Abstract
MicroRNAs (miRNAs) are a class of small, non-coding RNAs that play a pivotal role in the regulation of posttranscriptional gene expression in a wide range of eukaryotic organisms. Although DNA viruses have been shown to encode miRNAs and exploit the cellular RNA silencing machinery as a convenient way to regulate viral and host gene expression, it is generally believed that this pathway is not available to RNA viruses that replicate in the cytoplasm of the cell because miRNA biogenesis is initiated in the nucleus. In fact, among the >200 viral miRNAs that have been experimentally verified so far, none is derived from an RNA virus. Here, we show that a cytoplasmic RNA virus can indeed encode and produce a functional miRNA. We introduced a heterologous miRNA-precursor stem-loop sequence element into the RNA genome of the flavivirus tick-borne encephalitis virus, and this led to the production of a functional miRNA during viral infection without impairing viral RNA replication. These findings demonstrate that miRNA biogenesis can be used by cytoplasmic RNA viruses to produce regulatory molecules for the modulation of the transcriptome.
INTRODUCTION
MicroRNAs (miRNAs) are a class of short (∼21–25 nt), single-stranded non-coding RNA molecules that play a critical role in many cellular processes, such as cell development, differentiation, apoptosis and oncogenesis (1–3). They exert their suppressive activity on gene expression by guiding the RNA-induced silencing complex (RISC) to complementary RNA stretches within a specific messenger RNA. The biosynthesis of the vast majority of miRNAs involves the sequential action of two RNAse III enzymes, Drosha and Dicer (4). A larger RNA precursor containing a characteristic miRNA hairpin structure is transcribed in the nucleus. This so-called primary miRNA is recognized by Drosha (acting with cofactor DGCR8) and cleaved into a ∼70-nt stem-loop RNA intermediate (precursor-miRNA, premiRNA), which is then transported by exportin-5 to the cytoplasm, where it undergoes processing by Dicer, and gives rise to a mature, single-stranded miRNA (5–11).
The number of identified miRNAs in plants and animals is rapidly increasing, and >700 miRNAs are known so far in humans (12). The first miRNAs expressed from human viruses were discovered in 2004 (13). Since then, >200 miRNAs have been described, but up to now, all of them have been derived from viruses with a nuclear DNA stage in their replication cycle, mainly of the herpesvirus family (12,14). Although computer algorithms have also predicted the existence of miRNA hairpins in viruses with RNA genomes (15), a study involving the cloning of small RNAs from cells infected with two positive-strand RNA viruses, hepatitis C virus (HCV) and yellow fever virus (YFV), did not result in the identification of any RNA virus-derived small interfering RNAs (siRNAs) or miRNAs (16). A recent paper by Parameswaran et al. (17) identified virus-derived small RNAs (vsRNAs) in a wide range of vertebrate cells infected with RNA viruses, such as Dengue, West Nile and HCV. However, there was no evidence that these small RNAs included miRNAs, and precursor structures or physiological relevance of the discovered vsRNAs remain to be elucidated.
In the present study, we investigated whether RNA viruses are fundamentally capable of producing functional miRNA, by inserting a known miRNA element into the genome of an RNA virus whose life cycle is confined to the cytoplasm of the host cell. Thereby we show that a functional miRNA can indeed be produced during infection without imparing viral RNA replication.
MATERIAL AND METHODS
Viruses and cells
Western subtype TBEV strain Neudoerfl, which has been characterized in detail, including the determination of its entire genomic sequence (18,19) (GenBank accession no. U27495), was used as the wild-type control in all infection experiments, and all mutants described in this study were derived from this strain.
The Epstein-Barr virus (EBV) miR-BART2 sequence was obtained from genomic DNA of human herpesvirus 4 [EBV (20), GenBank accession no. V01555].
BHK-21 cells were grown at 37°C with 5% CO2 in Eagle’s minimal essential medium (Sigma) supplemented with 5% fetal calf serum (FCS), 1% glutamine and 0.5% neomycin (growth medium) and were maintained in Eagle’s minimal essential medium supplemented with 1% FCS, 1% glutamine, 0.5% neomycin and 15 mM HEPES, pH 7.4 (maintenance medium).
Mutant construction
All viral constructs described in this study are derivatives of pTNd/c, an infectious cDNA clone of TBEV strain Neudoerfl (21). To create variants containing the miR-BART2 precursor stem-loop and flanking EBV sequence, DNA was amplified from EBV B95-8 genomic DNA using primers 5′-ATCGACCGGTATGCCACCTCCCTGCCTG-3′ and 5′-CGATACCGGTGCGTGGCCCGTGGATCTG-3′. The PCR product was digested with AgeI and ligated with AgeI-digested reporter replicon construct C17, yielding C17 BART(+) or C17 BART(−), depending on the orientation of the insert. C17 is a modified version of the previously described construct C17fluc-TaV2 A (22) in which the firefly luciferase (fluc) start codon ATG was deleted. C17 matBART(+) was constructed by first annealing oligonucleotides 5′-CCGGACTATTTTCTGCATTCGCCCTTGCGT-3′ and 5′-CCGGACGCAAGGGCGAATGCAGAAAATAGT-3′, resulting in a double-stranded DNA fragment with AgeI overhangs and then ligating this fragment with AgeI-digested C17 plasmid DNA. Full-length infectious cDNA clones were constructed by excising an XbaI/NheI fragment from each C17 replicon and inserting it into the corresponding site of XbaI/NheI-digested pTNd/c.
