown in this way were of the orthorhombic form (6–7,23) and these were extremely difficult to redissolve for re-crystallization. Our STMV preparation now uses dialysis against 8–12% sodium chloride in water. The crystals so obtained from NaCl are of the cubic habit (24) and re-dissolve readily in water, permitting repeated re-crystallization before extraction with phenol.
Preparation of STMV RNA
STMV was dissolved in 50 mM Tris–HCL buffer at pH 7.0 and extracted three times with water-saturated phenol, and then twice with chloroform to remove residual phenol. For agarose gel electrophoresis, circular dichroism (CD) and differential scanning calorimetry (DSC) experiments, RNA was concentrated by precipitation with 1/10 volume of 3 M sodium acetate, pH 5.4 and three volumes 100% ethanol. The solution was incubated at −80° C for 18 h. The RNA was collected via centrifugation at 21 000 rpm for 5 min in an IEC Micromax microcentrifuge. The supernatant was removed and the pellet dried in a Labconco centrivap concentrator at 45°C for 5 min. The RNA was resuspended in DEPC-treated H2O (Research Genetics, an Invitrogen Corporation; Huntsville, AL, USA). Concentration and purity were evaluated based on optical density at 260 and 280 nm and their ratio using a Hewlett-Packard 845 UV–visible spectrophotometer.
Agarose gel electrophoresis
Electrophoresis was carried out using a MiniSubTM DNA Cell horizontal slab–gel system from BioRad with a Cabisco Biotechnology power supply. Agarose gels (1%) were made with 1× TBE buffer and run at 100 V for 45 min. The gels were visualized under UV illumination using a FOTO/phoresis light box from FOTODYNE. Photographs were recorded with a FOTODYNE Polaroid camera (Cat. No. 1-1440) and scanned using a HP ScanJet IIC/ADF scanner.
The reversible folding of STMV RNA visualized on agarose gels was demonstrated using a 50 μl master mix of RNA. A 3 μl aliquot at 4°C was removed and added to a tube containing 10% formamide in 37% formaldehyde (25–27). This was the initially closed, or folded conformational state. The formamide to formaldehyde ratio had previously been optimized so that the closed state remained folded in the presence of the formamide and formaldehyde reagent and the open state denatured. The master mix was subsequently heated at 65°C for 10 min and another 3 μl aliquot immediately removed and added to a tube containing the same optimized concentration of formamide and formaldehyde. This yields the open state. The master mix was allowed to cool to 4°C for 10 min, another 3 μl aliquot was removed and added to the formamide–formaldehyde reagent, thereby trapping the closed, refolded state and so on. The process was repeated six times. Upon completion, all solutions were placed on ice for 30 min and then examined using agarose gel electrophoresis as above.
Circular Dichroism (CD)
CD measurements were made on an OLIS-RSM 1000 Rapid Scanning Monochromator. A 2.0 ml cuvette with a path length of 1 cm was used for all experiments. Data were collected every 1 nm between 200 and 300 nm over the range of temperatures. A 1.0 s integration time was used throughout. We previously demonstrated that no incubation time is required, thus data were collected once the appropriate temperature was achieved. The ellipticity values obtained were normalized to molar ellipticity for comparison. The following equation was used to convert millidegrees to molar ellipticity values (deg × cm2/dmol):
where [θ] is molar ellipticity, θ is the ellipticity in millidegrees, M is the molecular weight of the RNA, c is the molar concentration, l is the cuvette path length and n is the number of bases. Once the data were collected, the 262 nm wavelength was used for analysis. Initial scans indicated this to be the peak. Though other scans under varying conditions yield peaks between 258 and 267 nm, there is no apparent difference in the profiles thus the 262 nm wavelength was utilized for all analyses.
Differential Scanning Calorimetry (DSC)
DSC analysis of STMV RNA was performed using a MicroCal (Northampton, MA, USA) Extended Range VP-DSC with a cell volume of 0.5 ml. Purified RNA was degassed by vacuum using the MicroCal accessory and the resulting solution was scanned at 1.5 C/min from 5°C to 80°C. DSC data were analyzed using the procedure of IGOR Pro (Wavemetrics, OR, USA) in which the baselines, heat capacity and melting temperatures were fitted.
Preparation of virus and RNA samples for AFM
Preparation of STMV RNA was as described previously. In some cases, the RNA was precipitated with 100% ethanol at −20°C and suspended in DPEC treated water; in other cases, RNA was analyzed immediately following treatment with chloroform. RNA from STMV was first transformed from the conformation initially present in the phenol extracted nucleic acid to a still highly condensed linear form by exposure to temperatures between 50°C and 75°C, but additional change was difficult to produce. Eventually, we were successful in further melting the structure of the RNA by heating it from 80°C to 85°C in the presence of 0.015 M EDTA, or 3 M sodium chloride. For AFM, heating with EDTA was the preferred approach in order to avoid fouling of the AFM substrate by crystalline salt. Heating the RNA at pH 9–10 was also attempted, but, as might have been expected, this led to rapid degradation of the RNA to small fragments as short single-stranded regions were swiftly hydrolyzed.
For AFM imaging of virus disruption and RNA release, a number of methods were applied which had previously performed well with other viruses (22,28). Most failed with STMV. High pH, heating to 80°C and higher, and combination of the two had little effect on the intact virus. STMV treated at 37°C with proteinase K or trypsin could still be crystallized in familiar crystal forms, indicating no significant damage. Although incubation of STMV with stoichiometric amounts of proteinase K at 60°C did produce degradation as shown by light scattering, we found that the best method for disruption of STMV, which permitted some degree of control, was exposure of the virus to buffer containing phenol at various concentrations and temperatures. The buffer–phenol solutions were made by simply diluting the upper layer in the conventional two phase system obtained in making phenol saturated with buffer.
