Abstract
It has been proposed that robust rhythmic gene expression requires clock-controlled elements (CCEs). Transcription of Per1 was reported to be regulated by the E-box and D-box in conventional reporter assays. However, such experiments are inconclusive in terms of how the CCEs and their combinations determine the phase of the Per1 gene. Whereas the phase of Per2 oscillation was found to be the most delayed among the three Period genes, the phase-delaying regions of the Per2 promoter remain to be determined. We therefore investigated the regulatory mechanism of circadian Per1 and Per2 transcription using an in vitro rhythm oscillation-monitoring system. We found that the copy number of the E-box might play an important role in determining the phase of Per1 oscillation. Based on real-time bioluminescence assays with various promoter constructs, we provide evidence that the non-canonical E-box is involved in the phase delay of Per2 oscillation. Transfection experiments confirmed that the non-canonical E-box could be activated by CLOCK/BMAL1. We also show that the D-box in the third conserved segment of the Per2 promoter generated high amplitude. Our experiments demonstrate that the copy number and various combinations of functional CCEs ultimately led to different circadian phases and amplitudes.
INTRODUCTION
The system regulating mammalian circadian timing is organized in a hierarchical manner, in which a master pacemaker in the suprachiasmatic nucleus (SCN) controls peripheral oscillators (1). The clock mechanism in the SCN and the peripheral oscillators are known to be similar at the molecular level (2–5), and comprise a network of transcriptional/translational feedback loops involving a set of clock genes (1,6).
In the primary feedback loop, the positive components include members of the basic helix–loop–helix-PAS transcription factor family, CLOCK and BMAL1. These heterodimerize and initiate the transcription of target genes containing E-box cis-regulatory elements, including Period (Per1 and Per2) and Cryptochrome (Cry1 and Cry2) (7–11). The translated PER and CRY proteins translocate into the nucleus to interfere with the CLOCK and BMAL1 heterodimer complex, thus blocking their transcription (6,12–14). This heterodimer also induces the transcription of Rev-erbA and Ror, which interact with Rev-erbA/ROR-binding elements (RREs) in the promoter of Bmal1, repressing or driving its transcription, respectively (15–19). DBP and E4BP4 are able to activate or suppress transcriptional activity, respectively, through the same sequence called as D-box (20).
It has been proposed that circadian timing of clock and clock-controlled genes may be dependent on conserved and functional clock-controlled elements (CCEs) during the morning (E/E′-boxes), daytime (D-boxes) and night (RREs) (21,22). For example, RREs play an important role in generating circadian night expression in phase with Bmal1 expression (19). On the other hand, circadian regulation of Per2 appears to be more complex, because multiple CCEs are involved in its regulation. Recent studies have shown that the E′-box located 20 bp upstream from the Per2 transcription start site (TSS) is essential to generate Per2 oscillation in vitro (23) and in vivo (24). In addition, the D-box (+197) located downstream from the Per2 TSS is also required for robust circadian expression of Per2 (25). In contrast to our understanding of rhythm generation, it is not sufficient to verify the phase-control elements. In particular, the phase-delaying regions of the Per2 promoter remain to be elucidated.
Transcription of mPer1 is regulated by the canonical E-box and D-box (9,26,27). However, it is important to note that circadian rhythm generation cannot be verified using a conventional transient reporter assay. A conventional reporter assay, although useful for the characterization of promoter activity and protein–DNA interactions, has not revealed the phase and amplitude of circadian transcription. In contrast, a previous study verified that the D-box of the Per3 promoter was sufficient to generate the phase of Per3 circadian oscillation using an in vitro rhythm oscillation-monitoring system (21). Therefore, we investigated the regulatory mechanism of circadian Per1 and Per2 transcription in detail.
The current report shows that the number of functional CCEs might play a significant role in determining the phase of the clock gene. Furthermore, we provide the evidence that the non-canonical E-box is responsible for delaying the phase of the clock gene.
MATERIALS AND METHODS
Plasmid construction
The mouse Per2 gene promoter region [chr1 (−): 93289505 bp-chr1: 93293019 bp in the Build 36 Assembly of the UCSC Genome Browser; http://genome.ucsc.edu/], which is essential for oscillation, was linked with destabilized firefly luciferase (dLuc; luciferase bound to the ornithine decarboxylase PEST sequence) based on the pGL3-basic vector (Promega, Madison, WI, USA), as described elsewhere (21). Site-directed mutagenesis of the reporter construct was performed with a QuikChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). The wild-type sequences were as follows: E′-box (CACGTT), E″-box (CATGTG) and D-box (TTATGTAA). The mutation sequences were as follows: mutated E′-box (ACCGGT), mutated E″-box (ACCGGT) and mutated D-box (CACCCGGC). Construction of the mPer1-dLuc reporter vector was as described elsewhere (21).