The non-replicating C17 variant C17 BART(+)GAA was constructed by replacement of the NS5 coding sequence of C17 BART(+) with an KpnI/XbaI-digested fragment of the previously described reporter replicon mutant NS5-GAA. The sequences of all constructs in this study were confirmed using an automated DNA sequencing system (PE Applied Biosystems, GA3100).
Reporter constructs and luciferase assays
To construct the luciferase reporter plasmids psiCHECK1-BART2 and psiCHECK2-BART2, oligonucleotides 5′-TCGAGCGCAAGGGCGAATGCAGAAAATAGT-3′ and 5′-GGCCACTATTTTCTGCATTCGCCCTTGCGC-3′, containing XhoI and NotI overhangs, were annealed and ligated into the multiple cloning site of psiCHECKTM-1 and psiCHECKTM-2 (Promega).
The Dual-GloTM Luciferase Assay System (Promega) was used according to the manufacturer’s instructions to measure both firefly and Renilla luciferase (Rluc) activity simultaneously. For experiments with infectious viruses, cells were electroporated (EP) with 6 µg psiCHECK2-BART2 plasmid DNA using a Gene Pulser apparatus (Bio-Rad). One day after electroporation, 60 000 cells/well were seeded in 96-well plates for subsequent infection in triplicate with wild-type and mutant virus at an MOI of 10. Renilla and fluc levels were determined immediately after infection and at 24 and 48 h post infection. For experiments using non-infectious replicons, cells were coelectroporated with equimolar amounts of in vitro-transcribed replicon RNA (corresponding to ∼1.2 × 1012 RNA copies) and 6 µg psiCHECK1-BART2 plasmid DNA. Procedures for RNA in vitro transcription and electroporation of BHK-21 cells were performed as reported in previous studies (21,22).
Immunofluorescence assay
Approximately 1 × 105 cells were seeded onto individual glass cover slips in 24-well plates prior to infection (TNd/c mutant set) or after transfection (C17 mutant set). Immunofluorescence (IF) staining was performed 24 h after infection with TNd/c mutants or 48 h after transfection with replicon RNA. Cells were permeabilized by acetone-methanol (1 : 1) fixation, and viral proteins were detected by incubation with rabbit anti-TBEV serum and a fluorescein-isothyocyanate-conjugated anti-rabbit antibody (Jackson ImmunoResearch Laboratories).
Northern blot analysis
miRNA-BART2 northern blotting
Total RNA was extracted using a mirVana miRNA Isolation Kit (Ambion) following the vendor’s recommendations. Of total RNA, 20 µg per sample was electrophoresed through a 12%-urea-polyacrylamide gel, which was stained with ethidium bromide for visualization of bands and then electroblotted to Hybond XL nylon membranes (GE Healthcare). A mirVana miRNA Probe Construction Kit (Ambion) and [alpha-32P] CTP (Hartmann Analytic) were used to construct a radioactive RNA probe (5′-GCAAGGGCGAAUGCAGAAAAUA-3′) complementary to EBV miR-BART2-5p. Blots were hybridized overnight in PerfectHybTM Plus (Sigma) hybridization buffer at 50°C and then washed twice with 5× SSC + 1% SDS and twice for 15 min with 1× SSC + 1% SDS prior to signal detection on a phosphorimager. Cells EP with synthetic miRNA (Pre-miR™ miRNA Precursor Molecules, Ambion) at a concentration of 30 nm were used as positive controls.
Northern blot of subgenomic RNA
1 µg of total RNA, extracted from BHK-21 cells using a mirVana miRNA isolation Kit (Ambion), was subjected to denaturing gel electrophoresis using a Northern MaxGly Kit (Ambion) and transferred to a positively charged nylon membrane (BrightStar Plus, Ambion) according to the manufacturer’s recommendations. Two different probes were constructed by in vitro transcription (MAXIscript T7 Kit, Ambion) using biotinylated UTP (Ambion). Templates for in vitro transcription were generated by PCR amplification of pTNd/c-BART(+) DNA using the following T7 promoter primers: 5′-CAGGGGTGAGGAATGCCCCCAGA-3′ and 5′-TAATACGACTCACTATAGGGCGGGTGTTTTTCCGAGTCAC-3′, yielding a probe complementary to the core 3′-NCR of TBEV and 5′-ATGCCACCTCCCTGCCTGGTGGAC-3′ and 5′-TAATACGACTCACTATAGGGCGTGGCCCGTGGATCTGTGAA-3′, yielding a probe specific for the miR-BART2 sequence insertion, including the EBV flanking regions. Hybridization and washing steps were performed according to the manual, and these were followed by a non-isotopic detection procedure (Bright Star Biodetect Kit, Ambion).
Time course analysis of miR-BART2 expression levels
Total RNA from infected cells was extracted using a mirVana miRNA Isolation Kit (Ambion). TaqManTM miRNA Assays (Applied Biosystems) specific for EBV BART2-5p and U6 snRNA (endogenous control) were performed according to the manufacturer’s protocol using an ABI 7300 Real-Time PCR System (PE Applied Biosystems). Relative quantification was carried out using the 2−ΔΔT method (23) for infected versus uninfected cells.
