Abstract
RNase P catalyzes the Mg2+-dependent 5′-maturation of precursor tRNAs. Biochemical studies on the bacterial holoenzyme, composed of one catalytic RNase P RNA (RPR) and one RNase P protein (RPP), have helped understand the pleiotropic roles (including substrate/Mg2+ binding) by which a protein could facilitate RNA catalysis. As a model for uncovering the functional coordination among multiple proteins that aid an RNA catalyst, we use archaeal RNase P, which comprises one catalytic RPR and at least four RPPs. Exploiting our previous finding that these archaeal RPPs function as two binary RPP complexes (POP5•RPP30 and RPP21•RPP29), we prepared recombinant RPP pairs from three archaea and established interchangeability of subunits through homologous/heterologous assemblies. Our finding that archaeal POP5•RPP30 reconstituted with bacterial and organellar RPRs suggests functional overlap of this binary complex with the bacterial RPP and highlights their shared recognition of a phylogenetically-conserved RPR catalytic core, whose minimal attributes we further defined through deletion mutagenesis. Moreover, single-turnover kinetic studies revealed that while POP5•RPP30 is solely responsible for enhancing the RPR’s rate of precursor tRNA cleavage (by 60-fold), RPP21•RPP29 contributes to increased substrate affinity (by 16-fold). Collectively, these studies provide new perspectives on the functioning and evolution of an ancient, catalytic ribonucleoprotein.
INTRODUCTION
RNase P, a ribonucleoprotein (RNP), is the endoribonuclease that catalyzes the removal of 5′-leaders in precursor tRNAs (pre-tRNAs) in all three domains of life (1–4). The bacterial variant is composed of 1 catalytic RNase P RNA (RPR) and 1 RNase P protein (RPP) cofactor. Eukaryal (nuclear) RNase P from yeast and human contain 1 RPR with 9 and 10 RPPs, respectively. Intermediate in complexity, the archaeal holoenzyme is associated with 1 RPR and at least 4 RPPs (POP5, RPP30, RPP21 and RPP29), which are homologous to eukaryal RPPs.
Several years after the remarkable finding that the bacterial RPR is a true RNA enzyme in the presence of Mg2+ and monovalent ions (5), archaeal and eukaryal RPRs were also shown to be catalytically active in vitro (6,7). This common attribute of evolutionarily divergent RPRs was anticipated from their shared ancestry, attested by sequence and structural similarity of their putative catalytic core (8–15). However, dramatic variations (106-fold) in catalytic potential indicate that not all RPRs are equal: activity of RPRs from bacteria > archaea > eukarya (5–7). Although RPRs display activity in vitro without RPPs, they are dependent on their cognate protein cofactors for cellular function. Interestingly, there is an inverse relationship between RPR activity and RNP composition—the protein:RNA mass ratio is 70% in eukaryal and 50% in archaeal RNase P compared to 10% in their bacterial counterpart. Thus, elucidating the intimate cooperation between the RNA and protein subunits of RNase P variants with differing RNP make-up offers a paradigm to understand how structural and functional attributes of RNAs might have been reassigned to protein cofactors during the evolutionary transition from an RNA to RNP world (1–3,15). Toward this goal, we have focused our efforts on the simpler and biochemically tractable archaeal version, especially as an experimental surrogate for the eukaryal relative which is yet to be reconstituted in vitro. Rapid advances in functional reconstitution (16–19) and structural studies (16,20–29) have validated this choice.
We have shown earlier that robust RNase P activity could be obtained from assembling recombinant subunits of Pyrococcus furiosus (Pfu; type A), and that the four RPPs functioned as two binary complexes (POP5•RPP30 and RPP21•RPP29) (18). Since the structural diversity of RNase P is exemplified even within archaea where the type A and M RPRs resemble the bacterial and eukaryal relatives, respectively (Figure 1) (13,15), we reasoned that heterologous reconstitution of types A and M archaeal RNase P variants will facilitate delineation of their conserved and divergent features, and thereby help to understand the dynamic co-evolution of the RNA and protein subunits in RNase P from all domains of life. Indeed, heterologous reconstitutions of archaeal RPPs with bacterial and organellar RPRs have helped define a minimal RNP catalytic core that is obscured by natural variations in the subunit make-up of RNase P. Our single-turnover studies also provide key insights into the division of labor among archaeal RPPs, and permit formulation of a simple kinetic framework to highlight this functional cooperation. These findings, together with the structures of the binary RPPs (25,28,29), should aid efforts to establish structure–function correlations in archaeal RNase P.
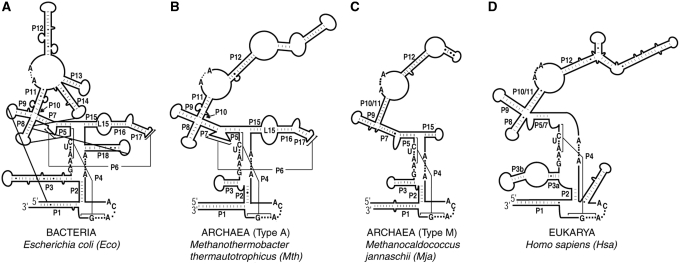
Figure 1.
Secondary structure representations of (A) Eco, (B) Mth, (C) Mja and (D) Hsa RPRs (12,15). Letters indicate universally conserved nucleotides (12). Paired helices (e.g. P1, P2) are numbered consecutively from 5′ to 3′ and according to the Eco RPR nomenclature (12). Alternative secondary structure representations (69) are provided in Supplementary Figure S1.
MATERIALS AND METHODS
Cloning, overexpression and purification of various RPRs and RPPs
Complete details are provided in the Supplementary Data.
RNase P assays
All assays were performed with Escherichia coli (Eco) pre-tRNATyr as the substrate, a trace amount of which was labeled with [α-32P] GTP. Although we provide below the details for the individual assemblies for different homologous and heterologous reconstitutions, we list here some common features. All reconstitutions and assays were performed in a thermal cycler. RPRs (unless otherwise stated) were folded as follows: incubation at 50°C for 50 min in water followed by 37°C for 30 min in 50 mM Tris–HCl (pH 7.5), 800 mM NH4OAc and 10 mM MgCl2. The optimal RPR:RPP ratios for each combination and the substrate concentrations were empirically determined. Optimal RNP assembly entailed successive incubations at 37 and 55°C for 5–10 min each. Assays were always initiated by adding pre-tRNATyr, which had been pre-incubated at 55°C for 2 min. Aliquots were removed at defined time intervals and added to an equal volume of stop solution [10 M urea, 5 mM EDTA, 0.05% (w/v) bromophenol blue, 0.05% (w/v) xylene cyanol, 10% (v/v) phenol] to terminate the reaction. For short incubations (e.g. 5 s), reactions were first terminated by immersing the reaction tubes in liquid nitrogen before adding stop solution. The reaction contents were separated using denaturing PAGE [8% (w/v) polyacrylamide, 7 M urea].
Multiple-turnover assays with Methanocaldococcus jannaschii, Methanothermobacter thermautotrophicus and Pfu RNase P
To facilitate qualitative comparisons with multiple-turnover assays reported for Pfu RNase P (18), we assayed Methanocaldococcus jannaschii (Mja) and Methanothermobacter thermautotrophicus (Mth) RNase P under conditions identical to those used for the Pfu counterpart. Partially reconstituted RNase P holoenzymes were assayed in 50 mM Tris–HCl (pH 7.5), 100 mM NH4OAc and 120 mM MgCl2 with 10 nM RPR + 100 nM POP5•RPP30 or 250 nM RPR + 625 nM RPP21•RPP29. For the holoenzyme with four RPPs, 10 nM folded RPR was incubated with 100 nM of all 4 RPPs in 50 mM Tris–HCl (pH 7.5), 800 mM NH4OAc and 30 mM MgCl2. In all cases, the resulting RNPs were incubated with 500 nM pre-tRNATyr for 30 min at 55°C.
Reconstitutions of Mja RPR ΔS and RPR ΔS Min were performed by combining 500 nM of each RNA with 1 µM of all four Mja RPPs in assay buffer [50 mM Tris–HCl (pH 7.5), 400 mM NH4OAc and 30 mM Mg(OAc)2]. For reconstitutions with binary RPPs, Mg(OAc)2 was increased to 120 mM. The resulting RNPs were incubated with 1 µM pre-tRNATyr for 15 min at 55°C.