Atomic Force Microscopy (AFM) analysis
AFM analysis (29–33) was generally carried out in air, though in some cases scanning was performed under buffer in a 75-µl fluid cell. Substrates were freshly cleaved mica, mica treated with magnesium chloride, or mica coated with poly-l-lysine. Nucleic acids usually adhered to magnesium ion treated surfaces that carry positive charges, while virus particles adhered firmly to mica coated with poly-l-lysine. Most details of the AFM analyses have been presented in earlier papers (22,34–39). Samples of 1.5–5 µl composed of 0.02 M HEPES at pH 7.0 made from DPEC water and containing virus, RNA or other viral components were applied to the substrate and allowed to sediment for 5–30 min, depending on the sample. Alternatively, the sample was simply allowed to dry in air onto the substrate. Excess liquid was removed on filter paper. The sample, on the substrate, was then exposed to 5% glutaraldehyde in buffer for 2 min and excess glutaraldehyde solution shaken off. The substrate was then washed two times with distilled water and dried in a stream of dry nitrogen gas.
Glutaraldehyde has proven necessary in most studies to rigidify biological samples sufficiently that they permit scanning at high magnification. Glutaraldehyde reacts with free amino groups of lysine residues of proteins, amino termini of proteins and the exocyclic amino groups of nucleic acid bases adenine, guanine and cytosine. Poly-l-lysine has proven to be a reliable adhesive for virtually all of the biological samples that we have investigated particularly cells, viruses and protein assemblies. While nucleic acids also adhere well to substrates coated with poly-l-lysine, the polymer produces a background level that often obscures the true heights of DNA and RNA, which protrude above the substrate only from 0.6 to 2.0 nm. Because nucleic acids also adhere well to substrates (mica, glass, plastic) treated with magnesium salts, such as magnesium chloride or magnesium acetate, that do not produce a high background, those are the preferred substrates for visualization of RNA and DNA.
AFM imaging was carried out using a Nanoscope III multi-mode instrument (Veeco Instruments, Santa Barbara, CA, USA). When scanned in liquids, virus and associated macromolecules were scanned at 26°C using oxide-sharpened silicon nitride tips in a 75 µl fluid cell containing buffer. For scanning in air, silicon tips were employed. The images were collected in tapping mode (40–41) with an oscillation frequency of 9.2 kHz in fluid and 300 kHz in air, with a scan frequency of 1 Hz. Vertical and lateral distances were calibrated using standards from Digital Instruments (Santa Barbara, CA, USA) of 10 nm height steps of pitch 200 nm, and standards from MCNC Analytical Labs with step height 10 nm and pitch of 1 µm. The AFM instrument was calibrated to small lateral distances by imaging the 111 face of a thaumatin protein crystal and using the known lattice spacings (42–43) as standard. Cantilevers for scanning in air were from Veeco (Santa Barbara, CA, USA) and were TAP150 with lengths of 115–135 µm and spring constants of 5 Nn. For solution scanning, they were 0TR4 with lengths of 100 µm and spring constants of 0.08 Nn.
In the AFM images presented here, height above substrate is indicated by increasingly lighter color. Thus points very close to the substrate are dark and those well above the substrate white. Because lateral dimensions are distorted due to an AFM image being the convolution of the cantilever tip shape with the surface features scanned, quantitative measures of size were based either on heights above the substrate, or on center to center distances on particle surfaces.
RESULTS
Biophysical experiments
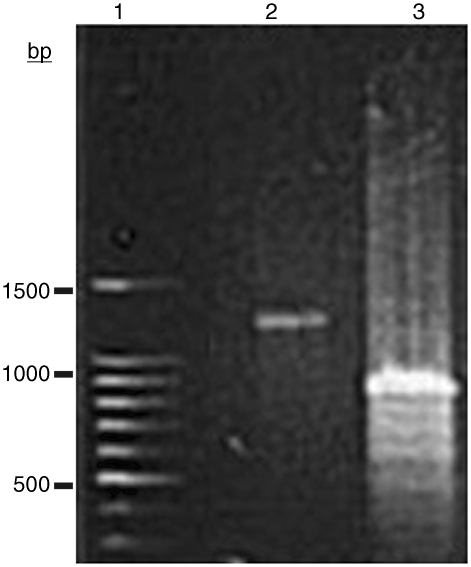
Figure 1 is an agarose electrophoretic gel of STMV RNA. Lane 2 is a sample of RNA that was heated to 65°C and then exposed to the formamide/formaldehyde reagent at that temperature. As a consequence of formaldehyde reaction with the nucleic acid, the reagent essentially traps the RNA in the conformation extant at the time of exposure. All of the RNA in the sample migrates as a single band corresponding to a double stranded DNA length of ∼800 bp. In lane 3, the RNA was heated to 65°C and allowed to cool in the absence of the reagent.
Figure 1.
Native agarose electrophoretic gel of STMV RNA produced by extraction of the virus with phenol. Double-stranded DNA of specific lengths was used to produce a molecular ladder as standards in lane 1. In lane 2, the RNA was heated for 10 min at 65°C and exposed to a reagent composed of 10% formamide and 37% formaldehyde before application to the gel. In lane 3, the RNA was heated to 65°C and simply allowed to cool to 4°C before application to the gel. The single band trapped at 65°C represents the ‘open’ conformation of the RNA. The predominant band that forms at low temperature after re-naturation corresponds to the ‘condensed’ state.