Quantification of TBEV genomic RNA and mature miR-BART2
To quantify the number of genomic TBEV RNA per cell in comparison to the number of mature miRNA-BART2 derived from these genomes, standardized real-time PCR was performed. Equal numbers of cells (5 × 104), as counted in a Casy TT cell counter (Schärfe Systems), were collected and total RNA was extracted using the mirVana miRNA Isolation Kit (Ambion). cDNA synthesis for qPCR of genomic TBEV RNA was performed using an iScript cDNA Synthesis Kit (BioRad) and real-time PCR was carried out on a BioRad iCycler. Primer, probe sequences and the temperature profile were selected as described in a previous study (24). Copy numbers of viral RNA were determined by comparison of the result to a standard curve that was prepared by using a serial 10-fold dilution of DNAse treated, in vitro transcribed full length RNA of known concentration.
In parallel, the same mirVana total RNA preparation was used to determine the copy number of mature miRNA-BART2 molecules in a TaqManTM miRNA Assay according to the manufacturer’s protocol. A 10-fold serial dilution of the RNA-oligonucleotide 5′-UAUUUUCUGCAUUCGCCCUUGC-3′ (corresponding to mature miR-BART2-5p) was used as a standard curve.
Virus production, passaging and focus assay
For the production of infectious virus particles, BHK-21 cells were transfected with equal amounts of in vitro-transcribed full-length RNA. The supernatant from the transfected cells was collected 48 h post infection and cleared by centrifugation. Various dilutions were applied to confluent cell monolayers to determine the number of focus-forming units (ffu). Fifty hours postinfection, cells were fixed with acetone-methanol (1 : 1) and treated with polyclonal rabbit anti-TBEV serum. Antibody-labeled cells were detected using an immunoenzymatic reaction consisting of successive incubations with goat anti-rabbit immunoglobulin G-alkaline phosphatase and the corresponding enzyme substrate (SigmaFast Red TR/Naphtol AS-MX tablets). To analyze the genomic stability of the viral mutants and to increase the titer, viruses for infection experiments were passaged at least five times, using a 1 : 5 000 dilution of the supernatant.
Drosha knockdown studies
Knockdown studies were performed using shRNA plasmids (Santa Cruz Biotechnology) in HeLa cells. Cells were transfected with 1 µg Drosha short hairpin plasmid sc-44080-SH or negative control shRNA plasmid-A sc-108060 and 0.5 µg reporter plasmid psiCHECK2-BART2 in 6-well plates using Lipofectamine 2000 (Invitrogen). Cells were trypsinized 24 h after transfection, pooled and seeded in 96-well plates. Infection with TNd/c or TNd/c BART(+) was initiated 48 h after the initial shRNA/reporter plasmid transfection. The luciferase assay was carried out as outlined above. Knockdown efficiency in parallel to the luciferase assay was determined at the protein level by western blotting using a rabbit polyclonal antibody to Drosha (Abcam) and a mouse monoclonal antibody to GAPDH (Ambion). Corresponding secondary antibodies for ECL detection (Amersham) were purchased from Jackson Immunoresearch.
RESULTS
Construction of a cytoplasmic RNA virus encoding a viral miRNA
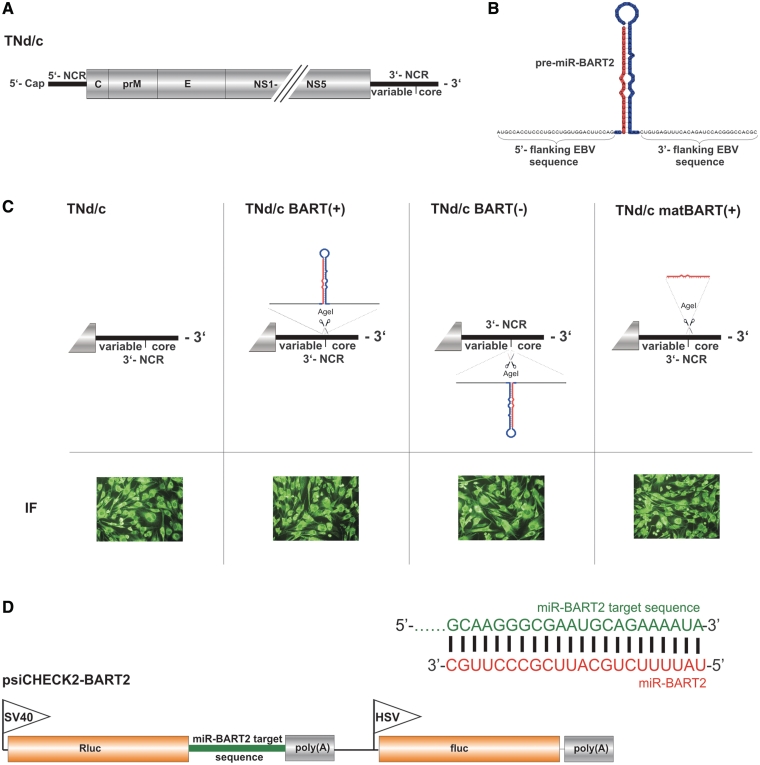
For these experiments, we used tick-borne encephalitis virus (TBEV), a human pathogen of the genus Flavivirus, family Flaviviridae. Like all members of this family of positive-strand RNA viruses, the replication cycle of TBEV takes place in the cytoplasm. Its genome consists of a single RNA molecule of ∼11 kb in length, which also serves as a messenger RNA, encoding a single polyprotein precursor that is processed to form three structural proteins (C, prM, E), which together with the genomic RNA compose the virion, and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5) that are necessary for replication (Figure 1A) (25–27).