In Figures 3 and 4, the archaeal RPRs were assayed under conditions as those used for the respective RPR + 4 RPPs.
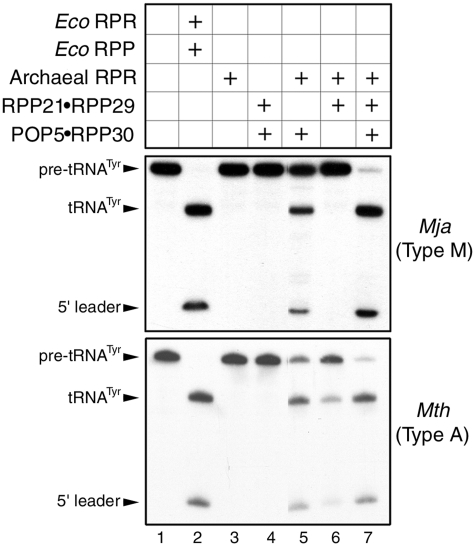
Figure 3.
Reconstitution of RNase P activity with Mja (top panel) or Mth (bottom panel) RPR and cognate RPPs purified as binary complexes (see text for details).
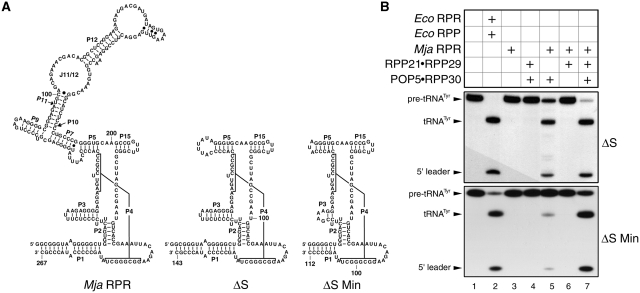
Figure 4.
Identification of the smallest functional RPR (yet reported) and a minimal catalytic RNP core of Mja RNase P. (A) Secondary structure models of Mja RPR and its deletion derivatives. (B) Reconstitution of RNase P activity using the two Mja RPR deletion derivatives and various combinations of Mja RPPs as indicated.
Multiple-turnover assays with heterologously-reconstituted RNase P holoenzymes
A total of 50 nM archaeal type A RPR and 250 nM type M RPPs (or type M RPR and type A RPPs) were reconstituted in 50 mM Tris–HCl (pH 7.5), 800 mM NH4OAc and 30 mM Mg(OAc)2. The resulting RNPs were incubated with 500 nM pre-tRNATyr for 15 min at 55°C.
A total of 50 nM of folded Eco (30), Bacillus subtilis (Bsu) (31) or Reclinomonas americana (Ram) mitochondrial (mt) (32) RPR was reconstituted with archaeal RPPs (500 nM) in 50 mM Tris–HCl (pH 7.5), 800 mM NH4OAc and 30 mM Mg(OAc)2. The resulting RNPs were incubated with 200 nM pre-tRNATyr at 55°C for 5 min (Eco and Bsu RPRs) or 15 min (Ram mt RPR).
Single-turnover kinetic studies with Mth RNase P
For these experiments, 50 mM 2-(N-morpholino)ethanesulfonic acid (MES)–HCl, pH 5.8 (unless otherwise indicated) was used instead of Tris–HCl in RPR folding and assays. The folded RPR was assayed either alone or with a 2-fold molar excess of RPPs using the optimal concentrations of MgCl2 and NH4OAc for each RNP (Table 1). In the case of RNPs, the amount of RPR in the assay was used as the concentration of enzyme based on the assumption that all of the RPR is assembled into RNPs under the conditions employed. To determine the maximal kobs (max. kobs) under single-turnover conditions at 55°C, we incubated ∼2 nM pre-tRNATyr with a range of enzyme concentrations: RPR, 0.3–20 μM; RPR + RPP21•RPP29, 0.5–3 μM; RPR + POP5•RPP30, 1–10 μM; and RPR + 4 RPPs, 0.3–3 μM.
Table 1.
Effect of Mth RPPs on the ionic requirements and rate of cleavage of pre-tRNATyr by Mth RPR at 55°Ca
| Assayed under the optimal condition for each catalytic entity |
Assayed under the optimal condition for the holoenzyme with four RPPs (30 mM Mg2+, 800 mM NH4+ |
|||||||
|---|---|---|---|---|---|---|---|---|
| Max. kobsa, min−1 | KM(STO)a, µM | [NH4+], M | [Mg2+], M | Relative | kobs, min−1 | Relative | ||
| max. kobs | kobs | |||||||
| Mth RPR | 0.13 ± 0.01 | 21.3 ± 2.6 | 2.0 | 0.50 | 1 | Mth RPR (3 µM) | b | – |
| Mth RPR + RPP21•RPP29 | 0.13 ± 0.01 | 1.4 ± 0.2 | 0.8 | 0.12 | 1 | Mth RPR + RPP21•RPP29 | b | – |
| Mth RPR + POP5•RPP30 | 7.9 ± 1 | 11.8 ± 2.3 | 0.8 | 0.12 | ∼60 | Mth RPR + POP5•RPP30 | 0.016 ± 0.0004 | 1 |
| Mth RPR + Both binary RPP complexes | 7.5 ± 0.14c | 1.2 ± 0.1 | 0.8 | 0.03 | ∼60 | Mth RPR + Both binary RPP complexes | 5.5 ± 0.18c | ∼344 |
aThe standard errors of the curve fits shown in Figure 7 are indicated in the estimates of max. kobs and KM(STO).
bReliable data could not be obtained due to weak or negligible activity.
cAll experiments were performed at pH 5.8, except for Mth RPR reconstituted with both binary RPP complexes, which was assayed at pH 5.4. After establishing a slope of approximately 1 in a plot of log kobs versus pH (see Supplementary Figure S6), we multiplied the maximal rate observed at pH 5.4 by 2.5 to obtain the rate that would have been observed at pH 5.8 should it have been measurable.
Because the Mth RPR + 4 RPPs reaction is too rapid at pH 5.8 (t1/2 ≤ 8 s) when [E] > 1μM, we could obtain reliable data only by decreasing the assay pH to 5.4. To establish the dependence of the rates of product formation catalyzed by Mth RPR + 4 RPPs on assay pH, the assays were performed in 20 mM instead of 30 mM Mg2+ and in the pH range 5.4–6.15 (Supplementary Figure S6). [In an earlier publication (19), the rates reported for a self-cleaving Mja (type M) RPR were at pH 5.4 and not pH 5.1 as was mistakenly noted].
Data analysis
After denaturing PAGE, the reaction products were visualized by phosphorimaging on the Typhoon (GE Healthcare). The resulting bands were quantitated by ImageQuant (GE Healthcare) to assess the extent of substrate cleaved. To obtain the rate of product formation (kobs) in single-turnover reactions, the percentage product formed at time t (Pt) was fit to Pt = P∞(1 − e−kt) using Kaleidagraph software (Synergy). For optimal curve fits, the amplitudes were defined based on experimentally observed values (Supplementary Figure S5). The individual curve-fit errors for kobs did not exceed 8%. For all kinetic studies, at least three replicates were performed to obtain the mean and standard deviation values.
The plot of kobs versus [E0] displayed hyperbolic dependence on the Mth RPR in the absence and presence of its RPPs (Figure 7). Kaleidagraph was used to fit these data to
to derive values for max. kobs and KM(STO).
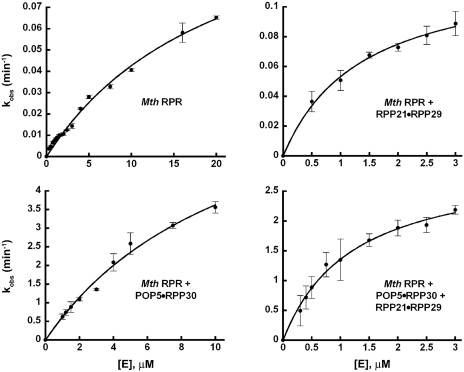
Figure 7.