As is evident from the gel, at 65°C, all of the RNA molecules in the sample have approximately the same or similar conformations. At room temperature, there is again a single major band on the gel, but corresponding to a length ∼25% less. That is, the RNA migrates significantly faster and, therefore, has assumed a more compact form. The gel shown here is readily reproducible and the same result has been observed with numerous samples of STMV RNA obtained by phenol extraction.
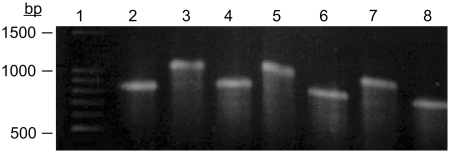
In the experiment illustrated by the agarose gel in Figure 2, the transition between the two conformational states, corresponding to the slow and fast bands on the gel of Figure 1, is shown to be reversible. Lane 2 is an aliquot of a larger sample of RNA at 25°C that was exposed to the formamide/formaldehyde reagent with no heating. The RNA parent sample was then heated to 65°C, as in the experiment of Figure 1, and an aliquot drawn, exposed to the reagent at that temperature and applied to lane 3. The parent sample of RNA was then cooled to 4°C, an aliquot drawn, exposed to the reagent and run on lane 4. The procedure of alternately heating and cooling the parent sample of RNA, drawing aliquots and exposing them to the reagent was repeated to produce the samples seen on lanes 5 through 8. The experiment was continued to greater numbers of cycles in other experiments with the same results.
Figure 2.
Native agarose electrophoretic gel showing the reversible transition of STMV RNA. The lanes contain formamide/formaldehyde fixed aliquots taken from a single sample of STMV RNA heated and cooled alternately between 4°C and 65°C. Lanes 2, 4, 6 and 8 correspond to aliquots of RNA cooled to 4°C and fixed, while lanes 3, 5 and 7 correspond to aliquots fixed at 65°C. Lane 1 is a DNA molecular weight standard ladder.
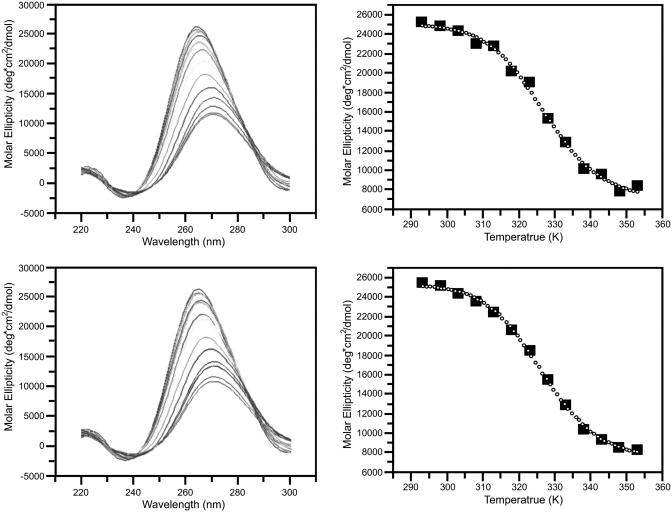
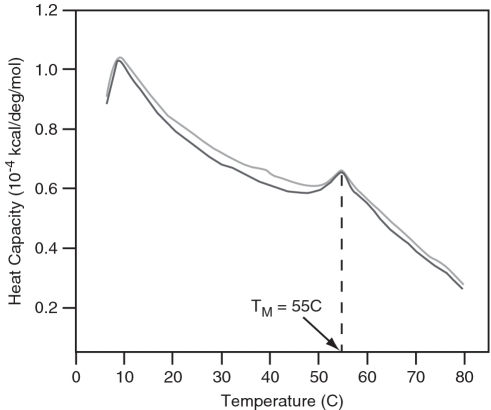
As is evident from Figure 2, the STMV RNA can be induced to repeatedly and reversibly transform between an ‘open’ and a ‘condensed’ conformational state, the transition occurring between 4°C and 65°C. A similar result was obtained by monitoring absorption ellipticity by CD at 260 nm as a sample of RNA was repeatedly heated and cooled. As seen in Figure 3, the course of the transformation is highly reproducible and the plots of ellipticity in the cooling and heating legs of the cycles are virtually superimposeable. The experiment illustrated by Figure 4 suggests that the transformation between the two states is relatively smooth and gradual, but marked by a discontinuity. The DSC curves for two different samples of STMV RNA are again practically congruent. The curves indicate that there is some discreet structural transition occurring at ∼55°C.
Figure 3.
CD of STMV RNA as a function of temperature. CD ellipticity profiles were collected at 5°C intervals between 20°C and 80°C. The top left panel shows the CD profile as a function of wavelength and temperature as the RNA unfolds. The darkest line represents the RNA at room temperature and the profile becomes broader and blue shifts as the temperature increases. The molar ellipticity of the unfolding process was plotted at 262 nm wavelength as a function of temperature at top right. In the lower left panel the folding process was monitored by measuring the CD ellipticity starting at 80°C. As the temperature decreased the profile progressively returns to that seen for 20°C. The molar ellipticity of the folding process is again shown, with the process initiated at high temperatures progressing towards room temperature (lower right).
Figure 4.
DSC analysis for STMV RNA. The RNA displays a broad melting endotherm. A distinct transition occurs between 42°C and 60°C with a midpoint corresponding to the melting temperature at 55°C. Two independent runs are shown.
