Abstract
eIF4E-binding proteins (4E-BPs) regulate translation of mRNAs in eukaryotes. However the extent to which specific mRNA targets are regulated by 4E-BPs remains unknown. We performed translational profiling by microarray analysis of polysome and monosome associated mRNAs in wild-type and mutant cells to identify mRNAs in yeast regulated by the 4E-BPs Caf20p and Eap1p; the first-global comparison of 4E-BP target mRNAs. We find that yeast 4E-BPs modulate the translation of >1000 genes. Most target mRNAs differ between the 4E-BPs revealing mRNA specificity for translational control by each 4E-BP. This is supported by observations that eap1Δ and caf20Δ cells have different nitrogen source utilization defects, implying different mRNA targets. To account for the mRNA specificity shown by each 4E-BP, we found correlations between our data sets and previously determined targets of yeast mRNA-binding proteins. We used affinity chromatography experiments to uncover specific RNA-stabilized complexes formed between Caf20p and Puf4p/Puf5p and between Eap1p and Puf1p/Puf2p. Thus the combined action of each 4E-BP with specific 3′-UTR-binding proteins mediates mRNA-specific translational control in yeast, showing that this form of translational control is more widely employed than previously thought.
INTRODUCTION
It has become increasingly evident that the translation process for a large number of mRNAs is controlled at the level of initiation in a wide variety of cells. The selection of mRNAs for translation is mediated by the binding of eukaryotic initiation factor (eIF) 4F complex to the 5′-end of each mRNA. The eIF4F complex is composed of eIF4E that binds to the 5′ 7-methyl guanosine mRNA cap, eIF4A an RNA helicase, and eIF4G a scaffolding protein capable of bridging contacts between eIF4E and 40S ribosomal subunits. eIF4G and the eIF4E-binding proteins (4E-BPs) share a motif (YXXXXLϕ, where X is any residue and ϕ is hydrophobic) that is critical for eIF4E interaction. Hence 4E-BP bound mRNAs exclude eIF4G and are translationally silenced (1,2). Mammalian 4E-BP1 phosphorylation status modulates its affinity for eIF4E. mTOR dependent hyperphosphorylation of 4E-BP1 causes its dissociation from eIF4E relieving repression and promoting eIF4F assembly, translation and cell growth (3,4). A second class of 4E-BP has been identified from studies of early development, e.g. in Xenopus and Drosophila where asymmetric expression of proteins is important for determining the body plan of the embryo (2,5). For these 4E-BPs additional protein–protein and protein–mRNA contacts tether the 4E-BP to specific mRNA targets. Thus Xenopus Maskin is a 4E-BP that also binds to the cytoplasmic polyadenylation element-binding (CPEB) protein which itself interacts with sequences within mRNA 3′-UTRs. Together and in combination with other proteins, these factors mediate circularization and repression of transcripts until developmentally regulated signalling events activate both cytoplasmic polyadenylation, disrupt the Maskin-eIF4E interaction and promote translation (2,5).
Saccharomyces cerevisiae possesses two 4E-BPs, Caf20p and Eap1p of 18 and 70 KDa, respectively. Neither protein shares obvious sequence homology with each other or 4E-BPs from non-yeast species, except for the eIF4E interaction consensus motif, however both yeast 4E-BPs interact with eIF4E to inhibit translation of capped reporter mRNAs (6,7). Although deletion of either 4E-BP gene confers no growth defect under standard laboratory conditions, caf20Δ alleviates growth defects of some translation factor mutations and conversely its overexpression exacerbates these phenotypes (8). eap1Δ causes rapamycin resistance (7) and partial resistance to translation inhibition caused by diamide (a thiol oxidant), and cadmium (a heavy metal) (9). In the yeast Σ1278b background deletion of either 4E-BP prevents pseudohyphal growth following nitrogen limitation (10). Similarly, eap1Δ and caf20Δ cells respond differently to membrane stress (11,12). Taken together the data are consistent with the idea that each 4E-BP interacts with eIF4E and competes with eIF4G to modulate the translation of a different subset of specific yeast genes.
Little progress has been made in identifying specific mRNA targets regulated by the yeast 4E-PBs. One possible candidate is the G1 cyclin CLN3 as it can be controlled by altering eIF4E activity. CLN3 mRNA contains a short up-stream ORF that plays a role repressing Cln3p expression (13). Temperature sensitive eIF4E mutations (e.g. cdc33-1) confer G1 arrest and reduce expression of CLN3 and enhanced CLN3 expression is sufficient to restore G1-S phase progression (14). Recently, a second mRNA target, POM34 mRNA was described, which encodes a membrane protein component of the nuclear pore complex. In cells bearing spindle-pole body duplication defects Eap1p was required for POM34 translation (15).