Effects of Mth RPPs on the single-turnover rate of Mth RPR-catalyzed pre-tRNA cleavage. The rates of product formation (kobs) by Mth RPR with and without RPPs were determined under single-turnover conditions and plotted as a function of the concentration of the respective catalytic entity to obtain the max. kobs and KM(STO) reported in Table 1.
RESULTS
While type A archaeal RPRs process pre-tRNAs in vitro in the absence of their cognate protein cofactors, type M counterparts display such an activity only when the substrate is provided in cis, perhaps reflecting their inability to bind substrates (6,19). Compared to bacterial and archaeal type A relatives, type M RPRs exhibit two striking structural changes (Figure 1) (13). First, they are missing P8, which is part of the P7-9 cruciform in bacterial RPRs that is involved in T-loop recognition and binding (33–36). Second, type M RPRs lack P6, P16, P17 and the loop L15 that connects P15 and P16; L15 interacts with the 3′-RCCA of the pre-tRNA substrate (37,38). If type M RPPs have evolved to compensate for these structural alterations in their cognate RPRs, then the effects of RPPs on RPR catalysis will be distinctive in type A and M archaeal RNase P. As a first step to elucidate such co-evolutionary trends, we sought to reconstitute RNase P from Mja (type M), and compare it with those from Pfu and Mth (type A).
Purification of archaeal RPPs as binary complexes and their functional validation
To expediently assemble different archaeal RNase P holoenyzmes in vitro, we explored a strategy that entailed (i) purifying the four RPPs as two binary RPP complexes, after their co-overexpression in Eco, and (ii) reconstituting these recombinant RPP pairs with their cognate (or another heterologous) RPR prepared by in vitro transcription. The rationale for this approach was based on several observations. First, biochemical and genetic studies had already established pair-wise interactions between POP5•RPP30 and RPP21•RPP29 (18,19,39,40). Second, NMR studies revealed significant chemical-shift perturbations in the HSQC spectrum of each RPP upon addition of its binary complex partner, indicating strong macromolecular interactions within the pairs even in the absence of the RPR (26,27,29). Also, the crystal structure of Pyrococcus horikoshii (Pho) POP5 was solved as a heterodimer with RPP30 (25). Third, there is a growing realization that multi-component protein complexes are better reconstituted in vivo than in vitro, presumably due to better protein folding (41,42). The interacting pair of proteins from such complexes could be expressed from either a bicistronic construct in one vector or from two compatible plasmids each of which harbors one ORF. Similar to the findings which we describe below, two yeast RPPs (Pop6 and Pop7) were isolated as a recombinant heterodimer and their crystal structure determined in complex with an RNA structural domain derived from the RNase MRP RNA (43,44).
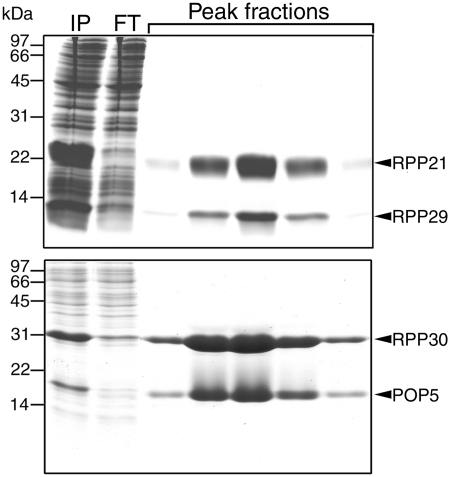
We cloned the genes encoding either type A or M POP5 and RPP30 (or RPP21 and RPP29) into compatible vectors that allow T7 RNA polymerase-based overexpression in Eco BL21(DE3) cells; we also cloned these pairs in tandem in a single overexpression vector. These tandem constructs were used in this study. We did not use affinity tags to facilitate purification since they interfered with assembly. By using ammonium sulfate fractionation and cation-exchange chromatography (which exploits the typically high pI values of RPPs, Supplementary Table S2), we were able to isolate to homogeneity RPP pairs from phylogenetically diverse archaea (Figure 2 and Supplementary Figure S2). In some cases (e.g. Mja), the individual RPPs could not be either overexpressed or purified to homogeneity in the absence of their partner.
Figure 2.
SDS–PAGE profiles depicting the purification of binary complexes of Mja RPPs: RPP21•RPP29 (top panel) and POP5•RPP30 (bottom panel). IP represents the ammonium sulfate-fractionated sample that was subjected to the SP-Sepharose column chromatography and FT denotes the flow through.
Before initiating biochemical studies, we investigated if there are any functional differences between the binary RPPs and those that were purified individually and assembled as pair-wise complexes in vitro. We therefore purified the four Pfu RPPs individually (18) and as two interacting pairs (Supplementary Figure S2). For the latter, we used SYPRO Ruby staining of an SDS–polyacrylamide gel followed by fluorescence-based quantitation to establish that the heterodimer stoichiometry was ∼1:1 (Supplementary Table S3). We then reconstituted Pfu RPR with either the four individually purified RPPs or the two binary RPP complexes and assayed their activity. The kcat and KM values for cleavage of pre-tRNATyr were indistinguishable for these two in vitro reconstituted holoenzymes (Supplementary Table S4), thus allaying concerns of possible artifacts arising from use of binary complexes.
Reconstitution of an archaeal type M RNase P and comparison with its type A relatives
When Mja POP5•RPP30 and RPP21•RPP29 were reconstituted with the Mja RPR, the Mja RNase P holoenzyme exhibited multiple turnover at 55°C with Eco pre-tRNATyr as the substrate. Since neither the Mja RPR nor the binary RPP complexes alone promote cleavage of pre-tRNATyr under similar assay conditions (Figure 3, top panel, lanes 3 and 4), it is clear that the RNP complex is the functional unit under these conditions.
Although Pfu (type A) RPR is capable of multiple turnover in the absence of RPPs, the presence of either RPP21•RPP29 or POP5•RPP30 increases activity particularly at lower substrate and Mg2+ concentrations, demonstrating that each partial holoenyzme constitutes a minimal functional complex (18). Compared to the reaction with RPR + 4 RPPs, these partial holoenzymes displayed 600- or 100-fold lower kcat/KM values for cleavage of pre-tRNATyr (18). To test whether these findings are qualitatively applicable for a type M RNase P, we assayed the partial RNP complexes assembled from Mja RPR and either Mja RPP21•RPP29 or POP5•RPP30. Under multiple-turnover conditions, activity was observed with Mja RPR + POP5•RPP30 but not RPP21•RPP29 (Figure 3, top panel, lanes 5 versus 6), a notable difference with Pfu RNase P. However, in both types A and M, the RPR + 4 RPPs has a 30 mM Mg2+ requirement compared to 120 mM Mg2+ needed by the RNPs assembled with only one of the binary RPPs.
The observed differences in behavior of the partially reconstituted Mja (type M) and Pfu (type A) RNase P complexes could be attributed either to fundamental differences in the structure and functioning of these two classes of archaeal RNase P or to the remote possibility that Pfu RNase P might somehow differ from other type A relatives. To rule out the latter, we purified RPPs from Mth, another member of the type A family and repeated the partial reconstitution studies. The results obtained with Mth RNase P (Figure 3, bottom panel) parallel those we reported for Pfu RNase P (18), confirming similar patterns for the two type A variants and highlighting the distinctive reconstitution results obtained with types A and M.
Reconstitution of Mja RPR deletion derivatives with Mja RPPs
Like protein enzymes, the bacterial RPR is modular comprising a specificity (S) and a catalytic (C) domain (45). While the S domain interacts with conserved nucleotides recognizing the T stem–loop of the pre-tRNA, the C domain cleaves the pre-tRNA while binding to the 5′-leader, acceptor stem and the 3′-RCCA (33,36,37,46). Emphasizing the critical role of the C domain is the finding that there are approximately 11 universally conserved nucleotides at nearly equivalent locations in its secondary structure (Figure 1) (12,15). The juxtaposition of the two domains in the bacterial RPR’s tertiary fold (47,48) highlights the cooperation between these two domains for efficient substrate binding and catalysis. When the S domain was removed from the bacterial RPR, its activity was nearly 25 000-fold weaker than the full-length RPR and this difference was narrowed to 40-fold upon addition of the bacterial RPP (49). Reminiscent of this scenario was our earlier finding that an archaeal type A (Pfu) RPR’s C domain was defective in the absence of POP5•RPP30 (18). To examine whether an isolated C domain from type M RPR behaves similarly, we deleted in Mja RPR the S domain, corresponding to nucleotide 67 through 192, to generate Mja RPR ΔS.