Figure 1.
TNd/c BART mutants and reporter plasmid psiCHECK2-BART2. (A) Genome organization of wild-type TBEV virus strain Neudoerfl (TNd/c). 5′-NCR, 5′-non-coding region; 3′-NCR, 3′-non-coding region. The positions of the ‘variable’ and conserved ‘core’ regions of the 3′-NCR are indicated. (B) EBV miR-BART2 precursor in the predicted secondary structure (miRBase accession number: MI0001068) flanked by 30 nt of EBV B95-8 sequence on either side. The sequence of the precursor of miR-BART2 is highlighted in red and blue, with the red region corresponding to the mature miR-BART2. (C) Top: Schematic diagram of the 3′-NCR of parental virus TNd/c and the derived TNd/c BART mutants. Below: IF staining of infected cells with rabbit anti-TBEV serum 24 h postinfection. (D) Mature miR-BART2 (red) and the corresponding target sequence in the 3′-UTR of the EBV BALF5 mRNA (green) that was inserted into the 3′-UTR of the Rluc mRNA gene of the parental vector psiCHECK-2. The fluc expressed from the HSV-TK promoter served as an internal standard. Promoters are indicated by flags: SV40, simian virus 40; HSV-TK, herpes simplex virus thymidine kinase. Diagrams are not drawn to scale.
The 3′-NCR of TBEV has been shown to tolerate the insertion of heterologous sequence elements (22). We took advantage of this flexibility to insert the miR-BART2 hairpin precursor from the herpesvirus EBV between the variable and core regions of the 3′-NCR of a wild-type TBEV strain (TNd/c, Figure 1B and C). miR-BART2 downregulates expression of the EBV DNA polymerase BALF5, and this is thought to prevent the transition from latent to lytic replication (16,28,29). To favor proper formation of the stem-loop secondary structure, which is crucial for recognition by the classical miRNA-processing machinery (6,30,31), we decided to add flanking EBV sequence on either side of the hairpin. We generated three different BART mutants. The EBV miR-BART2 precursor was introduced in the plus-strand (messenger-sense) orientation [TNd/c BART(+)] and also as the reverse complement [TNd/c BART(−)] to allow a potentially functional miRNA precursor to be formed at the 5′-end of the negative-strand (antisense) copy of the viral RNA during replication. Bioinformatic secondary structure analysis prior to construction of these mutants prompted us to add 30 nt of original genomic EBV sequence on either side of the hairpin, resulting in a tolerable maximum of 128 nt for the full-hairpin insertions. In a third construct, only the ‘mature’ miR-BART2 sequence, devoid of any EBV-derived flanking sequences or hairpin structure elements, was inserted [TNd/c matBART(+)]. To assess the capability of these TBEV BART mutants to replicate and be translated, RNA from the mutants and wild-type TBEV was made by in vitro transcription and introduced into BHK-21 cells by electroporation. IF staining with an anti-TBEV serum 24 h after transfection showed that all three mutants were able to replicate and express viral proteins (Figure 1C), indicating that the inserted miRNA hairpin structure elements did not impair viral RNA replication or protein expression.
Next, we examined the stability of the heterologous sequence elements in a passage experiment on BHK-21 cells. After five rounds of sequential infection with diluted supernatant from the previous passage, no sequence changes could be detected in the inserted miR-BART2 sequences or the flanking elements. The only mutations observed were ones that occurred in the viral envelope protein E and were of a type previously associated with cell culture adaptations (Supplementary Table S1) (32,33). Taken together, these results demonstrated that the insertion of a miRNA hairpin precursor in either orientation into the RNA genome of this flavivirus was genetically stable and compatible with viral growth.
Functional activity of the inserted miRNA
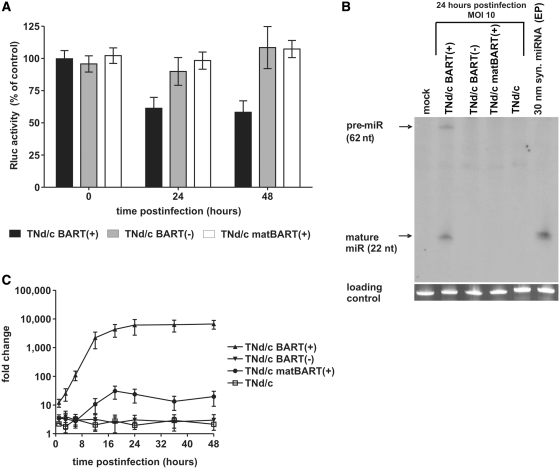
As a tool to measure the functional activity of miR-BART2, we established a dual-luciferase reporter assay. The original miR-BART2 target sequence was inserted downstream of the translational stop codon of the Rluc coding sequence of the siRNA evaluation vector psiCHECKTM-2, yielding a construct encoding an Rluc mRNA fused to a miR-BART2 target site, termed psiCHECK2-BART2 (Figure 1D). The fluc on the same plasmid, driven from a second promoter, served as an internal control. As expected, Rluc expression from this construct was suppressed in a dose-dependent manner upon transfection with synthetic miR-BART2 (Supplementary Figure S1A). To investigate whether the viral mutants TNd/c BART(+), TNd/c BART(−) and TNd/c matBART(+) were able to downregulate reporter gene expression, BHK-21 cells were transfected with psiCHECK2-BART2 and 24 h thereafter infected with BART mutants or wild-type virus. TNd/c BART(+) clearly suppressed reporter gene expression at 24 and 48 h post infection (Figure 2A). The observed reduction by ∼50% is equal to the effect seen with the natural miR-BART2 target BALF5 protein in EBV-infected cells with forced miR-BART2 expression (28). In contrast, infection of cells with TNd/c BART(−) or TNd/c matBART(+) did not decrease reporter gene expression.