Given the paucity of information concerning specific 4E-BP targets, we used a translational profiling approach to identify mRNAs whose translation is altered following loss of each yeast 4E-BP and found that both altered translation of a large fraction of yeast genes. Quantitative real-time reverse transcription-polymerase chain reaction (RT–PCR) and immunoblot analyses have validated the micro-array approach for selected targets and a phenotypic analysis indicates that the observed altered translation of several nitrogen metabolism genes may contribute to the sensitivity of the eap1Δ strain to growth on some nitrogen sources. Despite sharing a common interaction site on eIF4E, we find that most target mRNAs differ between the 4E-BPs suggesting that additional factors are necessary to impart 4E-BP mRNA specificity. To begin to address this mechanistic issue we compared the identities of mRNAs found by others to interact with specific RNA-interacting proteins, with our translational profiling data. Our analysis suggested that certain yeast PUF proteins may be among those important for mRNA specificity. As PUF proteins have been implicated in translational control in higher organisms, we tested this prediction directly and report that Puf4/5p-bound mRNA complexes associate with a fraction of Caf20p, while Puf1/2p associate with Eap1p. Taken together these studies reveal that ∼1000 genes are subject to translational control via the 4E-BPs and provides evidence that 4E-BP association with specific 3′-UTR-binding proteins imparts mRNA target specificity in yeast.
MATERIALS AND METHODS
Yeast strains and growth conditions
Isogenic yeast strains BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0), Y17334 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 caf20Δ::kanMX4) and Y17036 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 eap1Δ::kanMX4) were obtained from Euroscarf (Frankfurt, Germany) and grown in synthetic dextrose (SD) minimal medium containing auxotrophic supplements and grown to A600 = 0.7 for all array experiments.
For nitrogen source growth experiments auxotrophic markers were complemented with plasmid encoded gene copies [pRS313-HIS3; pRS314-TRP1; pRS315-LEU2 (16), and pRSK-URA3 LYS2 (a gift from M. Stark, University of Dundee)]. Wild-type or mutant EAP1 plasmids: pJF3896 (HA-EAP1 LEU2 CEN) and pBMK492 (HA-eap1-Y109A,L114A LEU2 CEN) replaced pRS315 where indicated (10). Cells were grown on SD medium containing a single indicated nitrogen source at the following concentrations: 0.1% w/v ammonium sulphate, allantion, 0.02% w/v l-isoleucine, l-leucine, l-tryptophan, l-valine, l-asparagine, l-aspartic acid, l-glutamine, l-glutamic acid, l-serine, 0.01% w/v ammonium chloride, 0.01M urea. All media were adjusted to pH 5.3. Synthetic complete (SC) contained 0.5% w/v ammonium sulphate and 0.2% w/v of a mix of all 20 amino acids, inositol, para-aminobenzoic acid, uracil, adenine as described earlier (17). Agar plates (2% w/v bacto agar) were incubated at 30°C for 2–5 days. Doubling times (Td) in liquid media were determined using 96-well microplate cultures grown in a temperature-controlled absorbance reader with shaking (BMG Labtech). A600 measurements were recorded every 5 min for 20 h. A previously described MS Excel macro was adapted and used to automate Td determinations (18).
PUF-TAP tagged strains were a kind gift from Gerber and Herschlag (19). CAF20-Myc and EAP1-Myc tagged versions of each PUF-TAP strain were made using pYM4 and PCR to integrate C-terminal 3Myc-KanMX6 marker as described earlier (20). Similarly, pYM19 (9Myc-HIS3MX6) and pYM13 (TAP-KanMX6) was used to create Myc-tagged and TAP-tagged versions, respectively, of CAF20, EAP1, PUF2, PUF3 and PUF5 in BY4742, Y17334 and Y17036 (20). PCR was used to verify genomic integrations.
Polysome fractionation, RNA preparation and microarray analysis
Cell extracts, polyribosome fractionation, RNA preparation and array bioinformatics analysis were done exactly as described earlier (21). Complete data sets are available at ArrayExpress (www.ebi.ac.uk/microarray/ using accession numbers caf20Δ: E-MEXP-1308; eap1Δ: E-MEXP-1309).
Quantitative RT–PCR analysis
RNA analysis by RT–PCR was carried out using the MyIQ single-colour real-time PCR detection system and IQ SYBR Green Supermix (BioRad Laboratories) as described earlier (21,22). Signals were quantified relative to actin mRNA control.
TAP affinity chromatography
PUF-TAP or 4E-BP-TAP and control strains were grown in YPD medium to a concentration of 6 × 106 cells per ml. Cultures were harvested by centrifugation (5500g, 10 min, 4°C), washed in ice cold 1 mM phenylmethlysulphonyl fluoride (PMSF) and resuspended in two cell volumes of Buffer A [20 mM Tris–HCl (pH 8.0)], 140 mM KCl, 1.8 mM MgCl2, 0.1% Nonidet P-40 (NP-40), 0.5 mM dithiothreitol (DTT), 1 mM PMSF, 1× complete EDTA-free protease inhibitor cocktail tablet [Roche], 100 U/ml RNasin [Promega]. Cell suspension was frozen and ground under liquid nitrogen and stored at −80°C. Crude cell lysates were cleared by centrifugation (5500g for 20 min, 4°C). Extracts (2 mg) were incubated with 400 µl (50% [v/v]) IgG Sepharose beads (GE Healthcare) for 2 h at 4°C and affinity purified as described earlier (19). PUF proteins were released from the IgG beads by heating at 65°C for 3 min in 2× non-reducing Protein Loading Buffer [62.5 mM Tris–HCl (pH 6.8), 2% SDS, 10% (v/v) glycerol, 0.002% (w/v) bromphenol blue].