Animals
Mice were housed under 12:12 h light/dark or constant dark conditions. Food and water were given ad libitum. SCN, livers and white adipose tissues (WATs) were obtained every 4 h over 2 days. Upon removal, tissues were immediately frozen in liquid nitrogen and stored at −80°C until they were processed for RNA analysis. We extracted total RNA from the 50 pooled SCNs, 4 pooled livers and 4 pooled WATs at each time point. All protocols for experiments using animals in this study were approved by the Yamanouchi and Astellas Pharma Inc., Animal Research Committee.
Real-time monitoring of bioluminescence and data analysis
Rat-1 cells were cultured as described in previous studies (21), transfected with several mutant promoter vectors of mPer1-dluc and mPer2-dluc and incubated for 72 h. Then, dibutyryl cyclic AMP (dbcAMP; Wako Pure Chemical Industries, Ltd.) was added to a final concentration of 1 mM. In the presence of 0.1 mM luciferin (Promega), light emission was measured and integrated for 1 min at intervals of 15 min with the PMT detector assembly (Hamamatsu Photonics), and the luminescence was observed continuously for ∼5 days at 30°C. The phase and amplitude were calculated as described in previous studies (28–31), using the Origin 6.1 program (Origin Lab, Corporation Northampton, MA, USA). Briefly, bioluminescence records were detrended by subtraction of the 24 h running average from the raw data, then smoothed with the 2 h running average. The phases were measured starting with the third peak after stimulation. Relative amplitude data were calculated from the third to the eighth peak of oscillation.
Luciferase assay
NIH3T3 cells were cultured as described in previous studies (21), and plated in 24-well plates at 4 × 104 cells/well the day before transfection. The NIH3T3 cells were transfected using Lipofectamine 2000 reagent (Invitrogen) with an internal control (pRL-TK; 2 ng) and the SV40-dLuc vector containing the wild-type or mutant CCE (10 ng), in the presence or absence of pCI-Bmal1 (100 ng), pCI-Clock (100 ng), pCI-Cry1 (100 ng) and pDNA3.1-Dec1 (100 ng) (32). A pCI-neo or pCMV-Sport6 plasmid was used to adjust the amount of DNA (412 ng). After 24 h transfection, the cells were harvested and assayed with the Dual-Luciferase Reporter Assay System (Promega).
Quantitative polymerase chain reaction
Quantitative polymerase chain reaction (Q-PCR) was performed as described previously (21). The glyceraldehydes-3-phosphate dehydrogenase (GAPDH) expression levels were quantified and used as an internal control. The oligonucleotide DNA primers for Q-PCR were as follows: Per1 (F: 5′-CGTCCTACCTCCTTTATCCAGA-3′, R: 5′-TGTTTGCATCAGTGTCATCAGC-3′); Per2 (F: 5′-CATTGAACTTGAGACTGAGGT-3′, R: 5′-AAGGGAACACACTGAGAGGAT-3′); Per3 (F: 5′-GAAGCGAGAGGCAGAAGCACAA-3′, R: 5′-GAAAAGAGGGGAGGAGATAAGG-3′); and Gapdh (F: 5′-CAAAATGGTGAAGGTCGGTGTG-3′, R: 5′-ATTTGATGTTAGTGGGGTCTCG-3′).
Statistical analysis
Multiple comparisons among group mean differences were checked using Dunnett’s test. A P < 0.05 (one asterisk) was considered to be statistically significant.
Peak time analysis
Estimation of molecular peak time was conducted as described by Ueda et al. (33). We prepared cosine curves of 24 h periodicity with peaks from 0 to 24 h in increments of 10 min, yielding a total of 144 test cosine curves, and calculated the correlation value of the best-fitted cosine curve for each probe set. We estimated the peak time of each cycling gene from the peak time of the best-fitted cosine curve and defined it as the molecular peak time.
RESULTS AND DISCUSSION
Temporal expression profiles of mouse Per1, Per2 and Per3 in central and peripheral tissues
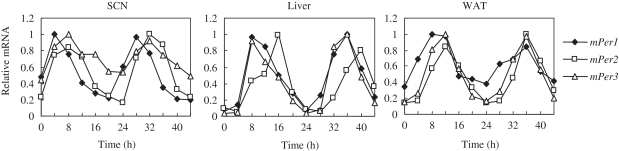
We confirmed the circadian expression profiles of mPer1, mPer2 and mPer3 in the SCN, liver and WAT under constant dark conditions. The expression levels of mPer1, mPer2 and mPer3 were normalized by Gapdh, as a house-keeping gene (Figure 1). The phases of mPer1 were 5.2 h (SCN), 11.3 h (liver) and 9.7 h (WAT). The phases of mPer2 were 8.7 h (SCN), 14.7 h (liver) and 12.8 h (WAT). The phases of mPer3 were 8.7 h (SCN), 11.3 h (liver) and 11.3 h (WAT).
Figure 1.
The expression rhythm of mouse Per1, Per2 and Per3 mRNAs. Total RNA was extracted from SCN, liver and WAT, and Q-PCR was performed to examine mPer1, mPer2 and mPer3 mRNA expression. The relative level of each mRNA was normalized to the corresponding Gapdh mRNA level. The maximal amount of mRNA was set to 1. Filled square, open square and open triangle lines represent mPer1, mPer2 and mPer3, respectively.