Figure 2.
A functional miRNA is produced during infection with TBEV mutant TNd/c BART(+). (A) Relative Rluc activity after infection with TNd/c BART mutants. BHK-21 cells were transfected with psiCHECK2-BART2 and 24 h thereafter infected with viral mutants at an MOI of 10. Values represent Rluc expression levels relative to the infection with wild-type virus. Error bars indicate SD of three experiments, each measured in triplicate. (B) Northern-blot analysis of intracellular RNA 24 h after infection using a radioactive complementary RNA probe specific for the mature miR-BART2 sequence. Cells EP with synthetic miRNA-BART2 (syn. miRNA) served as a positive control. (C) Time course analysis of miR-BART2 expression in BHK-21 cells infected at an MOI of 10. At the indicated time points, total RNA of infected cells was isolated and subjected to a TaqManTM miRNA Assay specific for mature miR-BART2. U6 snRNA served as an endogenous control. The ΔCT value of uninfected cells at time point 0 was used as a base value to calculate fold change in expression by the comparative CT method.
Time course of miR-BART2 generation during infection
To obtain direct evidence for the formation of miR-BART2, total RNA was isolated from infected cells and subjected to northern blot analysis 24 h after infection (Figure 2B). Consistent with the data from the luciferase assay, mature miRNA and the corresponding pre-miR were readily detected in cells infected with TNd/c BART(+), but not in samples of the other mutants or wild-type virus. To establish a time course of miRNA formation in infected cells, we performed a TaqManTM miRNA Assay (Figure 2C). In cells infected with TNd/c BART(+), levels of miR-BART2 increased within 24 h to a maximum plateau level, whereas the signal detected for TNd/c BART(−) and TNd/c matBART(+) remained at or close to background levels.
Dependence of miRNA production on replication of viral RNA
The experiments described above consistently indicated that significant amounts of functional miR-BART2 were generated with TNd/c BART(+) and prompted a more detailed investigation of the relationship between RNA replication and miRNA formation. For this purpose, we prepared an analogous set of replicons [C17 BART(+), C17 BART(−) and C17 matBART(+)] based on the TBEV reporter replicon C17fluc-TaV2A, a construct in which the viral structural proteins had been replaced by an in-frame insertion of the fluc gene (Figure 3A) (22). Additionally, we designed a replication-negative variant of C17 BART(+) with a GDD-to-GAA mutation in the active site of the viral polymerase, termed C17 BART(+)GAA (Figure 3A). IF staining with an anti-TBEV antiserum 48 h after transfection confirmed that all of the replicons encoding a functional RNA polymerase were able to replicate and express viral proteins (Figure 3A, bottom).
Figure 3.
RNA replication is necessary for but not affected by miRNA biogenesis. (A) Top: schematic diagram of parental luciferase reporter replicon C17, which contains the first 17 aa of the capsid protein and has the rest of the structural proteins replaced by fluc. The 3′-NCR of each replicon is identical to that of the corresponding TNd/c virus mutant (Figure 1 C). Replication-negative C17 BART(+)GAA differs from C17 BART(+) by a GDD-to-GAA mutation in the polymerase active site of the viral NS5 protein. Below: IF staining with rabbit anti-TBEV serum 48 h post transfection. (B) Replication efficiencies of the C17 BART mutant set monitored by fluc activity in BHK-21 cells. Error bars represent the SD of three independent experiments, each measured in triplicate. (C) Relative Rluc activity in BHK-21 cells co-transfected with miR-BART2 reporter plasmid psiCHECK1-BART2 and the C17 BART mutants C17 BART(+) and C17 BART(+)GAA. Rluc levels are shown as a percentage of the level obtained with the parental replicon C17. Error bars represent the SD calculated from a minimum of three independent experiments, each measured in triplicate. (D) Schematic diagram of the reporter plasmid psiCHECK1-BART2, which contains the miR-BART2 target sequence fused to the Rluc-coding sequence under the control of an SV40 promoter. (E) Relative Rluc activity of C17 BART mutants at 48 h post transfection (top) and northern blot analysis (below) of the same intracellular RNA preparation used in the Rluc assay. The positions of the precursor and the mature miRNA are indicated by arrows. Total RNA of cells EP with synthetic miR-BART2 (syn. miRNA) served as a control. Diagrams (A) and (D) are not to scale.