Immunoblotting
Whole cell extracts; Cells were harvested and resuspended in three times wet pellet volume of Buffer B [50 mM Tris–HCl pH 7.5, 100 mM NaCl 1 mM EDTA 1 mM PMSF and 1× Protease Inhibitors (Roche)] and ground in liquid Nitrogen. Samples were assayed for protein concentration (Bradford) and diluted to 1 ug protein per ml. TAP proteins were resolved by SDS–PAGE electrophoresis followed by transfer to a nitrocellulose membrane. Immunoblotting detection used horseradish peroxidase (HRP) conjugated primary antibodies to Protein A (Abcam) and c-Myc (9E10, Santa Cruz Biotechnology and 4A6, Millipore), and chemiluminescent detection (Pierce Biotechnology). Other primary antibodies used were caf20p (10), Gcd11p (23), Cic1p (24), Lsm8p (25), Taf3p, Taf7p (26), Sec9p (27), Pub1p (28), Ade2p and Arp2p (Santa Cruz Biotechnology) and were detected using appropriate HRP-conjugated secondary antibodies as indicated earlier.
RESULTS AND DISCUSSION
Microarray approach to determine roles for 4E-BPs
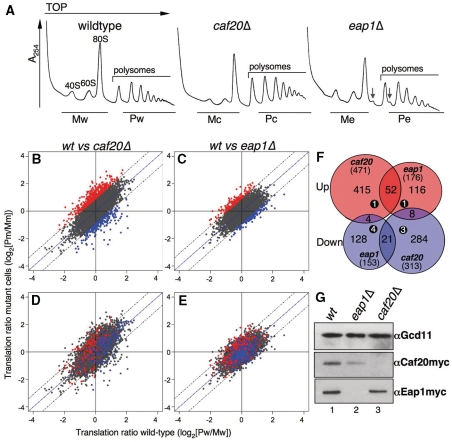
To determine whether translation of specific mRNAs was altered by elimination of Caf20p or Eap1p under steady state growth conditions, we used a microarray-based approach so that the translation state of all mRNAs could be surveyed simultaneously. We used isogenic strains produced by the systematic yeast gene deletion project for this study. Our initial growth characterization revealed that the caf20Δ and wild-type strains behaved identically in both rich and minimal medium, while the eap1Δ strain typically exhibited an initial growth lag when recovering from stationary phase, but then proliferated at a rate indistinguishable from the wild-type strain (data not shown). Polyribosome profile analysis revealed a minor halfmer phenotype for the eap1Δ strain, but no obvious global defect in the caf20Δ strain (Figure 1A). For array analysis the following samples were collected: (i) total RNA samples to control for changes in transcript abundance and (ii) polysome gradient fractionated samples. Fractionated samples were split into monosomal and polysomal fractions as indicated in Figure 1A. Each RNA sample was processed into complementary RNA and hybridized to Affymetrix yeast S98 gene chip arrays using standard protocols (21,22). All analyses were performed in duplicate and the resulting data were analysed and compared using standard bioinformatics procedures (as outlined in the Supplementary Data). MA plots and statistical analysis of variance tests (Supplementary Figure S1A, B, D, E, G and H and data not shown) indicated that there was only modest dispersal between biological replicate samples, confirming that the arrays are reliable reporters of relative transcript abundance in our samples. As anticipated, a greater dispersal of data points was seen between wild-type and mutant samples (Supplementary Figure S1C, F and I).
Figure 1.
Widespread translational changes are associated with each 4E-BP deletion. (A) A254 traces showing polysome profiles from exponential phase cultures for the strains used. RNA fractions collected were pooled into monosomal (M) or polysomal (P) fractions from wild-type (w) caf20Δ (c) and eap1Δ (e) cells for translational-profiling array experiments. (downward arrow) “halfmer” peaks. (B) Graphical representation of global changes in translation accompanying deletion of CAF20 or (C) EAP1. Mean polysome-to-monosome ratio intensities for each probeset from mutant (y-axis) is plotted against the wild-type (x-axis). Points above or below a log2 = 0.9 cut-off are considered significant changes (coloured red and blue, respectively). Plots (D) and (E) as B and C except spots representing translationally regulated genes (red and blue) from the eap1Δ plot (C) are shown on the caf20Δ plot (D), and regulated genes from the caf20Δ plot (B) on the eap1Δ plot (E). (F) Venn-style diagram showing the number of genes altered (in parenthesis) and the overlap between data sets. Numbers in black filled circles represent ‘potentiated’ genes (see text). (G) Immunoblots of Caf20p-Myc and Eap1-Myc levels in strains indicated. eIF2γ (GCD11) loading control from Caf20-Myc tagged cells is also shown.
4E-BPs have minimal impact on mRNA abundance
Under normal cellular conditions, changes in mRNA transcript abundance reflect changes in both mRNA synthesis and decay. In this study, overall transcript abundance changes were relatively minor. caf20Δ alters transcript abundance >2-fold for 100 genes (29 up and 71 down) and eap1Δ 99 genes (56 up and 43 down). A complete listing of significantly altered genes is shown in Supplementary Tables S1–S4. As expected the transcript signals for the deleted gene were down-regulated most in each data set. The small number of genes affected is consistent with 4E-BPs having no widespread direct transcriptional role, instead the changes observed could be an indirect consequence of altered translation of transcription factors (see below). There was also no evidence of ‘potentiation’. This is a term used to describe co-ordinated transcription and translation responses observed during the reprogramming of gene expression to adapt to some cellular stresses (21,29). Fewer than 10 genes were both transcriptionally and translationally up-/down-regulated across both experiments (Figure 1F; numbers in black filled circles).