Clock and clock-controlled genes have numerous conserved and functional CCEs for morning (E/E′-boxes), daytime (D-boxes) and night (RREs) (21). These CCEs play key roles in the circadian timing of clock genes (21,22). Our previous study revealed that mPer1 contains five E-boxes and one D-box in the promoter region, whereas mPer2 contains one E′-box and one D-box (21). In contrast, mPer3 has two D-boxes (21). These elements were conserved between human and mouse genomic sequences in regions 10 kb upstream and 2 kb downstream from the TSS. If the phase of clock genes is determined by three major CCEs (that is, the E-box, D-box and RRE) and their combinations, we predicted that mPer1 then mPer2 and mPer3 would have successively later phases. However, the phase of mPer2 oscillation was found to be the latest among the three Period genes in central and peripheral tissues. The phases of the mPer1 and mPer3 messenger RNA (mRNA) rhythms were largely in accordance with this hypothesis, but that of mPer2 was later than expected. Here, wave peak point is used interchangeably with the term ‘phase’. These observations led us to hypothesize a different mechanism in which other unknown elements were involved in generating the variety of phases of clock genes. Therefore, it was necessary to investigate each promoter in greater detail.
Phase control of mPer1 oscillation was governed by canonical E-boxes
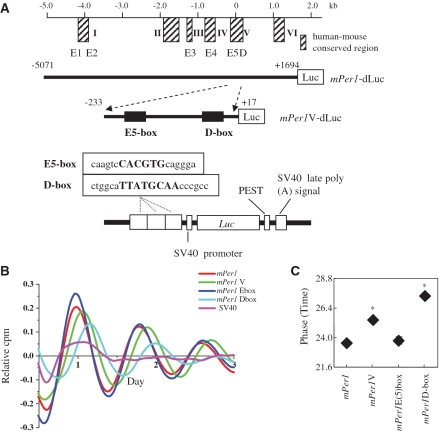
It has been reported that the transcription of mPer1 was regulated by the canonical E-box and D-box using a conventional transient reporter assay (9,26,27). However, that type of experiment is not conclusive in terms of how the CCEs and their combinations determine the phase of the Per1 gene. We constructed several mutant promoter vectors of the Per1 genes. Their roles were verified using an in vitro rhythm oscillation-monitoring system, in which cultured fibroblasts that were transiently transfected with reporter vectors exhibited robust circadian bioluminescence. Using a real-time bioluminescence system, we were able to investigate the relationship between phase control and CCE. The region of the inserted fragment in mPer1-dLuc (26) was shown to be sufficient to drive rhythmic expression in vivo (34). We constructed mPer1 V-dLuc (from −233 to +17) containing one E-box and one D-box. We also constructed SV40-driven dLuc reporters containing three tandem repeats of either the E(5)box or the D-box, designated mPer1 E(5)box SV40-dLuc or mPer1 D-box SV40-dLuc, respectively (Figure 2A). In our preceding work (21), we observed that reporter vectors with one CCE did not exhibit high-amplitude oscillations, while those with two and three CCEs exhibited circadian oscillations in comparable phases. Therefore, bioluminescence from three CCEs was used for the intrinsic phase.
Figure 2.
Real-time analysis of circadian expression of luciferase driven by the mouse Per1 promoter. (A) Schematic representation of reporter vectors for the mPer1 promoter. +1 corresponds to the TSS. (B) Transcriptional oscillation of mPer1 was monitored by a real-time monitoring system. Rat-1 cells were transfected with the mPer1 reporter constructs and stimulated with 1 mM dbcAMP. Bioluminescence records were detrended by subtraction of the 24 h running average from the raw data. (C) Phase data for mPer1, mPer1 V, mPer1 E(5)box and mPer1Dbox. The phases were measured from the third peak after stimulation; *P < 0.05.
Rat-1 cells were transfected with the constructs and stimulated with dbcAMP. We then measured the luminescence from these cells. To better calculate phase differences, data sets were detrended by subtracting the 24 h running average from the raw data (Figure 2B). The phases (measured from the third peak after stimulation) of mPer1-dLuc, mPer1V-dLuc, mPer1 E(5)box and mPer1 D-box were 23.5 ± 0.1 h, 25.4 ± 0.2 h, 23.8 ± 0.3 h and 27.3 ± 0.2 h (means ± SE, n = 3), respectively. The phase of mPer1-dLuc was similar to that of mPer1 E(5)box SV40-dLuc (Figure 2C). It was noteworthy that the phase of mPer1 V-dLuc, containing one E-box and one D-box, was in the middle of the expected phase of the E-box and D-box (Figure 2C). One possible explanation for these observations is that combinations of different types of CCEs might generate a new phase of circadian gene expression.