AFM analysis of STMV RNA
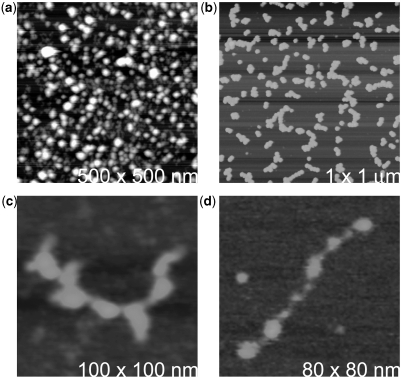
Figure 5 a contains AFM images of STMV RNA that has been extracted from virus using the conventional phenol procedure described above. The RNA was subsequently exposed to 4°C in the presence of ethanol to concentrate it, but the images were recorded at 25°C. As observed previously (21–22) and as seen in Figure 5a, the RNA has the form of condensed, roughly spherical particles with a fairly uniform diameter of 10 nm. The diameter corresponds closely with the inside diameter of the protein capsid calculated from the known X-ray crystallographic structure (8). It is reasonable to assume, therefore, that the RNA has retained its encapsidated conformation at 25°C. These particles correspond to the ‘condensed’ state observed in the experiments above, and must be a consequence of significant tertiary as well as secondary structural interactions within the particle.
Figure 5.
As the extracted RNA was heated from 25°C to 65°C the RNA gradually unfolded. (a) Spherical particles of highly condensed STMV RNA that result from phenol extraction of the virus with phenol at 4°C. (b) The spheres begin unfolding as the temperature is increased and begin to form short, thick chains. One of these is seen at higher magnification in (c). (d) With progressively higher temperature, the chains elongate as they further unfold. The height ranges for the images are (a) 0.0 to –25 nm; (b and c) 0.0–20 nm; and (d) 0.0–5 nm.
Figure 5b–d illustrates the result of heating the spherical RNA particles to 65°C and application of the RNA to the AFM substrate. At elevated temperature, the RNA appears as rather short, irregular, thick strands that correspond to chains of still highly condensed structural domains. At least some crucial tertiary interactions are lost in passing from the ‘condensed’ to the ‘open’ conformation, while secondary structural interactions are probably preserved. If the elongated, ‘open’ form of the RNA is cooled to 4°C, and the RNA visualized by AFM, then the spherical particles, the ‘condensed’ form, is again observed. Thus the transition, consistent with the biophysical experiments, is reversible.
The AFM images define the ‘condensed’ (Figure 5a) and ‘open’ (Figure 5b–d) states, but they suggest little about the secondary structure of the RNA, nor what conformations exist at and >65°C, and under more rigorous conditions. To explore this question, we exposed STMV RNA to higher temperatures, to a variety of denaturing agents, and combinations of the two. The objective was to further, but gradually unravel the RNA conformation and reveal something of the overall secondary structure. The approach was similar to our earlier AFM investigation of turnip yellow mosaic virus (35). The problem, however, turned out to be considerably more difficult than we had anticipated. This owed to the extraordinary stability of STMV RNA.
Heating of the RNA in neutral buffer from 65°C to 85°C produced only modest further lengthening of the RNA chains and revealed little more regarding the conformation. At temperatures approaching 100°C, the RNA began to fragment as short, single-stranded stretches hydrolyzed. High pH at temperatures of 75–90°C was similarly devastating because it accelerated hydrolysis. Eventually, we found that heating in the range of 75–85°C at neutral pH, but in the presence of 0.015 M EDTA or 3 M NaCl promoted significant further unfolding of the RNA. The effectiveness of EDTA and high-salt concentrations both suggested that residual tertiary interactions involved divalent cations, consistent with the general observation that these ions are important in determining RNA conformation.
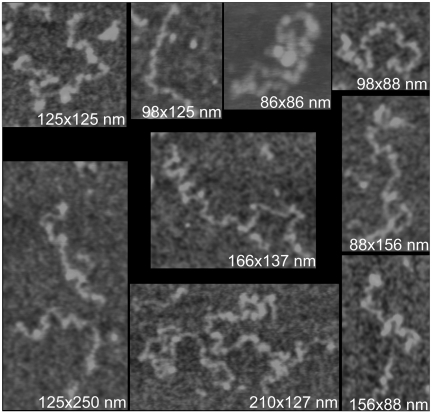
Figure 6 presents AFM images of RNA molecules that have further unfolded. In some cases, the contour lengths are between 200 nm and 280 nm. Along the lengths of the molecules the height varies from ∼0.5 to 3.0 nm above the substrate. A noteworthy feature of these molecules is that they are all essentially linear and unbranched. Occasionally, some small protrusions can be seen emanating from the chains, but these are relatively short. This suggests that the secondary structure does not contain major diversions involving disparate regions of the nucleotide sequence.
Figure 6.
Single molecules of STMV RNA recorded by AFM. The RNA was exposed from 75°C to 85°C in the presence of EDTA. The height range for all images is 0.0–2.5 nm.
AFM analysis of disrupted STMV particles
A question related to the conformation of the RNA, both encapsidated and free of the protein shell, is how the RNA departs the virion. To address this question, we carried out a series of experiments in which we attempted to disrupt the virus by various means while monitoring the results using AFM. Again, our efforts were frequently thwarted by the extraordinary stability of the STMV particles. Approaches that had been successful with turnip yellow mosaic virus, a T = 3 icosahedral plant virus of considerable fortitude (35), such as high pH, elevated temperature or non-ionic detergents, failed with STMV.
In the end, we found that we could disrupt virions sufficiently to affect release of RNA, or destroy capsids in their entirety, by exposing them to buffer containing various concentrations of phenol at temperatures in the range of 65–75°C. The concentration of the phenol had to be carefully adjusted and kept in balance with temperature. At one extreme nothing happened to the virus, at the other extreme of phenol concentration and temperature, the virions virtually exploded and the interval between the two extremes was narrow.