Replication of these replicons was monitored by measurement of fluc activity in transfected BHK-21 cells, and this revealed that the insertion of the miRNA sequence (in either orientation or in its mature form) did not alter the level of RNA replication relative to the parental C17 replicon (Figure 3B). As expected, and consistent with the IF results, C17 BART(+)GAA was negative for RNA replication. To confirm that miRNA formation from these replicons mirrored the findings with infectious virus, functional miR-BART2 activity was assayed by measuring Rluc reporter gene repression (Figure 3C; see Supplementary Figure S2 for a complete data set) using a modified reporter plasmid lacking fluc, psiCHECK1-BART2 (Figure 3D). C17 BART(+) clearly suppressed reporter gene activity, and the presence of the miRNA was additionally confirmed in a northern blot assay (Figure 3E). In contrast, no activity and no miRNA was observed for C17 BART(−), C17 matBART(+) or the parental control C17. Notably, the replication-deficient control C17 BART(+)GAA also did not suppress Rluc expression or generate detectable miRNA (Figure 3C and E).
miRNAs are produced at lower molar concentrations than viral RNA
Apparently, miRNA formation did not significantly impair viral RNA replication which by itself was necessary for efficient miRNA production. This raised the question to which extent the viral plus-stranded RNA molecules were processed into mature miR-BART2 molecules. To answer this question, concentrations of both viral RNA and miR-BART2 were determined by qPCR at various time points post infection. As depicted in Figure 4, viral RNA replication of TNd/c and TNd/c BART(+) was identical. The number of genomic TBEV RNA molecules of both the wild-type and the mutant viruses reached a value of ∼7 × 104 copies per cell 48 h after infection. In contrast, the copy number of mature miR-BART in cells infected with TNd/c-BART(+) reached a concentration of about 500 miRNA molecules per cell 48 h post infection. This indicates that <1% of the viral RNA needed to be processed to generate a functionally active concentration of miRNAs.
Figure 4.
Quantification of viral genomic RNA and derived mature miR-BART2. BHK-21 cells were infected with TNd/c and TNd/c BART(+) at an MOI of 10. At the indicated time points 5 × 104 cells were collected and total RNA was isolated. Copy numbers of the viral genome were determined by qRT-PCR using a standard curve of in vitro transcribed full-length RNA. Copy numbers of mature miR-BART2 were determined in parallel by employing a synthetic RNA oligonucleotide for the generation of a standard curve in a TaqManTM miRNA Assay specific for mature miR-BART2. Data points represent geometric mean values from duplicate experiments, with error bars indicating SD.
The role of Drosha in miRNA biogenesis from the TBEV genome
Pathways that completely bypass Drosha cleavage have been reported recently [reviewed in (34)]. To investigate whether miRNA generation from the TBEV genome involves Drosha, we performed a transient shRNA knockdown. We cotransfected human HeLa cells with the luciferase reporter plasmid psiCHECK2-BART2 and Drosha-specific or negative-control shRNA plasmids. As shown in Figure 5A, transfection with Drosha-specific shRNA plasmids resulted in strong downregulation of Drosha protein expression 72–96 h after transfection but was unaffected by the delivery of a negative control shRNA plasmid encoding a scrambled sequence. Monitoring reporter gene expression in transfected cells 24 and 48 h after infection with TNd/c BART(+) and the wild-type virus (corresponding to 72 and 96 h after the initial shRNA transfection) revealed a measurable difference in miRNA activity between the Drosha-knockdown and control cells. As observed with BHK-21 cells, reporter gene expression in HeLa cells expressing normal Drosha levels was clearly reduced upon infection with TNd/c BART(+). However, Rluc expression did not appear to be suppressed after knockdown of Drosha (Figure 5B) suggesting that Drosha was involved in miRNA formation from the flavivirus.
Figure 5.
The involvement of Drosha in the generation of miR-BART2 from TNd/c BART(+). HeLa cells were transfected with shRNA plasmids targeting human Drosha and a negative control shRNA plasmid encoding a scrambled sequence (shRNA neg. co.). (A) Western blot showing the expression of Drosha. GAPDH was used as a loading control. (B) Transfected cells were subjected to infection with TNd/c BART(+) and TNd/c wild-type virus with a viral dose corresponding to an MOI of 100 on BHK-21 cells. Time points 24 and 48 h post infection correspond to 72 and 96 h after shRNA transfection. Rluc expression levels in shRNA-transfected cells that were infected with TNd/c BART(+) are depicted relative to shRNA-transfected cells infected with wild-type virus. Error bars represent the SD of three independent experiments, each measured in triplicate.
DISCUSSION
This is the first study that describes the generation of a functional miRNA from a cytoplasmic RNA virus. This observation is unexpected since two theoretical barriers are generally thought to impede miRNA biogenesis for this class of viruses: first, in contrast to DNA and retroviruses, the replication cycle of cytoplasmic RNA viruses takes place in the cytoplasm, away from the nucleus where Drosha resides and initiates the processing of the miRNA hairpin structure for a majority of all known miRNAs. Second, even if the miRNA precursor could be liberated from an RNA virus genome, this excision event would be expected to destroy copies of the viral genome and thereby reduce RNA replication efficiency.
A detailed follow-up study has to be carried out now, to clarify, how and where the miRNA is generated in our system, and which key players of the classical miRNA biogenesis pathway are involved in this process. Regardless of the mechanism, however, our results demonstrate that these barriers are not absolute and that they can eventually be circumvented.