The role of each E-box in the mPer1 promoter was evaluated by a conventional reporter assay (26). This study revealed that each of the E-boxes functions as an enhancer for the transactivation of mPer1 by CLOCK/BMAL1. We confirmed that the phase of SV40-driven dLuc reporters containing first E-box × 3 and second E-box × 3 was similar to that of mPer1 E(5)box SV40-dLuc (fifth E-box × 3) (Supplementary Figure). Here, we compared four conditions (that is, mPer1-dLuc, mPer1 V-dLuc, mPer1 E(5)box and mPer1 D-box). As a whole, the phase of the Per1 gene rhythm was reflected by the E-box, despite the fact that it was adjusted by both the E-box and the D-box. Our finding led us to speculate that five E-boxes rather than one D-box might play a dominant role in controlling the phase of Per1 gene oscillation. These observations suggested that the number of functional CCEs might play a significant role in determining the phase of clock genes.
The third conserved region in mPer2 is responsible for mPer2 oscillation
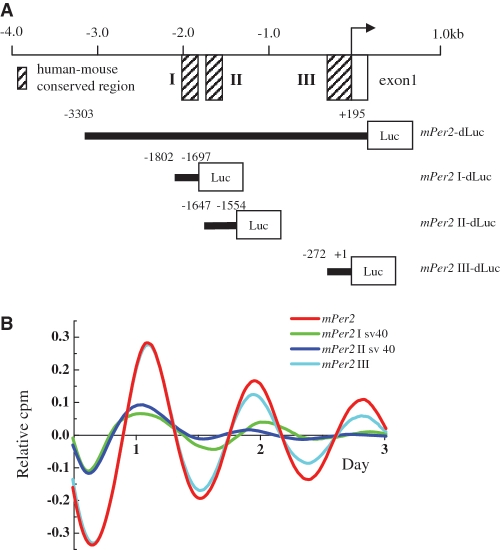
To analyze the mechanism controlling Per2 rhythm, we created a construct in which the mPer2 promoter reporter (mPer2-dLuc) was inserted into the −3303 to +195 region from the TSS fused to a dLuc reporter gene (Figure 3A). This mPer2-dLuc promoter reporter construct is sufficient to make circadian oscillation in vitro (19) and in vivo (35,36).
Figure 3.
Segment III, conserved between human and mouse Per2, is responsible for Per2 expression. (A) Schematic representation of deletion mutants of the mPer2 promoter. mPer2-dLuc and mPer2 III-dLuc were subcloned into the dLuc vector, and mPer2 I-dLuc and mPer2 II-dLuc were subcloned into the SV40-driven dLuc reporter. I, II and III are indicated in the conserved regions shared by human and mouse in the Per2 promoter. +1 corresponds to the TSS. (B) Transcriptional oscillation of mPer2 was monitored. Rat-1 cells were transfected with the mPer2 construct and stimulated with 1 mM dbcAMP. Bioluminescence records were detrended as in Figure 2.
The Per2 promoter reporter (–3303 to +195) consists of segments I (−1802 to −1697), II (−1647 to −1554) and III (−254 to +41) that are conserved between humans and mice. For the functional analysis of the mPer2 promoter, we generated deletion-mutant constructs (Figure 3A). Although segments I and II are highly conserved in the mPer2 promoter, these regions did not show basal promoter activities (data not shown). Therefore, mPer2 I-dLuc and mPer2 II-dLuc were subcloned into the SV40-driven dLuc reporter. Cells were transfected with these constructs, stimulated with dbcAMP and the bioluminescence was measured. Clearly, mPer2 III-dLuc showed oscillation similar to that of mPer2-dLuc, whereas mPer2 I-SV40-dLuc and mPer2 II-SV40-dLuc did not show such an oscillation (Figure 3B). The relative amplitudes of mPer2 I-SV40-dLuc, mPer2 II-SV40-dLuc and mPer2 III-dLuc compared to mPer2-dLuc were 9.8 ± 3.1%, 8.3 ± 2.1% and 75.3 ± 4.5% (means ± SE, n = 3), respectively. These results suggested that the third conserved region in mPer2 was responsible for mPer2 oscillation.
Characterization of the third conserved segment of mouse and human Per2 genes
The third conserved segment of 218 bp was positioned immediately upstream from the first exon and had 79% identity (Figure 4). This segment included an E-box like element, E″-box (CATGTG), E′-box (CACGTT), D-box (TTATGTAA) and two putative SP1 binding sites, but no canonical TATA-box (Figure 4). Recent studies have shown that the E′-box, which is activated by CLOCK/BMAL1, is responsible for generating Per2 oscillation in vitro (23) and in vivo (24). The E″-box (CATGTG) is classified as a CANNTG-type non-canonical E-box (a novel E-box-like element). However, whether this element is involved in the gene expression of Per2 remains unclear.
Figure 4.
Nucleotide sequence alignments of conserved segment III shared by human and mouse Per2. H and M represent human and mouse sequences, respectively. The colored letters represent the putative consensus sequence for transcription factors. A nucleotide sequence alignment of the 5′-proximal promoter of mouse Per2 is presented with putative cis-regulatory elements of the E″-box (from −162 to −157), D-box (−152 to −145), Sp1 (−100 to −92 and −47 to −40) and E′ box (−24 to −19). Promoter analysis was performed by TFSEARCH. Numbers indicate nucleotide distances from the predicted TSS.