Mja RPR ΔS by itself was inactive but was able to reconstitute with POP5•RPP30 (Figure 4B, top panel, lane 3 and 5). In contrast, under identical assay conditions, we did not observe any noticeable RNase P activity when RPP21•RPP29 was incubated with Mja RPR ΔS (Figure 4B, top panel, lane 6). This dichotomy is not unanticipated since (i) RPP21•RPP29 fails to activate even the full-length Mja RPR and (ii) footprinting studies showed that Mja POP5•RPP30 and RPP21•RPP29 interact with the RPR’s C and S domains, respectively (29). However, we were surprised that RPP21•RPP29 was able to stimulate the activity of Mja RPR ΔS + POP5•RPP30 (Figure 4B, top panel, lane 7), likely reflecting indirect effects arising from interactions between the two RPP pairs. Such an effect was not observed when we tested Pfu RPR ΔS with the two cognate binary RPP pairs (18).
Relative to a type A RPR, the C domain of a type M RPR is smaller due to the lack of certain structural elements (e.g. P6, P16 and P17) (13). To identify the smallest RPR variant that might yet generate a functional RNP, we trimmed the P1 and P3 helices and the length of the loop that caps P5 in Mja RPR ΔS. The resulting minimal Mja RPR ΔS Min (112 nt; Figure 4A) reconstitutes with POP5•RPP30 and is further activated upon addition of RPP21•RPP29, similar in behavior to Mja RPR ΔS (Figure 4B, lanes 5 and 7).
Heterologous reconstitutions within and across domains highlights a universal catalytic core
The results with Mja RPR ΔS Min revealed that the universally conserved catalytic core embedded in a minimal structural fold suffices for functional reconstitution with Mja RPPs. To test the premise that a common functional core might exist in all archaeal RNase P holoenzymes, we mixed type A RPRs with type M RPPs and vice versa. Indeed, heterologous assemblies did yield functional holoenzymes (Supplementary Figure S3). However, type A and M RPRs functioned more effectively with their cognate RPPs; such type-preferential reconstitution patterns might reflect the co-evolution of RPRs with their respective RPPs. These data nevertheless led us to examine if the bacterial and organellar RPR, which possess this conserved core, could also reconstitute with archaeal RPPs (minimally with POP5•RPP30).
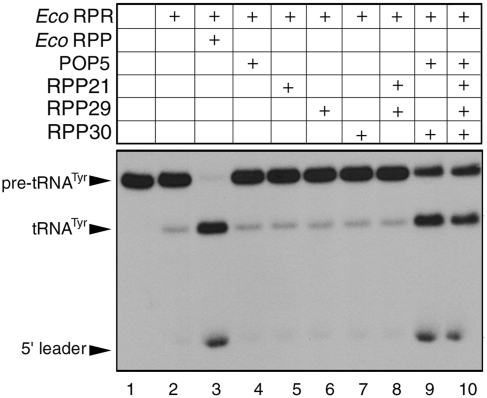
Under optimal conditions for reconstitution, both Eco (bacterial type A) and Bsu (bacterial type B) RPRs reconstituted with both type A (Pfu) and M (Mja) archaeal RPPs to generate functional RNase P holoenzymes [Figure 5, (50) and data not shown]. To examine which subset of RPPs suffices to promote bacterial RPR catalysis, we tested the Eco RPR with Pfu POP5+RPP30 or RPP21+RPP29 at 55°C and pH 7.5. We observed that while POP5+RPP30 could enhance the activity of Eco RPR, RPP21+RPP29 has no effect (Figure 5, lanes 8 and 9). Both type A and B RPRs were stimulated roughly 10- to 15-fold, respectively, by Pfu POP5•RPP30 [(50) and data not shown]. Given the remarkable tertiary structure similarity between POP5 and the bacterial RPP (26), we further examined if POP5 alone could stimulate the bacterial RPR’s pre-tRNA cleavage activity; however, this is not the case (Figure 5, lane 4). Taken together, these results suggest that POP5•RPP30 is functionally equivalent to the bacterial RPP.
Figure 5.
Heterologous reconstitution of an RNase P holoenzyme using Eco RPR and Pfu RPPs (in the combinations indicated).
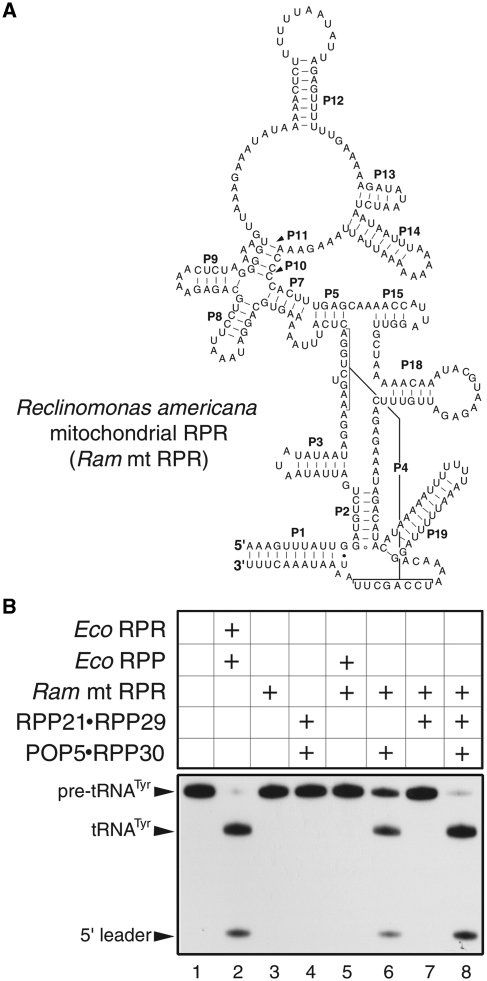
We then assessed whether Mja RPPs could support organellar RPR catalysis. We focused on the mitochondrially (mt)-encoded RPR of Ram, a jakobid flagellate. Ram mt RPR represents a highly reduced version of the bacterial RPR, lacking P6, P16, P17 and L15 (Figure 6A) and was found to be catalytically inactive under various conditions tested in vitro (32). Interestingly, Mja POP5•RPP30 (and not RPP21•RPP29) can heterologously reconstitute with Ram mt RPR (Figure 6B, lanes 6 and 7); this activity is enhanced upon addition of RPP21•RPP29 (Figure 6B, lane 8), in a manner reminiscent of the reconstitutions of Mja RPR ΔS or ΔS Min with Mja RPPs (Figure 4).
Figure 6.
Heterologous reconstitution of an RNase P holoenzyme using Ram mt RPR and archaeal (Mja) RPPs. (A) Secondary structure representation of Ram mt RPR. (B) Reconstitution of RNase P using Ram mt RPR and Mja RPPs (in the combinations indicated).
Single-turnover kinetic studies to elucidate the role of RPPs in aiding the cleavage step
From the various homologous and heterologous reconstitutions that we performed, it was clear that the two RPP pairs have different functional effects. Therefore, we sought to dissect these differences further using detailed rate measurements. Although we previously showed that only the addition of Pfu POP5•RPP30 to Pfu RPR increases its kcat (18), it remains unclear whether product release is rate limiting under multiple-turnover conditions [as demonstrated for bacterial and yeast RNase P (51,52)]. Hence, we undertook single-turnover assays to measure the RPR’s rate of pre-tRNA cleavage either alone or when aided by each archaeal RPP pair.
To facilitate manual single-turnover measurements, we slowed down the reaction rate by decreasing the assay pH from the typical 7.5 to 5.8, since the hydroxide nucleophile, which attacks the scissile phosphodiester linkage in the pre-tRNA, is believed to result from deprotonation of a hydrated Mg2+ ion in the RPR’s active site (53–56). To shorten incubation times, we used Mth in lieu of Pfu RNase P because the Mth RPR is more active in vitro (6).