Overview of translational controls
We measured polysome:monosome [P/M] ratios for each transcript (probe set), termed a ‘translational state’, calculated as log2[P/M] ratio for each mutant and wild-type control from mean normalized sample intensities. A ‘change in translational state’ was determined by comparing the ratios from mutant cells (m) to that from wild-type cells (w). These analyses are represented as scatter plots in Figure 1B and C and numerically (Supplementary Tables S5–S8). The plots reveal that the majority of mRNA probe sets fall on or around the diagonal, indicating that the translation of most genes is not affected by deletion of either gene. In line with previous studies using similar techniques, we applied a significance cut off of log2 = ±0.9 (i.e. >1.86-fold) change (21,29). This revealed that caf20Δ altered the translation of 784 mRNAs (471 up- and 313 down-regulated), while eap1Δ affected 329 mRNAs (176 up and 153 down) (Figure 1F). The fact that more mRNAs are Caf20p targets (784/1028 = 76%) than Eap1p agrees with our observations that the relative abundance of Myc tagged Caf20p is 2.8 ± 0.63-fold greater than Eap1 (data not shown). These data are consistent with Caf20p regulating translation of a greater number of mRNAs than Eap1p.
Eap1p acts as a translational repressor for mRNAs that are poorly associated with polysomes under normal growth conditions. This observation is consistent with the idea that Eap1p plays a role repressing translation of a subset of genes during normal growth. This is illustrated graphically in Figure 1D, where the spot location is derived from the caf20Δ experiment, but spots are coloured according to the eap1Δ data. It is evident that most eap1Δ up-regulated genes (red spots) are to the left half of the plot, while the down-regulated ones (blue) are on the right. Therefore the majority of genes which are up-regulated in eap1Δ cells (red spots in Figure 1C and D) are among those transcripts which are poorly associated with polysomes in wild-type cells (81% or 142/176 log2[Pw/Mw]<0). A reciprocal arrangement holds for down-regulated genes (14% or 21/153 log2[Pw/Mw]<0). This is much less evident for Caf20p regulated transcripts. Figure 1E illustrates where the caf20Δ regulated genes are highlighted on the eap1Δ plot. Although >50% of the genes up-regulated by caf20Δ are on the left in Figure 1E, the bias is less pronounced, suggesting distinct roles for Caf20p and Eap1p in repressing translation of genes under normal growth conditions.
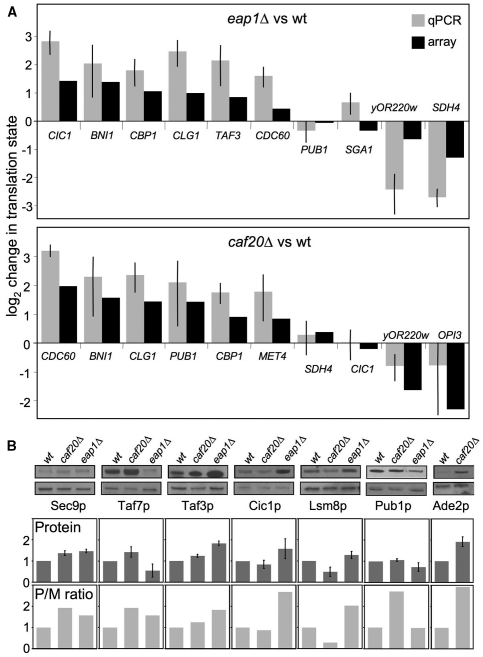
As an independent confirmation that the arrays accurately reported translation state changes, quantitative RT–PCR analysis was performed on polysome fractionated RNA samples with oligonucleotide primers specific for a subset of genes (Figure 2A). Genes were selected to cover a range of up- and down-regulated mRNAs and also some unregulated mRNAs. Despite differences between these techniques, a good correlation was observed between the array and RT–PCR approaches.
Figure 2.
qPCR and immunoblotting confirms translational profiling. (A) Quantitative reverse-transcription PCR analysis of translation state for qPCR (light grey bars ±SD, n = 3) and array data (black bars) for indicated transcripts (plotted on a log2 scale so that reduced polysome association in mutant cells is shown as a negative number). (B) Top: total protein levels of Sec9p, Taf7p, Taf3p, Cic1p, Lsm8p, Pub1p and Ade2p were analysed by immmunoblot analysis in indicated yeast strains. Experiments were done in triplicate. Loading control Arp2p (Actin Related Protein 2) is shown beneath each. Middle: densitometry analysis for each protein relative to Arp2p is beneath each panel (dark grey bars). Bottom: polysome/monosome [P/M] ratios from translational profiling (light grey bars).