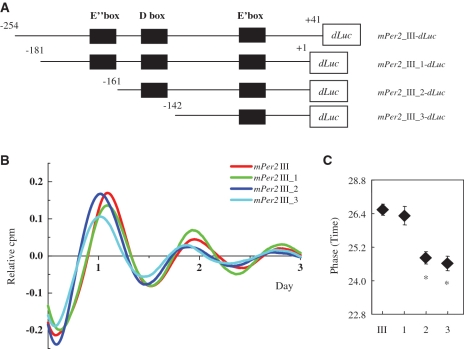
The 20-bp region containing the E″-box is responsible for the phase delay of mPer2 oscillation
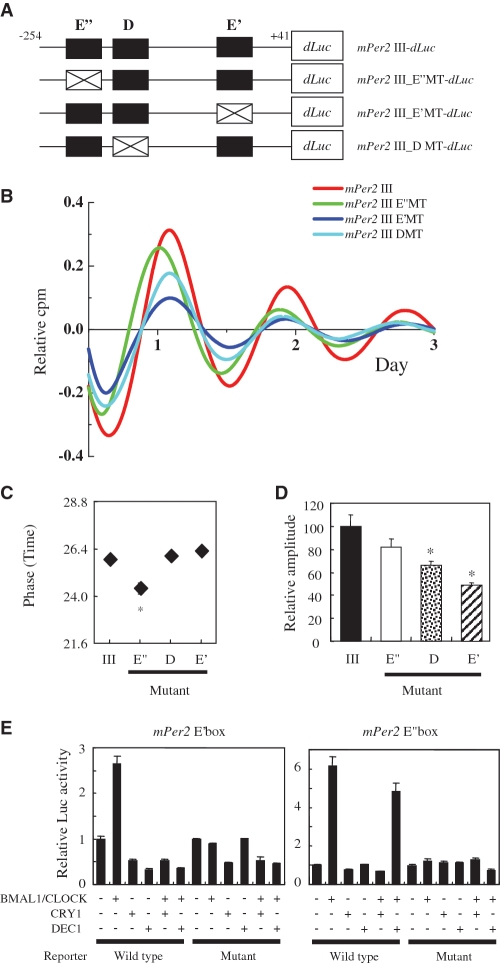
To analyze the function of these cis-elements in the third conserved segment, we generated various deletion-mutant reporters in the third conserved segment of mPer2 (Figure 5A). Rat-1 cells were transfected with these constructs, then stimulated with dbcAMP and the bioluminescence was measured. mPer2 III (−254 to +41)-dLuc and mPer2 III_1 (−181 to +1)-dLuc were observed to be in the same phase, whereas mPer2 III_2 (−161 to +1)-dLuc, and mPer2 III_3 (−142 to +1)-dLuc showed a phase advance against mPer2_III-dLuc (Figure 5B). The phases of mPer2 III-dLuc, mPer2 III_1-dLuc, mPer2 III_2-dLuc and mPer2 III_3-dLuc were 26.5 ± 0.2 h, 26.3 ± 0.3 h, 24.8 ± 0.2 h, and 24.6 ± 0.3 h (means ± SE, n = 4), respectively. mPer2 III_1 (−181 to +1)-dLuc constructs showed a 1.5 h phase delay compared with mPer2 III_2 (−161 to +1)-dLuc (Figure 5C). From these results, we conclude that the 20-bp region including the E″-box is responsible for the phase-delaying region.
Figure 5.
Deletion analysis of the third conserved region in the mPer2 gene. (A) Schematic diagrams of various mPer2 promoter constructs are shown and the names of the plasmids are listed. (B) De-trended bioluminescence data of mPer2 III and its deletion mutants fused to a dLuc reporter gene. Rat-1 cells were transfected with these constructs and stimulated with 1 mM dbcAMP. Bioluminescence records were detrended as in Figure 2. (C) Phase data for mPer2 III-dLuc, mPer2 III_1-dLuc, mPer2 III_2-dLuc and mPer2 III_3-dLuc. The phases were measured from the third peak after stimulation; *P < 0.05.
The E″-box contributes to delaying the phase of mPer2 oscillation
To analyze further the phase-delaying region, we constructed mutant reporter vectors of mPer2 III-dLuc by site-directed mutagenesis (Figure 6A). Rat-1 cells were transfected with these constructs, stimulated with dbcAMP, and the bioluminescence was measured. Similar phases were observed with the E′-box mutant (mPer2 III E′MT-dLuc) and the D-box mutant (mPer2 III DMT-dLuc), whereas a phase advance (1.5 h earlier) was detected only when using the E″-box mutant (mPer2 III E″MT-dLuc) construct (Figure 6B). The phases of mPer2 III-dLuc, mPer2 III E″MT-dLuc, mPer2 III DMT-dLuc and mPer2 III E′MT-dLuc were 25.9 ± 0.1 h, 24.4 ± 0.3 h, 26.0 ± 0.1 h and 26.3 ± 0.1 h (means ± SE, n = 3), respectively (Figure 6C). We therefore concluded that the E″-box plays a pivotal role in the phase delay of mPer2 oscillation.