An excess of enzyme over substrate ([E] = 0.3–20 µM, [S] ∼2 nM) was used in our single-turnover studies with Mth RNase P. We found that a single-exponential function describes the rate of product formation by the Mth RPR ± RPPs (Supplementary Figure S5). We use max. kobs to indicate rates determined at saturating concentrations of the RPR ± RPPs in the presence of optimal levels of NH4+ and Mg2+, which were established by screening different assay conditions. The term KM(STO) is used to refer to the KM calculated under single-turnover conditions.
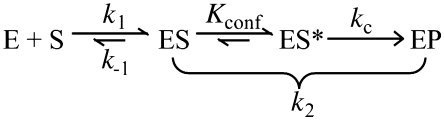
To justify using KM(STO) [(k–1 + k2)/k1] as a measure of KS (k–1/k1), we performed a pulse-chase experiment to investigate if the ES complex dissociates faster than substrate cleavage (i.e. k−1 >> k2; Scheme I) under the single-turnover conditions used. Through large dilution of a pre-formed Mth RNase P–pre-tRNATyr complex, we dissociated the substrate from the enzyme and expected a plateau in product formation post-dilution if k–1 >> k2 (due to the pre-tRNA’s inability to rebind for cleavage). Indeed, when the reactions catalyzed by Mth RPR ± RPP21•RPP29 were diluted a few minutes after mixing with pre-tRNATyr, product formation did not increase post dilution (Supplementary Figure S4).
Scheme I.
At pH 5.8 and 55°C, both Mth POP5•RPP30 and RPP21•RPP29 decreased the concentration of NH4+ and Mg2+ required for RPR-mediated pre-tRNATyr processing; the Mg2+ requirement decreased from 500 to 120 mM with either binary complex and to 30 mM with both (Table 1). The max. kobs for processing of pre-tRNATyr by Mth RPR increased 60-fold upon addition of POP5•RPP30 with no further change upon inclusion of RPP21•RPP29 (Table 1; Figure 7). Also, RPP21•RPP29 alone was unable to increase the RPR’s max. kobs. However, RPP21•RPP29 was able to reduce KM(STO) from 21.3 to 1.4 µM (to the same extent as four RPPs; Table 1). These results demonstrate the importance of POP5•RPP30 in cleavage and RPP21•RPP29 in substrate binding (however, see ‘Discussion’ section for additional comments on the role of POP5•RPP30).
The above studies were performed at the optimal Mg2+ concentration for each of the RNP complexes assembled with Mth RPR. We inquired if the results would be different if the Mth RPR was assayed with and without each binary complex at 30 mM Mg2+ and 800 mM NH4+, a condition optimal for the holoenzyme assembled with all four RPPs but not for the partial RNPs. There is no detectable activity in the RPR-alone reaction and weak (not reliably quantifiable) activity with RPR + RPP21•RPP29. In contrast, the RPR + POP5•RPP30 exhibits a kobs of 0.016 min−1, which increased to 5.5 min−1 (344-fold) upon addition of RPP21•RPPP29, suggesting that the latter significantly facilitates catalysis at lower Mg2+ concentrations.
Some rate comparisons are instructive. First, although Li et al. (57) did not report a max. kobs, they documented pre-tRNAGly cleavage by 10 µM Mth RPR at pH 6 with a rate of 0.034 min−1, which is similar to the 0.04 min−1 that we observed for cleavage of pre-tRNATyr by 10 µM Mth RPR at pH 5.8; this coincidence is reassuring given the different substrates and assay conditions used in these two studies. Second, the max. kobs for pre-tRNATyr cleavage by Mth RPR is 0.13 min−1 at pH 5.8 and 55°C (Table 1) compared to the bacterial RPR’s max. kobs of ∼5 min−1 at pH 6 and 37°C (57). These data, together with earlier studies on chimeric Mth-Eco RPRs (57), reaffirm the idea that the bacterial RPR structural elements which are missing in the type A archaeal RPR contribute to both substrate binding and cleavage rate.
DISCUSSION
We have shown that purification of RPPs as binary complexes allows expedient assembly of RNase P holoenzymes from different archaea. In addition to halving the protein preparation effort, co-overexpression was a necessary measure to obtain recombinant RPPs (e.g. Mja RPP30) that do not express by themselves. These facile assemblies represent an important first step to investigate the stoichiometry and oligomeric state of individual subunits, and to initiate high-resolution structural studies. The latter objective might also benefit from use of pared-down RPRs such as Mja RPR ΔS Min (112 nt), the smallest known RPR that is functional in the presence of POP5•RPP30. Importantly, these biochemical reconstitutions help uncover the mechanistic basis of protein-aided RNA catalysis in archaeal RNase P.
Delineating the roles of archaeal RPPs in aiding RPR catalysis
Hint of the role of POP5•RPP30 in promoting pre-tRNA cleavage first came from multiple-turnover studies on Pfu RNase P where it was shown to increase the RPR’s kcat by 25-fold (18). We then used an Mja RPR—pre-tRNATyr cis conjugate, a good model system for studying rate-limiting chemistry, and found that the rate of self-cleavage of this conjugate was accelerated ∼100-fold by Mja POP5•RPP30, but not at all by RPP21•RPP29 (19). However, if RPP21•RPP29 plays a role in substrate positioning that affects the rate of cleavage, such an effect would manifest during pre-tRNA cleavage in a trans rather than in a cis reaction where the substrate is already docked. The single-turnover studies with Mth RNase P now provide new insights on the division of labor between the two binary RPPs. Only RPP21•RPP29 decreased the KM(STO) of Mth RPR, while only POP5•RPP30 elicited a 60-fold increase in the max. kobs (Table 1 and Figure 7).
Our archaeal RNase P data could be interpreted by a framework similar to that used to describe the role of the bacterial RPP (60; Scheme I and Supplementary Figure S7). Various kinetic and structural studies on bacterial RNase P have indicated that subsequent to substrate binding, a conformational change converts ES to ES* (defined by the equilibrium constant Kconf) and optimally positions the pre-tRNA and catalytic Mg2+ ions for cleavage (at rate kc) (58–61). Adding to a growing body of evidence supporting such a two-step mechanism, data from recent stopped-flow kinetic studies (61) confirm an initial bi-molecular collision (E + S → ES) followed by a uni-molecular conformational change (ES→ES*). Thus, under single-turnover conditions with saturating concentrations of enzyme, max. kobs = kc (Kconf/1+Kconf). Based on kinetic studies with bacterial RPR ± RPP, Sun et al. (60) concluded that the RPP enhances the RPR’s rate of pre-tRNA cleavage by increasing Kconf and not kc, an inference supported by a recent finding that the bacterial RPP slows the reverse isomerization step (i.e. ES*→ES) (61).
Since Mth RPR is capable of pre-tRNA processing in the absence of RPPs, a direct increase in kc upon addition of Mth RPPs seems less likely since it would require an alternative mechanism for the RNP compared to the RPR, an unsupported premise. Therefore, we hypothesize that increased conversion of ES→ES* must underlie the ability of cognate POP5•RPP30 to elicit a 60- and 100-fold increase, respectively, in the Mth RPR’s max. kobs (Figure 7) and the pre-tRNATyr—Mja RPR self-cleavage rate (19). Because POP5•RPP30 significantly increases the rate of the cleavage step, we cannot assess if KM(STO)∼KS (as we did for RPP21•RPP29; Supplementary Figure S4). Thus, without determining the microscopic rate constants that contribute to KS, we cannot rule out the role of POP5•RPP30 in substrate binding. Since bacterial RPP and POP5 both adopt an RRM fold (26,62), and the central cleft in the α–β sandwich structure of the bacterial RPP binds the pre-tRNA leader (63,64), it is likely that POP5 (or POP5•RPP30) performs a similar role.
The failure of Mth RPP21•RPP29 to enhance the RPR’s max. kobs indicates that it does not favorably alter ES⇆ES*. However, it enhances the affinity of the RNP for Mg2+ as evidenced by its ability to increase by 344-fold the kobs of Mth RPR + POP5•RPP30 at 30 mM Mg2+ but not at 120 mM Mg2+ (Table 1). Its key role in substrate binding is borne out by its ability to decrease KM(STO) (i.e. KS; Table 1 and Supplementary Figure S4). In fact, human RPP21 has been shown to bind pre-tRNA (65), and the solution structure of Pfu RPP21•RPP29 reveals two electropositive surfaces, with the smaller one (exclusively in RPP21) postulated to bind pre-tRNA (29).