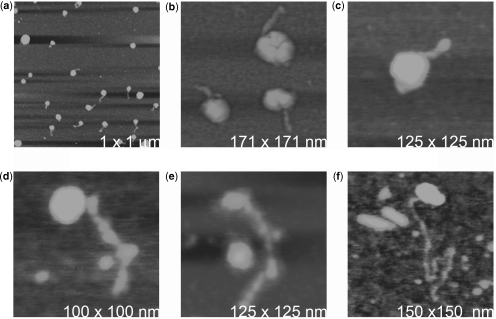
Under the mildest conditions that still produced RNA release, relatively intact virus exhibited ‘pigtails’ of RNA. AFM images of the ‘pigtails’ are seen at different times after exposure in Figure 7. The ‘pigtails’, as the name suggests, are linear, with bulges and narrows along their lengths, and they vary from barely perceptible as they first emerge, to as long as 200 nm upon elongation as in Figure 7f. Apparently, the capsid suffers a limited rupture at some point, or loses a pentagonal capsomere, and the RNA simply wanders outside. We also note that the end of the RNA generally seems to exhibit a bulge or knob, suggesting some leading domain or the presence of residual coat protein. We cannot, from the AFM images, know if this is the 3′- or 5′-end of the RNA, nor whether a similar exit mechanism is realized within a host cell.
Figure 7.
(a) When STMV particles were exposed to low concentrations of phenol at 75–85°C they developed ‘pigtails’ of emerging RNA. (b) The lengths of the pigtails within an observed population are relatively the same, as illustrated by the AFM image, suggesting that the initiation and rate of emission of the RNA is consistent among particles. (c) A pigtail has just started to emerge from a disrupted virion. Note the bulge at the end of the strand. (d) A pigtail has elongated, but swollen areas indicative of structural domains are evident along its length. (e and f) The RNA has assumed a fairly lengthy form. The ‘pigtail’ in (f) has a contour length of ∼200 nm. The height ranges for the images are (a) 0.0–5 nm; (b) 0.0–20 nm; (c) 0.0–5 nm; (d) 0.0–5 nm; (e and f) 0.0–8 nm.
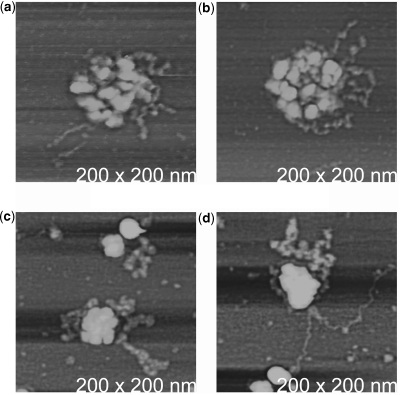
Under more extreme conditions of phenol concentration and temperature, the protein capsid disassembles in a disordered manner and the entire contents of the virus are left displayed as capsid remnants and RNA on the substrate. Figure 8 illustrates some of these occurrences. In these, it can be seen that the RNA makes no orderly exit, but simply splashes out of decomposed capsids.
Figure 8.
(a–d) STMV particles that were catastrophically disrupted on the AFM substrate so that the RNA was cast about in a disordered manner. The height ranges for the images are (a) 0.0–25 nm; (b) 0.0–20 nm; (c) 0.0–10 nm; and (d) 0.0–8 nm.
DISCUSSION
Biophysical experiments involving agarose gel electrophoresis, CD and DSC indicate that STMV RNA can exist in two distinct states that are dependent on temperature. A continuum of conformations likely exists between these two end states, but these are not stable and, with time, tend toward one of the two final states. These two states we refer to as the ‘open’ and ‘closed’ conformational state due to their differing mobilities on native agarose gels. The closed state gradually transforms into the open conformational state as the temperature is increased from 4°C to 65°C, with a marked discontinuity at 55°C.
The transformation is reversible and reproducible, with the path, based on CD and DSC, the same, independent of the direction of the transition. AFM images show that the closed state at 4°C corresponds to a condensed mass, roughly spherical, having a diameter of 10 nm, the inside diameter of the virion, that probably exhibits the same conformation that the RNA possesses when it is encapsidated within the virion. The diameter is the same as that of exhaustively proteolytically digested STMV, as measured by quasi-elastic light scattering, which still contains the amino terminal polypeptides of the coat protein (4).
The open conformation of the RNA, as judged from the AFM images, reflects the loss of important tertiary interactions, but probably not the loss of secondary structural interactions. The RNA at 65°C exists as short, thick chains of still highly condensed nucleic acid, suggesting that secondary structure is maintained, but also significant tertiary interactions as well. The more extensive elongation of RNA molecules seen by AFM at higher temperatures, but only in the presence of EDTA or high-NaCl concentrations, suggests that the residual tertiary interactions likely involve the participation of divalent cations.
With both elevated temperature and EDTA exposure, additional interactions are lost and the RNA extends to maximum lengths of ∼280 nm. This, nonetheless, still represents a substantial compression. The molecules remain intact at those lengths. Because single-stranded, extended RNA polynucleotides hydrolyze rapidly under the conditions required to attain an extended state, it seems safe to conclude that most of the linear chain is still involved in secondary structure that is considerably more resistant to cleavage.
Cautious treatment of virus with phenol at elevated temperature promoted the emergence of linear chains of RNA from otherwise intact particles. Presumably, one or more pentameric capsomeres were lost or loosened enough to permit an end of the encapsidated RNA chain to escape the virion. These were not isolated events confined to individual particles. That is, the emergence of the RNA to make the ‘pigtails’ was not a stoichastic process, as all of the virus in the sample produced the RNA ‘pigtails’ in a synchronous fashion, and all of the ‘pigtails’, at least initially, were of about the same length. In Figure 7a, for example, the particles were exposed simultaneously to the phenol and the AFM image records the lengths of the ‘pigtails’ at an arbitrary later time. The average length of the ‘pigtails’ is ∼45 nm with a standard deviation of 12 nm. The deviation represents only ∼4% of the full length of the RNA molecules presented in Figure 6.