Figure 6.
The E″-box (a novel E-box-like element) and the D-box in the third conserved segment of the Per2 promoter are functional cis-elements. (A) Schematic representation of point mutagenesis of putative cis-elements in the conserved segment III (mPer2 III-dLuc). The wild-type is indicated by a closed box and the mutant type is indicated by a white box (christcross). The mutations were as follows: mutated E′-box (ACCGGT), and mutated E″-box (ACCGGT), and mutated D-box (CACCCGGC). (B) Detrended bioluminescence data of mPer2 III-dLuc and its point mutants. Rat-1 cells were transfected with the mPer2 III-dLuc and its point mutants, and stimulated with 1 mM dbcAMP. Bioluminescence records were detrended as in Figure 2. (C) Phase data for mPer2 III, mPer2 III E″MT, mPer2 III DMT and mPer2 III E′MT. The phases were measured from the third peak after stimulation; *P < 0.05. (D) Relative amplitude data were calculated from the third to the eighth peak of oscillation. Relative amplitudes of mPer2 III_E″MT, mPer2 III_DMT and mPer2 III_E′MT to mPer2 III are shown; *P < 0.05. (E) Transcriptional activation by CLOCK and BMAL1, and transcriptional repression by CRY1 or DEC1 of the reporters containing three tandem repeats of either the wild-type or the mutants. The presence (+) or absence (−) of the expression plasmid is denoted. Each value is the mean ± SE of three replicates for a single assay.
CLOCK, BMAL1, CRY1 and DEC1 are transcription factors that regulate the E-box (21). CRY1 interacts with CLOCK/BMAL1, thus blocking their transcription. DEC1 serves as a transcriptional repressor for CLOCK/BMAL1-enhanced promoter activity, through binding competition for the E-box (22).
We compared the effects of CRY1 and DEC1 on transcriptional activities from E′-box and E″-box-carrying promoters. CRY1 abolished the CLOCK/BMAL1-enhanced transcriptional activities of both the E′-box and the E″-box (Figure 6E). In contrast, DEC1 abolished the CLOCK/BMAL1-enhanced transcriptional activities of the E′-box, whereas the E″-box-containing construct was weakly affected by DEC1 (Figure 6E). It is known that DEC1 directly binds to canonical E-box of clock genes and modulates the circadian phase, whereas it weakly affects clock genes containing the non-canonical E-box because of weak binding to it (37). While the mechanism by which the E″-box delays the phase of the Per2 oscillation has not been fully clarified, impaired transcriptional regulation of DEC1 at the E″-box might play a pivotal role in this system. Recently, the pulses of prolactin promoter activity were shown to depend on the noncanonical E-box (CATTTG) that binds to the CLOCK/BMAL1 complex (38). Our data, together with previous reports, suggest that non-canonical E-boxes might not only play a role in circadian enhancers, but might also generate various phases.
The D-box in the third conserved segment of mPer2 promoter is a functional cis-element that increases amplitude
The relative amplitudes of mPer2 III E″MT-dLuc, mPer2 III DMT-dLuc and mPer2 III E′MT-dLuc compared to mPer2-dLuc were 82.2 ± 6.8 %, 66.0 ± 3.4 % and 49.1 ± 1.6 % (means ± SE, n = 3), respectively (Figure 6D). We confirmed that the E′-box was essential to generate mPer2 oscillation, which is consistent with the observation by Akashi et al. (23) and Yoo et al. (24). We also showed, for the first time, that the D-box in the third conserved segment generated high amplitude.
Ohno et al. (25) reported that the D-box in the third conserved region is not a prerequisite for the circadian expression of Per2. Nevertheless, we have shown that the D-box in the third conserved segment of the Per2 promoter is a functional cis-element that increases amplitude. The circadian oscillations of Per2 expression exhibited stable during Day 3 in the current study (Figure 6B). On the other hand, the circadian oscillations of Per2 expression persisted but attenuated during this period in the published work (25). Therefore, sustained and longer circadian oscillation might help to generate the high amplitude in the D-box of the third conserved region of the Per2 promoter.