One possible way in which this could occur is that viral infection might lead to a partial breakup of cellular compartmentalization or to protein retrafficking, allowing the Drosha complex to act in the cytoplasm. Another possibility is that a fraction of the flavivirus RNA containing the miRNA-BART2 precursor enters the nucleus. Many positive- and negative-stranded RNA viruses with a cytoplasmic replication cycle have been reported to alter nuclear-cytoplasmic trafficking to expedite viral growth (35), and the presence of flaviviral RNA in the nucleus has been suggested in a recent study (36). First experiments, performed to unravel the role of Drosha, point in this direction and do not suggest a Drosha-independent pathway. As it is becoming clear since recently that also non-canonical pathways can lead to the generation of functional miRNAs (5,34), it remains possible, however, that the activity of this enzyme is bypassed. One such pathway, in which miRNA precursors are fed into the miRNA maturation pathway without a contribution of Drosha, is the mirtron pathway (37–39). In this case, pre-miRNAs are formed from debranched introns during mRNA splicing.
One of the most important findings of our study is that while active viral replication was required for production of miRNA-BART2, the presence of the functional miRNA precursor element in the TBEV genome did not have a measurable negative impact on RNA replication. Remarkably, in a recent study, it was found that several flaviviruses produce a 3′-terminal non-coding RNA fragment that accumulates in infected cells due to incomplete degradation of viral genomic RNA (40). This highly structured, nuclease-resistant RNA was shown to increase both virus replication and virus-induced cytopathogenicity, although the reason for this is still unknown. This observation supports the idea that fragmentation processes can also be beneficial and that RNA viruses can indeed use such mechanisms to generate smaller RNA molecules with biological activity without the need for sub-genomic promoters. In further experiments, we found that TBEV-infected cells also contain relatively large amounts of a 3′-terminal sub-genomic RNA and that the amount of this RNA present in the cell is not altered by the insertion of the BART2 sequence element (Supplementary Figure S3). Regardless of the mechanism of miRNA precursor processing from the genomic viral RNA molecule, our data indicate that miRNA copy numbers were at least 100-fold lower than the number of genomic RNAs present in the cell. This provides an explanation why RNA replication was not impaired by the insertion of the miRNA precursor sequence because as little as 0.5–1% of the pool of genomes present in the cell would suffice to give rise to the observed levels of mature miRNA. Due to the fact that many RNA viruses, including flaviviruses, replicate to very high copy numbers, this mechanism allows production of physiologically active levels of miRNA without having to sacrifice a relevant proportion of the genomic RNA molecules. We also found that although insertion of the natural miRNA-BART2 hairpin precursor, with flanking sequences from the EBV genome, into the TBEV 3′-NCR in the plus-strand (messenger-sense) orientation led to production of a mature, functional miRNA, insertion of only the portion corresponding to mature miRNA-BART2 did not. This further emphasizes that the resulting mature miRNA was not simply a remaining piece of randomly degraded viral RNA but indeed the product of an active miRNA processing pathway.
Interestingly, the insertion of the same miRNA-BART2 precursor in the antisense orientation did not result in miRNA production. Since all of the constructs tested were genetically stable over multiple passages and apparently had no effect on the efficiency of viral RNA replication, we conclude that there must be specific mechanistic reasons for the requirement of the precursor element to be in the positive-sense orientation. First, the replication of flaviviruses is asymmetrical, meaning that viral plus strands accumulate in around 10-fold excess over the corresponding genome-length minus strands (25). Second, the minus-strand RNA template may be inaccessible to the miRNA machinery when it is in the ER-associated viral replication complex. Recent biochemical analysis has suggested that the flavivirus replication complex resides in virus-induced double-layered membrane compartments (41). Therefore, it seems reasonable that the minus strand remains shielded from other cellular compartments and enzymes. The plus strand, in contrast, is forced to leave the site of replication to gain access to the cellular translation machinery.
The results of this study demonstrate that, contrary to current assumptions, RNA viruses can carry functional miRNA elements. Future studies will have to address whether similar results can be obtained with different viral or cellular miRNAs, when inserted into various genomic locations or into various other viral species derived from the spectrum of RNA virus families. A result published during the review process of our manuscript, provides compelling evidence that this may indeed be possible. Varble et al. (42) engineered a celluar miRNA hairpin into the genome of influenza virus and reported the synthesis of a functional miRNA without a negative impact on viral replication. Orthomyxoviruses, such as influenza, are segmented, negative-stranded RNA viruses which employ the host cell nucleus for replication. Although mechanisms of miRNA formation likely may differ, this report supports the notion that RNA viruses can encode miRNAs. It is tempting to speculate that among the vast assortment of diverse RNA viruses found in nature, some may indeed naturally encode miRNAs and use them as part of their replication strategy to regulate viral or host gene expression.
A more comprehensive search for natural miRNAs among many different RNA viruses seems warranted in the light of the current findings. Regardless of whether or not miRNAs are found to play a physiological role in their biology, our study demonstrates that their generation is mechanistically possible. This knowledge opens up new avenues for the rational design of RNA virus mutants and vectors encoding this important class of regulatory molecules.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Austrian ‘Fonds zur Foerderung der wissenschaftlichen Forschung (FWF)’ (grant no. P19411-B11).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to F.A. Graesser and T. Pfuhl (Homburg, Germany) for introduction to miRNA northern blotting, and U. Blaesi and A. Resch (MFPL, Vienna) for providing their radioactive facility. We thank Franz X. Heinz for stimulating discussions, G. O’Riordain for participation in mutant construction, C. Taucher, V.M. Hoenninger and K. Ramsauer for critical review of the manuscript, and S.L. Allison for his invaluable assistance in preparation of this article.