The 4E-BPs interact with eIF4E via a common motif, and eIF4E itself binds the cap structure common to all mRNAs. Therefore it seems unlikely that these features alone impart the observed mRNA specific translational controls. Therefore our data suggest that there must be other factors, which work in concert with the yeast 4E-BPs to mediate control of translation (see below). It seemed likely to us that these other factors may bind to specific sequences within each mRNA and contribute to the observed control. As such, it was important to verify whether protein levels vary as anticipated by the translational profiling data. Almost all efficient strategies for epitope-tagging yeast genes involve disrupting either the 3′-UTR or replacing elements 5′ of the ORF. The 5′- and 3′-UTRs are common sites used by RNA-binding factors to mediate mRNA specific controls in many eukaryotes, so we reasoned that using a tagging approach was not appropriate here. Instead we searched the literature to identify available protein specific antibodies. We screened the antibodies available and identified several that functioned with our whole-cell extracts. The antibodies were used and compared with an actin loading control that did not vary between strains (Figure 2B). Antibody signals varied between the strains tested in a manner that mirrored the trends seen in polysome/monosome ratios from our arrays. Thus Taf7p levels varied and were increased in caf20Δ and decreased in eap1Δ while both Cic1p and Lsm8p were more abundant in eap1Δ cells. Thus our arrays do appear to reflect the translational controls—at least for those candidates independently verified by RT–PCR and/or western blotting. This includes CIC1 and TAF3, which are both up-regulated translationally by eap1Δ in our microarrays, by RT–PCR and western blotting. An exception is Pub1p where RT–PCR confirmed the microarray data suggesting enhanced PUB1 polysome association in caf20Δ cells, but steady state protein levels were not significantly altered in this strain. These data suggest additional factors act to control Pub1p levels.
A significant overlap between caf20 and eap1 data sets
Although the majority of mRNA targets differ between the array experiments, we observed a statistically significant overlap between the mRNAs similarly up-regulated by both eap1Δ and caf20Δ (52 genes P = 9.8 × 10−23; Figure 1D–F, Supplementary Table S9). In contrast, there are very few reciprocally regulated genes (e.g. only four genes up-regulated by caf20Δ are down-regulated by eap1Δ; Figure 1F). During the course of our analyses, immunoblotting with anti-Caf20p anti-serum revealed that the steady state expression levels of Caf20p are reduced in eap1Δ cells (data not shown). As this observation may explain the observed overlap in translational control between these factors, we examined carefully the expression levels of both 4E-BPs. Myc-epitope tags were added by homologous recombination to the C-termini of CAF20 and EAP1 in separate strains. Immunoblotting confirmed that Caf20p expression was reduced ∼50% in eap1Δ cells, while Eap1p levels were maintained in caf20Δ cells (Figure 1G). Therefore the observed reduced Caf20p levels may cause lower affinity Caf20p target mRNAs to lose normal translational control in eap1Δ cells and contribute to the overlap in array data sets.
4E-BPs repress genes controlling gene expression
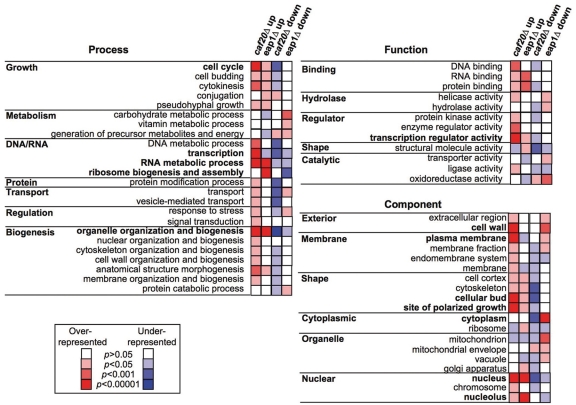
To gain insight into the functions of the translationally regulated transcripts, each regulated gene was placed into a functional class based upon classifications included within the GOslim mapper (Saccharomyces Genome Database) and annotations at the Yeast Proteome Database (Figure 3 and Supplementary Tables S1–S8 and S10). This analysis revealed that many transcripts with enhanced [P/M] ratios are themselves involved in gene expression. Deletion of either 4E-BP enhances translation of many transcription factors and chromatin remodelling enzymes. These changes did not cause large changes in transcript abundance as arrays of total mRNA showed that few genes are altered by >2-fold (Supplementary Tables S1–S4). We assume that these cells have adapted to the loss of each factor and that the changes to the translation profile observed have normalized overall transcription as far as possible under the standard growth conditions examined. RNA metabolism is another abundant class of up-regulated genes for both 4E-BPs. In addition, Eap1p represses translation of many ribosome biogenesis factors involved in synthesis/assembly of both ribosomal subunits. This may account for the apparent modest halfmer phenotype observed in eap1Δ cells (arrowed in Figure 1A). Overall these data suggest that regulation of genes involved in RNA metabolism is an important function of these 4E-BPs and that each protein regulates specific targets involved in the same broad pathways/processes.
Figure 3.
Functional classification of regulated mRNAs. Yeast ‘GO Slim’ summary of significantly enriched and under-represented gene Ontology classes calculated using the hypergeometric distribution (performed at www.yeastgenome.org), see Supplementary Table S10 for more details including all calculated P-values. Bold text indicates most over-represented classes (P < 0.00001) in one or more experiment.
Caf20p represses translation of CLN genes and those important for polarised growth
The cell cycle, budding and site of polarised growth GO Slim categories are particularly over-represented within the translationally up-regulated caf20Δ mRNAs. Cyclins CLN1, CLN2, CLN3, CLB1, and CLB4 were among many cell cycle associated mRNAs. As indicated in the introduction, one of the best-characterized examples of a gene regulated by eIF4E availability in yeast is the G1 cyclin CLN3. A temperature sensitive mutation in yeast eIF4E (cdc33-1) has a G1-S phase cell cycle defect that is overcome by elevated expression of CLN3 (14). We interpret our array data as confirming the importance of translational controls for regulating cyclin expression and further suggest that an eIF4E-Caf20p interaction is important for translational repression of CLN3.