The region between the E′-box and the TSS (–12 to +1) tightly regulates mPer2 oscillation
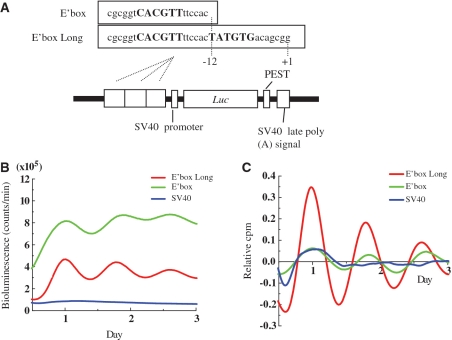
To better understand the mechanism driving oscillation of mPer2, we constructed deletion mutant reporters including E′-box, which is essential to generate mPer2 oscillation (Figure 7A). We transfected the deletion mutant vectors into Rat-1 cells, then stimulated them with dbcAMP, and measured the bioluminescence. The E′-box SV40-dLuc displayed a higher bioluminescence than did the E′-box Long SV40-dLuc (Figure 7B). However, when these data were de-trended by subtracting the 24 h running average from the raw data, the E′-box Long SV40-dLuc showed higher amplitude than the E′-box SV40-dLuc (Figure 7C). These results indicate that the −12 to +1 region enhances the amplitude of mPer2 oscillation.
Figure 7.
Analysis of the mPer2 tightly-interlocked region. (A) Schematic diagrams of various mPer2 promoter constructs are shown and the names of the plasmids are listed. The E′-box Long regions contains a direct repeat of the E-box-like elements, which is described previously (39). (B) Transcriptional oscillations of E′-box and E′-box long-SV40-dLuc were monitored. Rat-1 cells were transfected with these constructs and stimulated with 1 mM dbcAMP. (C) The signals obtained in (B) were detrended.
Recent studies have shown that the direct repeat of the E-box-like elements is the minimal required element for the generation of cell-autonomous transcriptional oscillation (39). In the present study, we also showed that the region (−12 to +1) adjacent to the E′-box containing the direct repeat of the E-box-like elements (39) serves to generate more robust oscillation in the Per2 gene.
Kumaki et al. (40) revealed that the importance of the affinity balance between transactivators and transrepressors in generating high-amplitude circadian transcriptional output. In the case of E-box, lowest affinity for CLOCK/BMAL1 was those giving the highest amplitude of oscillation. Additionally, we showed that the bioluminescence of the E′-box region itself was greater than that displayed by the E′-box long regions containing a direct repeat of the E-box-like elements. However, based on the de-trended data, we demonstrated that the E′-box long regions displayed a higher amplitude than the E′-box regions. These observations suggested that the E′-box long regions displayed a lower bioluminescence were attributable to the lowest affinity for CLOCK/BMAL1, as well as generating high amplitude.
CONCLUSIONS
A key issue concerning the logic of the mammalian circadian clock is how the expression peaks of circadian oscillating genes are determined. It has recently been proposed that the robust rhythmic expression requires CCE (E-box, D-box and RRE). While three major CCE might be capable of controlling circadian timing, not all phase have so far proved amenable. For example, although the recent application of a ‘synthetic molecular biology’ approach successfully generated many fundamental circadian phases, it has not yet been able to generate the basic morning phase (22). Therefore, it is necessary to investigate each promoter in further detail using an in vitro rhythm oscillation-monitoring system.
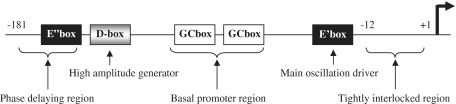
Here, we focused on the differential circadian timing of Per1 and Per2 transcription. These genes are E-box and D-box regulated genes, but in central and peripheral tissues peak Per2 mRNA expression is delayed by several hours relative to that of Per1. In the case of the Per1 gene, the copy number of the E-box might play an important role in determining the phase of Per1 oscillation. Deletion and point mutation analyses led to the identification of critical sequences responsible for the phase delay of Per2 oscillation. This sequence was classified as a CANNTG-type non-canonical E-box. Transfection experiments confirmed that the non-canonical E-box could be activated by CLOCK/BMAL1 and repressed by CRY1, whereas it was weakly affected by DEC1. Our observations revealed that CLOCK/BMAL1-mediated transcriptional regulation might not only play a role in circadian enhancers, but also generate various phases. We also showed that the D-box in the third conserved segment of the Per2 promoter generated high amplitude. Figure 8 summarizes the schematic models of the molecular mechanisms of Per2 transcription. Our study demonstrated that the copy number and various combinations of functional CCEs ultimately led to different circadian phase and amplitude.
Figure 8.
Schematic representation of molecular mechanisms of mPer2 circadian transcription.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Research and development project of the Industrial Science and Technology Program supported by the New Energy and Industrial Technology Development Organization (NEDO).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank H. Oda for reading the article and giving insightful comments. We are grateful to Y. Kato for providing the mDec1 expression vector.