Some of the ‘pigtails’ spontaneously elongated and produced extended chains as long as 200 nm. This was remarkable given the difficulty we experienced in inducing RNA extracted with phenol to unravel and produce such long chains. The fact that the capsids opened some portal to allow emission of an end of the RNA suggests that there may be some unique pentamer or other structural feature immediately adjacent to the wandering end of the RNA molecule when it is encapsidated. Alternatively, it could be that the close proximity of the wandering end on the inside of the capsid weakens, or unlocks a capsomere and causes it to dissociate from the virion, i.e. the RNA forces its way out.
We do not know if the wandering end is the 5′- or the 3′-end, but we favor the 3′-end. The 3′-end contains a tRNA-like structure, and in AFM images we observe an enlargement at the distal terminus of the ‘pigtail’. We also know from previous experiments that the tRNA-like structure is accessible on phenol extracted RNA since it can be amino acylated with histidine (18). Thus it must be on the outside of the condensed, spherical mass of RNA in the encapsidated conformation. It is possible that the 5′-end also has an exposed position adjacent to the interior surface of the capsid, but we have no evidence of that.
A fully extended single-stranded RNA would have an average distance between consecutive phosphate groups of ∼0.55 nm. Thus a fully extended STMV RNA would have an expected length of ∼1058 × 0.55 nm = 582 nm. The maximum contour length of the STMV RNA in the AFM images is about half that. In addition, from previous work (38), we know how extended RNA appears in AFM images, and the height to which it rises above the substrate plane. We also know from that work the appearance of condensed or multi-stranded nucleic acid. The RNA molecules illustrated in Figure 6, even those of greatest extent, do not have the length, height or appearance of fully extended RNA. Clearly, the molecules we visualized, even after extensive attempts to denature them, still contain a substantial amount of secondary structure and are still condensed to ∼50% of fully extended length. The question that this poses is: what is the nature of the residual secondary structure?
A plausible explanation is that the RNA exists as a chain of small domains, e.g. short stem–loops and pseudo knots that appear in a linear manner along the length of the RNA. We suggested this arrangement previously (11) and pointed out that it is consistent as well with the expectations of earlier investigators (12–15) and with X-ray crystallographic results (25–27). If many of these small domains remained, then they would explain the length, compression and the heights above the substrate of the RNA molecules. This might explain as well the general appearance of the RNA and its continued resistance to hydrolysis. We do not, however, see stem–loops protruding or branching from the molecules in Figure 6, which we might have expected. On the other hand, those expectations do strain the capabilities and resolution of the AFM technique.
We cannot ignore, however, a second possibility that might be favored on other grounds. Predictions of secondary structure based on optimization of base pairing predict a different conformation in which disparate parts of the RNA molecule come together to form extensive runs of base pairs (27). Such predictions lead to a considerably more self-involved and complex-secondary structure with numerous branches. It has been argued that branch structures should be favored because they are intrinsically compact and would be more easily packaged than extended structures (44). Most of the secondary structures of STMV RNA based on energy minimization (or base pairing rules) have a long, central trunk amounting to double-stranded RNA with numerous branches and associated domains.
Such models might possibly also account for the lengths, degree of compression and heights that are observed by AFM in Figure 6. What weighs against such secondary structural arrangements is that the molecules seen in the AFM images exhibit no prominent branches, protrusions or accessory domains. We feel fairly sure that were branches of significant size present, they would be visible in the AFM images. We see no branches or features suggestive of major domains, not only in the phenol extracted RNAs of Figure 6, but in the long strands of RNA emerging from disrupted virions in Figure 7.
The two conformational possibilities have profound implications for the assembly of STMV, and for the 3D structure of both the free and the encapsidated genomes. In the first case, a linear string of small domains, which might otherwise experience constant rearrangement as it sought a more favorable energy state, could only be secured in a unique and stable conformation if coat protein molecules (which are double-stranded RNA-binding proteins) bound to the stem–loops and pseudo knots as they were synthesized, folded and emerged from the replication complex. Such a mechanism, as we discussed elsewhere (11), leads to a rather straightforward icosahedral virion assembly pathway that is consistent with the double-stranded RNA distribution seen in the X-ray crystallographic structure of the virus. Such an assembly process, which would be favored on kinetic grounds, also produces a metastable, rather than a minimum energy conformation for the encapsidated RNA, a seeming physiological necessity. It also eliminates the intrusion of kinetic traps, false energy minima that might otherwise impede or prevent the RNA from reaching the conformation necessary for encapsidation.
Both McPherson (45) and Devkota et al. (46) have emphasized the importance of interactions between the coat protein and the RNA in determining the folding pathway and the final encapsidation structure of the RNA in small icosahedral viruses. They suggested an assembly pathway in which binding of polycationic protein tails causes the RNA to collapse, thereby concentrating the proteins’ globular domains in a shell outside the RNA. This would facilitate otherwise weak protein–protein interactions and lead to formation of the mature capsid.
The second option, which would be favored by thermodynamic considerations, involves long-range base pairing interactions. It would require that the STMV RNA be fully synthesized, and that it mature by passing through intermediate conformational states until it reached a minimum energy structure. That structure would then have to present double-stranded RNA-binding sites to the coat protein that would then allow the two kinds of macromolecules to co-condense into an icosahedral virion. While none of this is implausible, it does seem to us less probable. Nevertheless, if in fact the RNA is produced in the absence of protein and undergoes extensive folding before assembly, then presumably this would have to be the operative pathway. Given the state of our knowledge of the in vivo folding of long single-stranded RNA molecules, it is difficult to disregard either mechanism.