In metazoa correct intracellular localization of specific transcripts is important for both early developmental decisions and memory functions in neurons. 4E-BPs provide a mechanism to repress translation of delocalized mRNAs e.g. Drosophila 4E-BP Cup acts with Bruno to repress translation of oskar mRNA (30). Polarised growth in budding yeast requires asymmetric localization of components to the growing bud. Several mRNAs have also been found to be bud-localized. A survey of yeast genes uncovered 22-bud localized transcripts, of which 10 were asymmetrically localized to the bud (31). Intriguingly, six of these 10 genes (EGT2, IST2, MMR1, SRL1, TCB3 and TPO1) are translationally up-regulated in caf20Δ cells. GO Slim mapping analysis suggests that bud, cell wall, and site of polarized cell growth-localized proteins are among those enriched in the caf20Δ data set. Together, these observations suggest that Caf20p may play a role repressing translation of these mRNAs in the mother cell, but that localized transcripts could become translatable. Bud-localized activation of ASH1 mRNA translation has been studied directly and two alternative hypotheses have been proposed to account for its translational repression involving Khd1p, eIF4G and the kinase Yck1p (32) or involving eIF5B and Puf6p (33). Our data is consistent with and suggestive that the yeast 4E-BPs could play a role in spatial regulation of translation for specific transcripts in yeast but elucidation of molecular mechanisms is beyond the scope of this article.
Nitrogen and amino acid metabolism defects
Filamentous or pseudohyphal growth on poor nitrogen sources in Σ1278b yeast strains requires reprogramming of cellular growth. We recently showed that each 4E-BP was required for this response (10) highlighting a role for translational control in the response to nitrogen limitation. Others found that both CLN1 and STE12 translation is induced following induction of filamentous growth on poor nitrogen sources in Σ1278b yeast (34). Our arrays were performed on S288c derived cells that do not undergo this developmental switch because they lack the FLO8 transcription factor required for induction of some developmentally regulated genes (35), and on cells grown in a nitrogen sufficient medium. Nevertheless, both CLN1 and STE12 are translationally up-regulated in our arrays, (CLN1 by both factor deletions and STE12 by eap1Δ) suggesting that 4E-BP mediated repression of certain filamentous growth genes may occur during growth on rich media. Our array experiments also predicted that both caf20Δ and eap1Δ cells would have unbalanced nitrogen and amino acid metabolism gene expression due to altered translation of ammonia and amino acid transporters and metabolic enzymes. A large number of genes in this group are down-regulated in eap1Δ cells (Supplementary Table S8).
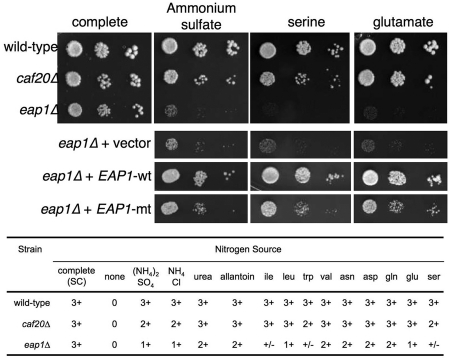
To assess if these observations had phenotypic consequences, auxotrophic markers in each strain were complemented and cells were plated on a variety of media and growth scored. A summary of the results is shown in Figure 4. When grown on standard complete media containing ammonium sulphate and a mixture of all amino acids all cells grew well. The eap1Δ cells in particular grew very poorly on less complex media containing sole nitrogen sources. Measurements of maximum doubling times in liquid cultures confirmed that caf20Δ also showed defects with some nitrogen sources albeit to a lesser extent than seen with eap1Δ (Table 1). As the glutamate/aspartate transporter DIP5 (36) transcript levels were 2-fold reduced in eap1Δ cells (Supplementary Table S4) it was important to ascertain whether these phenotypes were dependent upon translational controls. We found that the wild-type EAP1 plasmid rescued growth on alternate nitrogen sources, while a plasmid bearing missense mutations that disrupt the eIF4E interaction motif in Eap1p (EAP1-mt in Figure 4) shown to eliminate eIF4E binding (10) was significantly impaired in this response, suggesting that eIF4E-binding contributes to this phenotype. These data show that 4E-BP-mediated translational controls are important for normal amino acid metabolism in yeast in addition to their role in pseudohyphal development in Σ1278b yeast cells (10).
Figure 4.
eap1Δ and caf20Δ cells have nitrogen utilization defects. Cells complemented for auxotrophic markers were grown in SC complete medium to A600 = 0.6 washed and diluted to A600 = 0.1, then 10-fold serially diluted and spotted (3 µl) onto the indicated media. Growth was scored on a scale of 0 (none), +/− (minimal) to 3+ (wild-type).
Table 1.
Maximum doubling times of strains in liquid medium with indicated nitrogen sources
| Strain | Medium |
||
|---|---|---|---|
| Complete | Serine | Glutamate | |
| Wild-type | 118 ± 2a | 148 ± 20 | 153 ± 10 |
| Caf20Δ | 124 ± 5 | 182 ± 20 | 189 + 21 |
| eap1Δ | 140 ± 15 | 212 ± 19 | 233 ± 35 |
aMean maximum doubling times in min ± SD (n = 9 cultures) for cultures grown in liquid media equivalent to solid media shown in Figure 4.