REFERENCES
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr. Biol. 2000;10:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- 3.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 4.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown SA, Fleury-Olela F, Nagoshi E, Hauser C, Juge C, Meier CA, Chicheportiche R, Dayer JM, Albrecht U, Schibler U. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol. 2005;3:e338. doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato TK, Yamada RG, Ukai H, Baggs JE, Miraglia LJ, Kobayashi TJ, Welsh DK, Kay SA, Ueda HR, Hogenesch JB. Feedback repression is required for mammalian circadian clock function. Nat. Genet. 2006;38:312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 9.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 10.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 12.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 13.Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, Muijtjens M, Hoeijmakers JH, van der Horst GT. Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science. 1999;286:2531–2534. doi: 10.1126/science.286.5449.2531. [DOI] [PubMed] [Google Scholar]
- 14.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 15.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 16.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 17.Triqueneaux G, Thenot S, Kakizawa T, Antoch MP, Safi R, Takahashi JS, Delaunay F, Laudet V. The orphan receptor Rev-erbalpha gene is a target of the circadian clock pacemaker. J. Mol. Endocrinol. 2004;33:585–608. doi: 10.1677/jme.1.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- 19.Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 20.Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes. Dev. 2001;15:995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 22.Ukai-Tadenuma M, Kasukawa T, Ueda HR. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat. Cell Biol. 2008;10:1154–1163. doi: 10.1038/ncb1775. [DOI] [PubMed] [Google Scholar]
- 23.Akashi M, Ichise T, Mamine T, Takumi T. Molecular mechanism of cell-autonomous circadian gene expression of Period2, a crucial regulator of the mammalian circadian clock. Mol. Biol. Cell. 2006;17:555–565. doi: 10.1091/mbc.E05-05-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo SH, Ko CH, Lowrey PL, Buhr ED, Song EJ, Chang S, Yoo OJ, Yamazaki S, Lee C, Takahashi JS. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl Acad. Sci. USA. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohno T, Onishi Y, Ishida N. A novel E4BP4 element drives circadian expression of mPeriod2. Nucleic Acids Res. 2007;35:648–655. doi: 10.1093/nar/gkl868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hida A, Koike N, Hirose M, Hattori M, Sakaki Y, Tei H. The human and mouse Period1 genes: five well-conserved E-boxes additively contribute to the enhancement of mPer1 transcription. Genomics. 2000;65:224–233. doi: 10.1006/geno.2000.6166. [DOI] [PubMed] [Google Scholar]
- 27.Yamaguchi S, Mitsui S, Yan L, Yagita K, Miyake S, Okamura H. Role of DBP in the circadian oscillatory mechanism. Mol. Cell. Biol. 2000;20:4773–4781. doi: 10.1128/mcb.20.13.4773-4781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Effects of aging on central and peripheral mammalian clocks. Proc. Natl Acad. Sci. USA. 2002;99:10801–10806. doi: 10.1073/pnas.152318499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Circadian rhythms in isolated brain regions. J. Neurosci. 2002;22:350–356. doi: 10.1523/JNEUROSCI.22-01-00350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl Acad. Sci. USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawaguchi S, Shinozaki A, Obinata M, Saigo K, Sakaki Y, Tei H. Establishment of cell lines derived from the rat suprachiasmatic nucleus. Biochem. Biophys. Res. Commun. 2007;355:555–561. doi: 10.1016/j.bbrc.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 33.Ueda HR, Chen W, Minami Y, Honma S, Honma K, Iino M, Hashimoto S. Molecular-timetable methods for detection of body time and rhythm disorders from single-time-point genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2004;101:11227–11232. doi: 10.1073/pnas.0401882101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 35.He PJ, Hirata M, Yamauchi N, Hashimoto S, Hattori MA. The disruption of circadian clockwork in differentiating cells from rat reproductive tissues as identified by in vitro real-time monitoring system. J. Endocrinol. 2007;193:413–420. doi: 10.1677/JOE-07-0044. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida K, He PJ, Yamauchi N, Hashimoto S, Hattori MA. Up-regulation of circadian clock gene Period 2 in the prostate mesenchymal cells during flutamide-induced apoptosis. Mol. Cell. Biochem. 2010;335:37–45. doi: 10.1007/s11010-009-0238-7. [DOI] [PubMed] [Google Scholar]
- 37.Nakashima A, Kawamoto T, Honda KK, Ueshima T, Noshiro M, Iwata T, Fujimoto K, Kubo H, Honma S, Yorioka N, et al. DEC1 modulates the circadian phase of clock gene expression. Mol. Cell. Biol. 2008;28:4080–4092. doi: 10.1128/MCB.02168-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leclerc GM, Boockfor FR. Pulses of prolactin promoter activity depend on a noncanonical E-box that can bind the circadian proteins CLOCK and BMAL1. Endocrinology. 2005;146:2782–2790. doi: 10.1210/en.2005-0100. [DOI] [PubMed] [Google Scholar]
- 39.Nakahata Y, Yoshida M, Takano A, Soma H, Yamamoto T, Yasuda A, Nakatsu T, Takumi T. A direct repeat of E-box-like elements is required for cell-autonomous circadian rhythm of clock genes. BMC Mol. Biol. 2008;9:1. doi: 10.1186/1471-2199-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumaki Y, Ukai-Tadenuma M, Uno KD, Nishio J, Masumoto KH, Nagano M, Komori T, Shigeyoshi Y, Hogenesch JB, Ueda HR. Analysis and synthesis of high-amplitude Cis-elements in the mammalian circadian clock. Proc. Natl Acad. Sci. USA. 2008;105:14946–14951. doi: 10.1073/pnas.0802636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.