FUNDING
National Institutes of Health [grants GM08112 (to A.M.) and M074899 to the Center for High Throughput Structural Biolog]. Funding for open access charge: National Institutes of Health (grants GM080412).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to thank Dr John Shriver for his assistance in the CD and DSC measurements and Mr Aaron Greenwood for preparation of figures.
REFERENCES
- 1.Francki RI. Plant virus satellites. Annu. Rev. Microbiol. 1985;39:151–174. doi: 10.1146/annurev.mi.39.100185.001055. [DOI] [PubMed] [Google Scholar]
- 2.Kassanis B. Properties and behaviour of a virus depending for its multiplication on another. J. Gen. Microbiol. 1962;27:477–488. doi: 10.1099/00221287-27-3-477. [DOI] [PubMed] [Google Scholar]
- 3.Pritsche C, Mayo X. In: Plant Viruses. Mandahar CL, editor. Vol. 1. Boca Raton: CRC Press; 1989. pp. 289–321. [Google Scholar]
- 4.Ban N, Larson SB, McPherson A. Structural comparison of the plant satellite viruses. Virology. 1995;214:571–583. doi: 10.1006/viro.1995.0068. [DOI] [PubMed] [Google Scholar]
- 5.Dodds JA. Structure and function of the genome of satellite tobacco mosaic virus. Can. J. Plant Pathol. 1991;13:192. [Google Scholar]
- 6.Valverde RA, Dodds JA. Evidence for a satellite RNA associated naturally with the U5 strain and experimentally with the U1 strain of TMV. J. Gen. Virol. 1986;67:1875. [Google Scholar]
- 7.Valverde RA, Dodds JA. Some properties of isometric virus particles which contain the satellite RNA of TMV. J. Gen. Virol. 1987;68:965. [Google Scholar]
- 8.Larson SB, Day J, Greenwood A, McPherson A. Refined structure of satellite tobacco mosaic virus at 1.8 A resolution. J. Mol. Biol. 1998;277:37–59. doi: 10.1006/jmbi.1997.1570. [DOI] [PubMed] [Google Scholar]
- 9.Larson SB, Koszelak S, Day J, Greenwood A, Dodds JA, McPherson A. Three-dimensional structure of satellite tobacco mosaic virus at 2.9 A resolution. J. Mol. Biol. 1993;231:375–391. doi: 10.1006/jmbi.1993.1289. [DOI] [PubMed] [Google Scholar]
- 10.Larson SB, Koszelak S, Day J, Greenwood A, Dodds JA, McPherson A. Double-helical RNA in satellite tobacco mosaic virus. Nature. 1993;361:179–182. doi: 10.1038/361179a0. [DOI] [PubMed] [Google Scholar]
- 11.Larson SB, McPherson A. Satellite tobacco mosaic virus RNA: structure and implications for assembly. Curr. Opin. Struct. Biol. 2001;11:59–65. doi: 10.1016/s0959-440x(00)00166-4. [DOI] [PubMed] [Google Scholar]
- 12.Fresco JR, Alberts BM, Doty P. Some molecular details of the secondary structure of ribonucleic acid. Nature. 1960;188:98–101. doi: 10.1038/188098a0. [DOI] [PubMed] [Google Scholar]
- 13.Onoa B, Tinoco I., Jr RNA folding and unfolding. Curr. Opin. Struct. Biol. 2004;14:374–379. doi: 10.1016/j.sbi.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Spirin BA. Intra-organ lymphatic system of the human penis. Arkh. Anat. Gistol. Embriol. 1963;44:68–80. [PubMed] [Google Scholar]
- 15.Witz J, Stazielle C, editors. Viral RNAs. New York: Marcel Dekker; 1973. [Google Scholar]
- 16.Freddolino PL, Arkhipov AS, Larson SB, McPherson A, Schulten K. Molecular dynamicds simulations of the complete Satellite Tobacco Mosaic Virus. Structure. 2006;14:1–13. doi: 10.1016/j.str.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Zucker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felden B, Florentz C, McPherson A, Giege R. A histidine accepting tRNA-like fold at the 3′-end of satellite tobacco mosaic virus RNA. Nucleic Acids Res. 1994;22:2882–2886. doi: 10.1093/nar/22.15.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dodds JA. Structure and function of the genome of satellite tobacco mosaic virus. Can. J. Plant Pathol. 1991;13:192–195. [Google Scholar]
- 20.Mirkov TE, Mathews DM, Du Plessis DH, Dodds JA. Nucleotide sequence and translation of satellite tobacco mosaic virus RNA. Virology. 1989;170:139–146. doi: 10.1016/0042-6822(89)90361-9. [DOI] [PubMed] [Google Scholar]
- 21.Day J, Kuznetsov YG, Larson SB, Greenwood A, McPherson A. Biophysical studies on the RNA cores of satellite tobacco mosaic virus. Biophys. J. 2001;80:2364–2371. doi: 10.1016/S0006-3495(01)76206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuznetsov YG, Daijogo S, Zhou J, Semler BL, McPherson A. Atomic force microscopy analysis of icosahedral virus RNA. J. Mol. Biol. 2005;347:41–52. doi: 10.1016/j.jmb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Koszelak S, Dodds JA, McPherson A. Preliminary analysis of crystals of satellite tobacco mosaic virus. J. Mol. Biol. 1989;209:323–325. doi: 10.1016/0022-2836(89)90281-7. [DOI] [PubMed] [Google Scholar]