Comparing archaeal type A and M RNase P
There are similarities in the overall functioning of the type A (Mth, Pfu) and M (Mja) RNase P variants that we characterized. First, the rate of cleavage observed under saturating single-turnover conditions with Mth RPR and pre-tRNATyr (∼0.13 min−1 at pH 5.8; Table 1) is nearly identical to that of a cis cleaving Mja RPR–pre-tRNATyr (19). Thus, when the substrate binding defects of the type M RPR are overcome by providing the substrate in cis, its activity coincides with the type A relative. In fact, the fully reconstituted type A and M holoenzymes also display similar rates (Table 1) (19). Such a comparison takes into consideration the fact that the rates reported for the self-cleaving Mja (type M) RPR were at pH 5.4 and not pH 5.1 as was mistakenly reported in (19). Second, the C domains of both Pfu and Mja RPRs are functional with their respective POP5•RPP30 and not RPP21•RPP29, consistent with the footprints of these binary complexes on the C and S domains, respectively (18,29). Although one would predict the functional contribution of RPP21•RPP29 to be negligible in the absence of the S domain, this was the case only in type A RNase P.
While RPP21•RPP29 had no effect on the rate of cleavage of Pfu RPR’s C domain + POP5•RPP30 (18), its addition to Mja RPR’s C domain (i.e. ΔS) + POP5•RPP30 increased activity and lowered the Mg2+ requirement for optimal activity from 120 to 30 mM Mg2+ (Figure 3). How could Mja RPP21•RPP29 potentiate cleavage of pre-tRNAs by Mja ΔS RPR + POP5•RPP30 even when its binding site (S domain) is absent? The reason is likely due to protein–protein interactions between the two type M binary RPPs that create a structural platform, which partly exploits the ability of RPP21•RPP29 to directly bind pre-tRNA and position the substrate for optimal cleavage by Mja RPR + POP5•RPP30. Such an effect is redundant when the pre-tRNA is conjugated in cis as revealed by the inability of RPP21•RPP29 to influence the cis cleaving pre-tRNATyr–Mja ΔS RPR (19). Because the type M RPR is smaller than the type A RPR (13), it offers fewer possibilities for nucleating independent, cooperative interactions with multiple RPPs, thus accentuating the need for protein-protein interactions in type M RNase P.
Our studies on type A and M RNase P indicate that the substrate- and metal ion-positioning effects of RPP21•RPP29 contribute to an increased rate when enabled by POP5•RPP30 already bound to the C domain. However, it is intriguing that type M RPP21•RPP29 alone is unable to alleviate the substrate binding (and consequent catalytic) defects in type M RPRs (Figure 3). While this inference might be substrate identity dependent, it does reflect the acute dependence of type M but not type A RPRs on POP5•RPP30 for activity. Unraveling such type A versus M nuances will have to wait for an examination of their tertiary structure differences.
Functional parallels in RNase P from the three domains of life
Despite the distinct evolutionary paths traversed by bacterial, archaeal and eukaryal RNase P, as reflected in their strikingly different RNP compositions, some common attributes are beginning to emerge. First, as might be expected of a ribozyme remnant from a hypothetical RNA world, catalysis rests with the RPR in all three domains of life (5–7,15). Second, non-homologous RPPs recruited independently to play analogous roles are functionally interchangeable (at least partly), an attribute consistent with their shared function to help their cognate RPRs overcome similar catalytic limitations. We elaborate below observations from this study and others to provide a thematic basis for understanding the diversity of RNase P.
High-resolution structures reveal that the bacterial RPR is arranged as two one-helix thick layers, with the larger layer 1 juxtaposing the S domain’s substrate recognition elements and the C domain’s active site (47,48). Intra-molecular braces in layer 2 are vital for the precise orientation of the S and C domains, whose cooperation is essential for efficient catalysis. Although the bacterial RPR is capable of generating a functional tertiary fold in vitro at high ionic strength, association with a single RPP enhances its activity at lower (physiological) Mg2+ concentrations and broadens its substrate specificity likely due to a gain of new substrate recognition determinants (60,66,67). One might expect that some of the archaeal/eukaryal RPP(s) would exhibit equivalent catalytic functions as the bacterial RPP. In addition, since the bacterial RPR’s layer 2 struts are missing in archaeal/eukaryal RPRs, some archaeal/eukaryal RPPs would be predicted to substitute for these missing RNA–RNA tertiary contacts and play structural roles (15). Moreover, some other archaeal/eukaryal RPPs might have been recruited to fulfill a need for coordination with other macromolecular machineries, finer regulation and substrate specificity (4).
Our finding that archaeal POP5•RPP30 could reconstitute with bacterial and Ram mt RPRs (Figures 5 and 6) establishes a clear functional correspondence with the bacterial RPP. An interesting reciprocal relationship is also evident in the ability of the bacterial RPP to functionally reconstitute with select archaeal RPRs (6). While the overlapping footprints of bacterial RPP and archaeal POP5•RPP30 on their respective RPR’s C domains (18,29) and the structural similarity (despite weak sequence homology) of bacterial RPP and archaeal POP5 (26) collectively support the idea of convergent evolution, more data are required to validate parallels in their mechanisms of action.
We postulate that one function of archaeal RPP21•RPP29 is to substitute for the intra-molecular struts found in bacterial RPR. Such a premise is consistent with its inability to alter the rate or Mg2+ requirement of the reaction catalyzed by bacterial RPRs, which already have these tertiary contacts. Results from an independent substrate recognition study (V. Gopalan and L. A. Kirsebom, unpublished data) examining the ratios of correct:aberrant cleavages of model substrates indicate that binding of RPP21•RPP29 to the archaeal RPR’s S domain is critical for optimal recognition of the T stem–loop (TSL) region in the pre-tRNA. Akin to the bacterial RPR scenario, this productive TSL-S domain interaction might elicit a conformational change that aids catalysis by positioning the chemical groups and Mg2+ near the cleavage site in the C domain (33–35,58).
While we observe weak activity from heterologous reconstitutions, it is unlikely that evolutionarily distant, non-cognate RPRs and RPPs will generate a robust functional holoenzyme in vivo, an expectation based on the strong co-evolution of RNA and protein subunits in an RNP. In fact, a genetic complementation approach revealed that both type A and M archaeal RPPs (expressed either individually or as binary complexes) were inadequate in rescuing the bacterial RPP defect in vivo (68). This result is consistent with our finding that an RNP made up of a bacterial RPR + archaeal POP5•RPP30 is ∼50-fold weaker than the bacterial RNase P holoenzyme (50).
Elucidating commonalities in the structure and function of archaeal and eukaryal RNase P holoenzymes is a logical follow-up to the work described here. Although eukaryal RPRs share a conserved catalytic core with archaeal/bacterial RPRs, we were unable to reconstitute, under a few conditions tested, either yeast or human nuclear RPR with RPPs derived from thermophilic archaea (not shown). We are testing if mesophilic archaeal RPPs fare better in this regard.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (R01 GM067807) to Mark P. Foster and V.G.; National Science Foundation (MCB-0843543) to V.G.; American Heart Association Pre-Doctoral Fellowships to D.K.P. (0515218B) and H-Y.T. (0315171B). Funding for open access charge: National Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are extremely grateful to Lien Lai for her invaluable critique and suggestions that significantly improved the manuscript, Mark Foster for his useful suggestions, and Hue Lai for his efforts during the initial stages of the cloning experiments.