Overlap between 4E-BP regulated and PUF-bound mRNAs
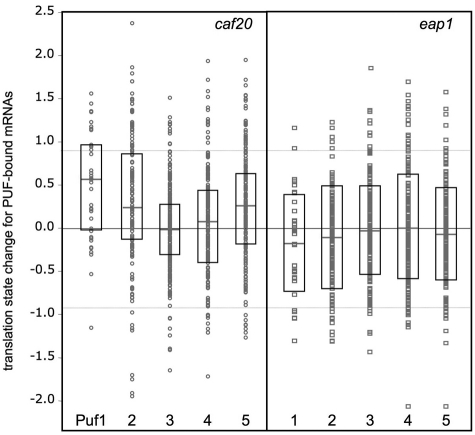
One paradigm for the action of 4E-BPs is that translational repression is mediated in part via protein–protein interactions between proteins bound to the 5′-cap and those attached to the 3′-UTR of specific mRNAs (2). As our array data reveal that Eap1p and Caf20p largely regulate translation of distinct genes (Figure 1), it seemed likely that additional mRNA-binding factors may contribute to the control mechanism. Several studies have used microarrays to identify RNAs associated with specific RNA-binding proteins following immune-precipitation of epitope tagged proteins (RIp-chip). We used statistical comparisons to assess if there were any significant overlaps between the identities of our 4E-BP regulated transcripts and transcripts identified by others as specifically associated with a range of mRNA-binding proteins in different RIp-chip experiments (19,37–39). The question addressed was: are genes identified as enriched with specific mRNA binding proteins also those identified as translationally controlled by the 4E-BPs or are the associations random? This analysis considers each transcript equally and does not account for mRNA abundance or any biases in the data sets. We found that there was a potentially significant overlap between genes up-regulated by caf20Δ (i.e. translation state change >0.9) and mRNAs associated with Puf1p, Puf2p, and Puf5p (P ≤ 0.0001) (Supplementary Table S9). The full distribution is shown graphically in Figure 5, where the translational state change for each PUF-bound mRNA is shown for the caf20Δ data. This additionally shows that the mean translational states for Puf1p, Puf2p and Puf5p are each shifted positively away from zero, while the means for Puf3p and Puf4p are not. As Gerber and colleagues found fewer mRNAs associated with Puf1p and Puf2p than Puf3-5p, the apparent enrichment in our caf20Δ up-regulated genes associated with Puf5p appeared most significant. Overlaps between genes translationally up-regulated by eap1Δ and bound by PUF proteins were also seen but only Puf4p showed a statistically significant enrichment (Supplementary Table S9). However, when plotted graphically (Figure 5) the mean distributions were not shifted >0, suggesting that only a few Puf-associated transcripts are potentially translationally regulated by Eap1p.
Figure 5.
Translation state change for Puf1-5p associated mRNAs in caf20Δ and eap1Δ cells. A comparison of the change in translation state for mRNAs identified by Gerber and colleagues as bound to Puf1p (35 mRNAs), Puf2p (142), Puf3p (219), Puf4p (202) and Puf5p (203 mRNAs) is plotted (open circle, open rectangle) for caf20Δ and eap1Δ, respectively. The median (grey horizontal bar) and upper and lower quartiles are indicated by the box-plot. Statistical analyses are shown in Supplementary Table S9.
A more recent RIp-chip study has identified mRNAs associated with a wide range of RNA-binding proteins (39). Equivalent comparative statistical analyses revealed potential overlap between caf20Δ up-regulated genes and mRNA-binding proteins including translational regulators (Khd1p, Mrn1p, Scp160p), factors involved in nuclear export of mRNA (Gbp2 and Nab2p) as well as Pub1p and Ssd1p. Similar comparisons with our eap1Δ data revealed overlaps with Nab2, Nrd1p, and the cytoplasmic polyA binding protein Pab1p (Supplementary Table S9). In addition, we found other relationships between the data sets, including correlations between mRNAs bound by specific proteins and those translationally down-regulated by the 4E-BP deletions. For example Mrn1p and Pub1p-bound mRNAs are both enriched among mRNAs down-regulated by eap1Δ and up-regulated by caf20Δ. We also noted that there were examples where fewer than expected mRNAs in common between specific data sets (Supplementary Table S9). While not conclusive, these comparisons suggest that different proteins binding to specific mRNAs could combine with 4E-BPs to regulate their translation in yeast in a manner similar to that proposed for multicellular eukaryotes. Because there are several yeast PUF proteins, we decided to examine experimentally whether there were any interactions between the 4E-BPs and the PUF proteins that could help to explain the polysome microarray data.