REFERENCES
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Bushati N, Cohen SM. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. Embo. J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 6.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 8.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 9.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 11.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 14.Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li SC, Shiau CK, Lin WC. Vir-Mir db: prediction of viral microRNA candidate hairpins. Nucleic Acids Res. 2008;36:D184–D189. doi: 10.1093/nar/gkm610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 17.Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, Ansel KM, Heissmeyer V, Einav S, Jackson W, et al. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 6:e1000764. doi: 10.1371/journal.ppat.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandl CW, Heinz FX, Kunz C. Sequence of the structural proteins of tick-borne encephalitis virus (western subtype) and comparative analysis with other flaviviruses. Virology. 1988;166:197–205. doi: 10.1016/0042-6822(88)90161-4. [DOI] [PubMed] [Google Scholar]
- 19.Mandl CW, Heinz FX, Stockl E, Kunz C. Genome sequence of tick-borne encephalitis virus (Western subtype) and comparative analysis of nonstructural proteins with other flaviviruses. Virology. 1989;173:291–301. doi: 10.1016/0042-6822(89)90246-8. [DOI] [PubMed] [Google Scholar]
- 20.Baer R, Bankier AT, Biggin MD, Deininger PL, Farrell PJ, Gibson TJ, Hatfull G, Hudson GS, Satchwell SC, Seguin C, et al. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 21.Mandl CW, Ecker M, Holzmann H, Kunz C, Heinz FX. Infectious cDNA clones of tick-borne encephalitis virus European subtype prototypic strain Neudoerfl and high virulence strain Hypr. J. Gen. Virol. 1997;78 (Pt 5):1049–1057. doi: 10.1099/0022-1317-78-5-1049. [DOI] [PubMed] [Google Scholar]
- 22.Hoenninger VM, Rouha H, Orlinger KK, Miorin L, Marcello A, Kofler RM, Mandl CW. Analysis of the effects of alterations in the tick-borne encephalitis virus 3′-noncoding region on translation and RNA replication using reporter replicons. Virology. 2008;377:419–430. doi: 10.1016/j.virol.2008.04.035. [DOI] [PubMed] [Google Scholar]
- 23.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 24.Kofler RM, Hoenninger VM, Thurner C, Mandl CW. Functional analysis of the tick-borne encephalitis virus cyclization elements indicates major differences between mosquito-borne and tick-borne flaviviruses. J. Virol. 2006;80:4099–4113. doi: 10.1128/JVI.80.8.4099-4113.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindenbach BD, Thiel H-J, Rice CM. In: Fields Virology. 5th edn. Knipe DM, Howley PM, editors. Philadelphia, PA.: Lippincott Williams & Wilkins Co.; 2007. pp. 1101–1152. [Google Scholar]
- 26.Wengler G, Wengler G. Terminal sequences of the genome and replicative-from RNA of the flavivirus West Nile virus: absence of poly(A) and possible role in RNA replication. Virology. 1981;113:544–555. doi: 10.1016/0042-6822(81)90182-3. [DOI] [PubMed] [Google Scholar]
- 27.Wengler G, Wengler G, Gross HJ. Studies on virus-specific nucleic acids synthesized in vertebrate and mosquito cells infected with flaviviruses. Virology. 1978;89:423–437. doi: 10.1016/0042-6822(78)90185-x. [DOI] [PubMed] [Google Scholar]
- 28.Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grasser FA. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666–675. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J. Biol. Chem. 2005;280:27595–27603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- 31.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kroschewski H, Allison SL, Heinz FX, Mandl CW. Role of heparan sulfate for attachment and entry of tick-borne encephalitis virus. Virology. 2003;308:92–100. doi: 10.1016/s0042-6822(02)00097-1. [DOI] [PubMed] [Google Scholar]
- 33.Mandl CW, Kroschewski H, Allison SL, Kofler R, Holzmann H, Meixner T, Heinz FX. Adaptation of tick-borne encephalitis virus to BHK-21 cells results in the formation of multiple heparan sulfate binding sites in the envelope protein and attenuation in vivo. J. Virol. 2001;75:5627–5637. doi: 10.1128/JVI.75.12.5627-5637.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 35.Hiscox JA. The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication. Virus Res. 2003;95:13–22. doi: 10.1016/S0168-1702(03)00160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uchil PD, Kumar AV, Satchidanandam V. Nuclear localization of flavivirus RNA synthesis in infected cells. J. Virol. 2006;80:5451–5464. doi: 10.1128/JVI.01982-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host. Microb. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Uchil PD, Satchidanandam V. Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 2003;278:24388–24398. doi: 10.1074/jbc.M301717200. [DOI] [PubMed] [Google Scholar]
- 42.Varble A, Chua MA, Perez JT, Manicassamy B, Garcia-Sastre A, tenOever BR. Engineered RNA viral synthesis of microRNAs. Proc. Natl Acad. Sci. USA. 2010;107:11519–11524. doi: 10.1073/pnas.1003115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.