REFERENCES
- 1.Liu F, Altman S. Ribonuclease P. Protein Reviews Series. Springer-Verlag: New York; 2010. [Google Scholar]
- 2.Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends Biochem. Sci. 2006;31:333–341. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Walker SC, Engelke DR. Ribonuclease P: the evolution of an ancient RNA enzyme. Crit. Rev. Biochem. Mol. Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai LB, Vioque A, Kirsebom LA, Gopalan V. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects. FEBS Lett. 2010;584:287–296. doi: 10.1016/j.febslet.2009.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 6.Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW. RNase P RNAs from some Archaea are catalytically active. Proc. Natl Acad. Sci. USA. 1999;96:7803–7808. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kikovska E, Svard SG, Kirsebom LA. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc. Natl Acad. Sci. USA. 2007;104:2062–2067. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas ES, Banta AB, Harris JK, Pace NR, Brown JW. Structure and evolution of ribonuclease P RNA in Gram-positive bacteria. Nucleic Acids Res. 1996;24:4775–4782. doi: 10.1093/nar/24.23.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haas ES, Armbruster DW, Vucson BM, Daniels CJ, Brown JW. Comparative analysis of ribonuclease P RNA structure in Archaea. Nucleic Acids Res. 1996;24:1252–1259. doi: 10.1093/nar/24.7.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown JW, Nolan JM, Haas ES, Rubio MA, Major F, Pace NR. Comparative analysis of ribonuclease P RNA using gene sequences from natural microbial populations reveals tertiary structural elements. Proc. Natl Acad. Sci. USA. 1996;93:3001–3006. doi: 10.1073/pnas.93.7.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siegel RW, Banta AB, Haas ES, Brown JW, Pace NR. Mycoplasma fermentans simplifies our view of the catalytic core of ribonuclease P RNA. RNA. 1996;2:452–462. [PMC free article] [PubMed] [Google Scholar]
- 12.Brown JW. The ribonuclease P database. Nucleic Acids Res. 1999;27:314. doi: 10.1093/nar/27.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris JK, Haas ES, Williams D, Frank DN, Brown JW. New insight into RNase P RNA structure from comparative analysis of the archaeal RNA. RNA. 2001;7:220–232. doi: 10.1017/s1355838201001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marquez SM, Harris JK, Kelley ST, Brown JW, Dawson SC, Roberts EC, Pace NR. Structural implications of novel diversity in eucaryal RNase P RNA. RNA. 2005;11:739–751. doi: 10.1261/rna.7211705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gopalan V. Uniformity amid diversity in RNase P. Proc. Natl Acad. Sci. USA. 2007;104:2031–2032. doi: 10.1073/pnas.0611193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boomershine WP, McElroy CA, Tsai HY, Wilson RC, Gopalan V, Foster MP. Structure of Mth11/Mth Rpp29, an essential protein subunit of archaeal and eukaryotic RNase P. Proc. Natl Acad. Sci. USA. 2003;100:15398–15403. doi: 10.1073/pnas.2535887100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kouzuma Y, Mizoguchi M, Takagi H, Fukuhara H, Tsukamoto M, Numata T, Kimura M. Reconstitution of archaeal ribonuclease P from RNA and four protein components. Biochem. Biophys. Res. Commun. 2003;306:666–673. doi: 10.1016/s0006-291x(03)01034-9. [DOI] [PubMed] [Google Scholar]
- 18.Tsai HY, Pulukkunat DK, Woznick WK, Gopalan V. Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proc. Natl Acad. Sci. USA. 2006;103:16147–16152. doi: 10.1073/pnas.0608000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pulukkunat DK, Gopalan V. Studies on Methanocaldococcus jannaschii RNase P reveal insights into the roles of RNA and protein cofactors in RNase P catalysis. Nucleic Acids Res. 2008;36:4172–4180. doi: 10.1093/nar/gkn360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sidote DJ, Hoffman DW. NMR structure of an archaeal homologue of ribonuclease P protein Rpp29. Biochemistry. 2003;42:13541–13550. doi: 10.1021/bi030170z. [DOI] [PubMed] [Google Scholar]
- 21.Numata T, Ishimatsu I, Kakuta Y, Tanaka I, Kimura M. Crystal structure of archaeal ribonuclease P protein Ph1771p from Pyrococcus horikoshii OT3: an archaeal homolog of eukaryotic ribonuclease P protein Rpp29. RNA. 2004;10:1423–1432. doi: 10.1261/rna.7560904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sidote DJ, Heideker J, Hoffman DW. Crystal structure of archaeal ribonuclease P protein aRpp29 from Archaeoglobus fulgidus. Biochemistry. 2004;43:14128–14138. doi: 10.1021/bi048578z. [DOI] [PubMed] [Google Scholar]
- 23.Takagi H, Watanabe M, Kakuta Y, Kamachi R, Numata T, Tanaka I, Kimura M. Crystal structure of the ribonuclease P protein Ph1877p from hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biochem. Biophys. Res. Commun. 2004;319:787–794. doi: 10.1016/j.bbrc.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 24.Kakuta Y, Ishimatsu I, Numata T, Kimura K, Yao M, Tanaka I, Kimura M. Crystal structure of a ribonuclease P protein Ph1601p from Pyrococcus horikoshii OT3: an archaeal homologue of human nuclear ribonuclease P protein Rpp21. Biochemistry. 2005;44:12086–12093. doi: 10.1021/bi050738z. [DOI] [PubMed] [Google Scholar]
- 25.Kawano S, Nakashima T, Kakuta Y, Tanaka I, Kimura M. Crystal structure of protein Ph1481p in complex with protein Ph1877p of archaeal RNase P from Pyrococcus horikoshii OT3: implication of dimer formation of the holoenzyme. J. Mol. Biol. 2006;357:583–591. doi: 10.1016/j.jmb.2005.12.086. [DOI] [PubMed] [Google Scholar]
- 26.Wilson RC, Bohlen CJ, Foster MP, Bell CE. Structure of Pfu Pop5, an archaeal RNase P protein. Proc. Natl Acad. Sci. USA. 2006;103:873–878. doi: 10.1073/pnas.0508004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amero CD, Boomershine WP, Xu Y, Foster M. Solution structure of Pyrococcus furiosus RPP21, a component of the archaeal RNase P holoenzyme, and interactions with its RPP29 protein partner. Biochemistry. 2008;47:11704–11710. doi: 10.1021/bi8015982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honda T, Kakuta Y, Kimura K, Saho J, Kimura M. Structure of an archaeal homolog of the human protein complex Rpp21-Rpp29 that is a key core component for the assembly of active ribonuclease P. J. Mol. Biol. 2008;384:652–662. doi: 10.1016/j.jmb.2008.09.056. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y, Amero CD, Pulukkunat DK, Gopalan V, Foster MP. Solution structure of an archaeal RNase P binary protein complex: formation of the 30-kDa complex between Pyrococcus furiosus RPP21 and RPP29 is accompanied by coupled protein folding and highlights critical features for protein-protein and protein-RNA interactions. J. Mol. Biol. 2009;393:1043–1055. doi: 10.1016/j.jmb.2009.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vioque A, Arnez J, Altman S. Protein-RNA interactions in the RNase P holoenzyme from Escherichia coli. J. Mol. Biol. 1988;202:835–848. doi: 10.1016/0022-2836(88)90562-1. [DOI] [PubMed] [Google Scholar]
- 31.Waugh DS, Pace NR. Gap-scan deletion analysis of Bacillus subtilis RNase P RNA. FASEB J. 1993;7:188–195. doi: 10.1096/fasebj.7.1.7678561. [DOI] [PubMed] [Google Scholar]
- 32.Seif E, Cadieux A, Lang BF. Hybrid E. coli–Mitochondrial ribonuclease P RNAs are catalytically active. RNA. 2006;12:1661–1670. doi: 10.1261/rna.52106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan T, Loria A, Zhong K. Probing of tertiary interactions in RNA: 2'-hydroxyl-base contacts between the RNase P RNA and pre-tRNA. Proc. Natl Acad. Sci. USA. 1995;92:12510–12514. doi: 10.1073/pnas.92.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loria A, Pan T. Recognition of the T stem-loop of a pre-tRNA substrate by the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry. 1997;36:6317–6325. doi: 10.1021/bi970115o. [DOI] [PubMed] [Google Scholar]