- 24.Kuznetsov YG, Larson SB, Day J, Greenwood A, McPherson A. Structural Transitions of STMV Particles. Virology. 2001;284:223–234. doi: 10.1006/viro.2000.0914. [DOI] [PubMed] [Google Scholar]
- 25.Hayat MA. Principles and Techniques of Electron Microscopy: Biological Applications. 4th edn. Cambridge, UK: University of Cambridge Press; 2000. [Google Scholar]
- 26.Feldman MY. Reaction of formaldehyde with nucleotides and ribonucleic acid. Biohim. Biophys. Acta. 1967;149:20–34. doi: 10.1016/0005-2787(67)90687-9. [DOI] [PubMed] [Google Scholar]
- 27.Moller K, Rinke J, Ross A, Buddle G, Brimacombe R. The use of formaldehyde in RNA-protein cross linking studies with ribosomal subunits from E. coli. Eur. J. Biochem. 1977;76:175–187. doi: 10.1111/j.1432-1033.1977.tb11583.x. [DOI] [PubMed] [Google Scholar]
- 28.Kuznetsov YG, McPherson A. Atomic force microscopy investigation of turnip yellow mosaic virus capsid disruption and RNA extrusion. Virology. 2006;352:329–337. doi: 10.1016/j.virol.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Binning G, Quate CF. Atomic force microscope. Phys. Rev. Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 30.Bustamante C, Keller D. Scanning force microscopy in biology. Phys. Today. 1995;48:32–38. [Google Scholar]
- 31.Goldsbury CS, Scheuring S, Kreplak L. 2009. Introduction to Atomic Force Microscopy in Biology. Vol. Curr. Protocols in Protein Science. [DOI] [PubMed] [Google Scholar]
- 32.Jena BP, Heinrich Horber JK, editors. Atomic Force Microscopy in Cell Biology. NY: Elsevier, Academic Press; 2002. [Google Scholar]
- 33.Morris VJ, Kirby AR, Gunning AP. Atomic Force Microscopy for Biologists. 2nd edn. London: Imperial College Press; 2010. [Google Scholar]
- 34.Kuznetsov YG, Malkin AJ, Lucas RW, Plomp M, McPherson A. Imaging of viruses by atomic force microscopy. J. Gen. Virol. 2001;82:2025–2034. doi: 10.1099/0022-1317-82-9-2025. [DOI] [PubMed] [Google Scholar]
- 35.Kuznetsov YG, McPherson A. Atomic force microscopy investigation of Turnip Yellow Mosaic Virus capsid disruption and RNA extrusion. Virology. 2006;352:329–337. doi: 10.1016/j.virol.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 36.Kuznetsov YG, Datta S, Kothari NH, Greenwood A, Fan H, McPherson A. Atomic force microscopy investigation of fibroblasts infected with wild-type and mutant murine leukemia virus (MuLV) Biophys. J. 2002;83:3665–3674. doi: 10.1016/S0006-3495(02)75366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuznetsov YG, Victoria J, Low GA, Robinson JEW, Fan H, McPherson A. AFM Imaging of Retroviruses HIV and MuLV. SCANNING. 2004;26:209–216. doi: 10.1002/sca.4950260409. [DOI] [PubMed] [Google Scholar]
- 38.Kuznetsov YG, McPherson A. Identification of DNA and RNA from retroviruses using ribonuclease A. Scanning. 2006;28:278–281. doi: 10.1002/sca.4950280506. [DOI] [PubMed] [Google Scholar]
- 39.Malkin AJ, Plomp M, McPherson A. In: Virus Structure and Imaging, DNA Viruses, Methods and Protocols. Lieberman PM, editor. Totowa, New Jersey: Humana Press; 2004. pp. 85–108. [Google Scholar]
- 40.Hansma HG, Hoh JH. Biomolecular imaging with the atomic force microscope. Annu. Rev. Biophys. Biomol. Struct. 1994;23:115–139. doi: 10.1146/annurev.bb.23.060194.000555. [DOI] [PubMed] [Google Scholar]
- 41.Hansma HG, Pietrasanta L. Atomic force microscopy and other scanning probe microscopies. Curr. Opin. Chem. Biol. 1998;2:579–584. doi: 10.1016/s1367-5931(98)80086-0. [DOI] [PubMed] [Google Scholar]
- 42.Ko TP, Day J, Greenwood A, McPherson A. Structures of three crystal forms of the sweet protein thaumatin. Acta. Crystallogr. D. Biol. Crystallogr. 1994;50:813–825. doi: 10.1107/S0907444994005512. [DOI] [PubMed] [Google Scholar]
- 43.Kuznetsov Yu G, Malkin AJ, McPherson A. AFM studies on the mechanisms of nucleation and growth of macromolecular crystals. J. Cryst. Growth. 1999;196:489–502. [Google Scholar]
- 44.Yoffe AM, Prinsen P, Gopal A, Knobler CM, Gelbart WM, Ben-Shul A. Predicting the sizes of large RNA molecules. Proc. Natl Acad. Sci. USA. 2008;105:16153–16158. doi: 10.1073/pnas.0808089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McPherson A. Micelle formation and crystallization as paradigms for virus assembly. Bioessays. 2005;27:447–458. doi: 10.1002/bies.20196. [DOI] [PubMed] [Google Scholar]
- 46.Devkota B, Petrov AS, Lemieux S, Boz MB, Tang L, Schneemann A, Johnson JE, Harvey SC. Structural and electrostatic characterization of pariacoto virus: implications for viral assembly. Biopolymers. 2009;91:530–538. doi: 10.1002/bip.21168. [DOI] [PMC free article] [PubMed] [Google Scholar]