- 35.Brannvall M, Kikovska E, Wu S, Kirsebom LA. Evidence for Induced Fit in Bacterial RNase P RNA-mediated Cleavage. J. Mol. Biol. 2007;372:1149–1164. doi: 10.1016/j.jmb.2007.07.030. [DOI] [PubMed] [Google Scholar]
- 36.Kirsebom LA, Trobro S. RNase P RNA-mediated cleavage. IUBMB Life. 2009;61:189–200. doi: 10.1002/iub.160. [DOI] [PubMed] [Google Scholar]
- 37.Kirsebom LA, Svard SG. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 1994;13:4870–4876. doi: 10.1002/j.1460-2075.1994.tb06814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh BK, Pace NR. Interaction of the 3'-end of tRNA with ribonuclease P RNA. Nucleic Acids Res. 1994;22:4087–4094. doi: 10.1093/nar/22.20.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall TA, Brown JW. Interactions between RNase P protein subunits in archaea. Archaea. 2004;1:247–254. doi: 10.1155/2004/743956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kifusa M, Fukuhara H, Hayashi T, Kimura M. Protein-protein interactions in the subunits of ribonuclease P in the hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biosci. Biotechnol. Biochem. 2005;69:1209–1212. doi: 10.1271/bbb.69.1209. [DOI] [PubMed] [Google Scholar]
- 41.Tan S. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr. Purif. 2001;21:224–234. doi: 10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- 42.Finkelstein J, Antony E, Hingorani MM, O'Donnell M. Overproduction and analysis of eukaryotic multiprotein complexes in Escherichia coli using a dual-vector strategy. Anal. Biochem. 2003;319:78–87. doi: 10.1016/s0003-2697(03)00273-2. [DOI] [PubMed] [Google Scholar]
- 43.Perederina A, Esakova O, Koc H, Schmitt ME, Krasilnikov AS. Specific binding of a Pop6/Pop7 heterodimer to the P3 stem of the yeast RNase MRP and RNase P RNAs. RNA. 2007;13:1648–1655. doi: 10.1261/rna.654407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perederina A, Esakova O, Quan C, Khanova E, Krasilnikov AS. Eukaryotic ribonucleases P/MRP: the crystal structure of the P3 domain. EMBO J. 2010;29:761–769. doi: 10.1038/emboj.2009.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loria A, Pan T. Domain structure of the ribozyme from eubacterial ribonuclease P. RNA. 1996;2:551–563. [PMC free article] [PubMed] [Google Scholar]
- 46.Zahler NH, Christian EL, Harris ME. Recognition of the 5′ leader of pre-tRNA substrates by the active site of ribonuclease P. RNA. 2003;9:734–745. doi: 10.1261/rna.5220703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. Crystal structure of a bacterial ribonuclease P RNA. Proc. Natl Acad. Sci. USA. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 49.Loria A, Pan T. The cleavage step of ribonuclease P catalysis is determined by ribozyme-substrate interactions both distal and proximal to the cleavage site. Biochemistry. 1999;38:8612–8620. doi: 10.1021/bi990691f. [DOI] [PubMed] [Google Scholar]
- 50.Pulukkunat DK. 2008. Biochemical studies on archaeal ribonuclease P reveal thematic convergence in protein-facilitated RNA catalysis. Ph.D. thesis. The Ohio State University, Columbus, OH. [Google Scholar]
- 51.Kurz JC, Niranjanakumari S, Fierke CA. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor-tRNAAsp. Biochemistry. 1998;37:2393–2400. doi: 10.1021/bi972530m. [DOI] [PubMed] [Google Scholar]
- 52.Hsieh J, Walker SC, Fierke CA, Engelke DR. Pre-tRNA turnover catalyzed by the yeast nuclear RNase P holoenzyme is limited by product release. RNA. 2009;15:224–234. doi: 10.1261/rna.1309409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith D, Pace NR. Multiple magnesium ions in the ribonuclease P reaction mechanism. Biochemistry. 1993;32:5273–5281. doi: 10.1021/bi00071a001. [DOI] [PubMed] [Google Scholar]
- 54.Warnecke JM, Furste JP, Hardt WD, Erdmann VA, Hartmann RK. Ribonuclease P (RNase P) RNA is converted to a Cd(2+)-ribozyme by a single Rp-phosphorothioate modification in the precursor tRNA at the RNase P cleavage site. Proc. Natl Acad. Sci. USA. 1996;93:8924–8928. doi: 10.1073/pnas.93.17.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warnecke JM, Held R, Busch S, Hartmann RK. Role of metal ions in the hydrolysis reaction catalyzed by RNase P RNA from Bacillus subtilis. J. Mol. Biol. 1999;290:433–445. doi: 10.1006/jmbi.1999.2890. [DOI] [PubMed] [Google Scholar]
- 56.Cassano AG, Anderson VE, Harris ME. Analysis of solvent nucleophile isotope effects: evidence for concerted mechanisms and nucleophilic activation by metal coordination in nonenzymatic and ribozyme-catalyzed phosphodiester hydrolysis. Biochemistry. 2004;43:10547–10559. doi: 10.1021/bi049188f. [DOI] [PubMed] [Google Scholar]
- 57.Li D, Willkomm DK, Hartmann RK. Minor changes largely restore catalytic activity of archaeal RNase P RNA from Methanothermobacter thermoautotrophicus. Nucleic Acids Res. 2009;37:231–242. doi: 10.1093/nar/gkn915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loria A, Pan T. Recognition of the 5' leader and the acceptor stem of a pre-tRNA substrate by the ribozyme from Bacillus subtilis RNase P. Biochemistry. 1998;37:10126–10133. doi: 10.1021/bi980220d. [DOI] [PubMed] [Google Scholar]
- 59.Pomeranz Krummel DA, Altman S. Multiple binding modes of substrate to the catalytic RNA subunit of RNase P from Escherichia coli. RNA. 1999;5:1021–1033. doi: 10.1017/s1355838299990416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun L, Campbell FE, Zahler NH, Harris ME. Evidence that substrate-specific effects of C5 protein lead to uniformity in binding and catalysis by RNase P. EMBO J. 2006;25:3998–4007. doi: 10.1038/sj.emboj.7601290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsieh J, Fierke CA. Conformational change in the Bacillus subtilis RNase P holoenzyme–pre-tRNA complex enhances substrate affinity and limits cleavage rate. RNA. 2009;15:1565–1577. doi: 10.1261/rna.1639409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stams T, Niranjanakumari S, Fierke CA, Christianson DW. Ribonuclease P protein structure: evolutionary origins in the translational apparatus. Science. 1998;280:752–755. doi: 10.1126/science.280.5364.752. [DOI] [PubMed] [Google Scholar]
- 63.Niranjanakumari S, Stams T, Crary SM, Christianson DW, Fierke CA. Protein component of the ribozyme ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proc. Natl Acad. Sci. USA. 1998;95:15212–15217. doi: 10.1073/pnas.95.26.15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsai HY, Masquida B, Biswas R, Westhof E, Gopalan V. Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenzyme. J. Mol. Biol. 2003;325:661–675. doi: 10.1016/s0022-2836(02)01267-6. [DOI] [PubMed] [Google Scholar]
- 65.Jarrous N, Reiner R, Wesolowski D, Mann H, Guerrier-Takada C, Altman S. Function and subnuclear distribution of Rpp21, a protein subunit of the human ribonucleoprotein ribonuclease P. RNA. 2001;7:1153–1164. doi: 10.1017/s1355838201010469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koutmou KS, Zahler NH, Kurz JC, Campbell FE, Harris ME, Fierke CA. Protein-precursor tRNA contact leads to sequence-specific recognition of 5' leaders by bacterial ribonuclease P. J. Mol. Biol. 2010;396:195–208. doi: 10.1016/j.jmb.2009.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun L, Campbell FE, Yandek LE, Harris ME. Binding of C5 protein to P RNA enhances the rate constant for catalysis for P RNA processing of pre-tRNAs lacking a consensus G(+1)/C(+72) pair. J. Mol. Biol. 2010;395:1019–1037. doi: 10.1016/j.jmb.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gosringer M, Hartmann RK. Function of heterologous and truncated RNase P proteins in Bacillus subtilis. Mol. Microbiol. 2007;66:801–813. doi: 10.1111/j.1365-2958.2007.05962.x. [DOI] [PubMed] [Google Scholar]
- 69.Massire C, Jaeger L, Westhof E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 1998;279:773–793. doi: 10.1006/jmbi.1998.1797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.