4E-BPs physically associate with specific PUFs
PUF proteins are found in diverse eukaryotes (as are 4E-BPs) and have been implicated in post-transcriptional control of gene expression in early development, germ cells and neuronal cells (40,41). PUFs have been implicated both in the control of mRNA stability and translational repression (40). Unlike the 4E-BPs, PUFs are sequence specific RNA-binding proteins that contain eight repeats of an ∼40 residue motif, the PUF motif (42). PUF proteins often associate with 3′-UTRs (19) where they can form complexes with other proteins (43). In yeast, Pufs 1 and 2 preferentially bind mRNAs encoding membrane-associated proteins, Puf4p rRNA-processing factor mRNAs and Puf5p chromatin modifier mRNAs (19). Puf6p was found to bind the ASH1 3′-UTR and repress its translation in mother cells. ASH1 mRNA is normally localized to the distal bud tip and both mRNA localization and translation were affected in puf6Δ cells (44). Most studies to date point to a function in mRNA stability, with each PUF protein having distinct targets, although few mRNA targets have been studied in any detail. Puf3p preferentially binds mitochondrial-localized transcripts e.g. COX17 to stimulate the latter’s degradation (45,46). In another study, the stability of 38 potential PUF-target mRNAs was assessed and only two of these (HXK1 and TIF1) were found to have altered mRNA half-lives in pufΔ strains under the conditions studied (47). One favourable explanation for a lack of effects on mRNA stability for certain targets is that PUF protein binding can mediate translational control. In support of this idea, the Wickens group has shown that both Puf4p and Puf5p bind HO mRNA and regulate post-transcriptional processes including mRNA stability and translational regulation in vitro (48,49). Intriguingly, we found that HO polysome association is significantly up-regulated in caf20Δ cells (Supplementary Table S5). These observations combined with our statistical analysis suggested to us that specific Caf20p-PUF interactions may determine in part which mRNAs are translationally regulated by 4E-BPs.
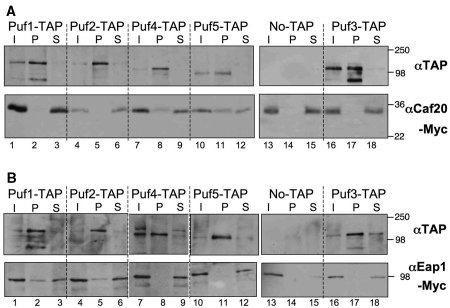
To determine if any of the statistical observations reflected real interactions, we obtained TAP-tagged PUF strains and incorporated tandem Myc epitopes at the C-terminus of CAF20 in each. Affinity chromatography was performed on each TAP strain and an untagged control in the presence of RNase inhibitors but in the absence of any crosslinker, as described (19) and the reactions were probed with anti-TAP and anti-Myc sera. Figure 6A shows that in each case the TAP-tagged protein was efficiently captured by this procedure. We found that a portion of Caf20p was associated with TAP-tagged Puf5p and to a lesser extent Puf4p, while none was associated with Pufs1-3 or the untagged control. These associations required RNA because they were not identified when RNase inhibitors were omitted from the buffers used (data not shown). We made an equivalent set of strains bearing C-terminally Myc-tagged EAP1 and repeated the TAP-affinity chromatography experiments to identify whether any Eap1p-PUF complexes formed (Figure 6B). Remarkably these experiments revealed a complementary set of interactions, as a fraction of Eap1p was associated with Puf1p and Puf2p, but not the other PUFs tested. As each set of experiments identified different protein complexes it provides evidence that they are not artifactual. Because the Protein A component of the TAP tag bound our Myc antibodies, we were not able to complete reciprocal precipitation experiments. Instead, we made new strains bearing Myc-tagged PUF proteins (PUF2, PUF3 and PUF5) and TAP tagged 4E-BPs and repeated the TAP-affinity chromatography experiments. In agreement with our original experiment, a small fraction of Puf5p-Myc was associated with Caf20p-TAP and Puf2p-Myc with Eap1p-TAP (Supplementary Figure S2). Importantly, no associations were seen between Puf3p-Myc and either 4E-BP-TAP protein or between Myc-tagged proteins and the protein A resin in control strains lacking a TAP tag, although a tiny fraction of Puf2p-Myc was additionally found associated with Caf20p-TAP.
Figure 6.
Specific PUF protein 4E-BP complexes are isolated from cells. Affinity chromatography purifying indicated PUF–TAP complexes from cells bearing the indicated TAP-tagged PUF protein or an untagged control and Myc tagged (A) Caf20p or (B) Eap1p. Upper panels probed with anti-protein A (αTAP) and lower with anti-Myc antibodies. I, input [10 µg total protein, P = Pellet (0.5mg) S = supernatant (10 µg unbound fraction)]. The immunoblots are representative of three independent experiments.
These experimental data do not entirely agree with the in silico predictions. The predicted Caf20p-Puf5p interaction was found. However the Caf20p-Puf4p and Eap1p-Puf1/2p interactions we identified were not predicted by our statistical comparisons. There are several possible explanations for this including the fact that our statistical treatment did not take into consideration mRNA abundances or the relative affinities of the interactions, as these are not known. Our experiments represent a ‘snap-shot’ of cells from non-stressed log cultures. Under different growth stresses, phases or synchronous states the associations may differ. However our data has shown that there is value using this type of approach to devise hypotheses to test experimentally. Importantly, when taken together, these experiments indicate that each yeast 4E-BP can form mRNA–protein complexes with specific PUF-bound mRNAs.
In summary, by using a translational profiling approach, we have identified that translation of a significant fraction of yeast mRNAs is regulated by the yeast eIF4E-BPs Caf20p and Eap1p. Many of the regulated transcripts are poorly translated in wild-type cells during exponential growth and these mRNAs are enriched in function in gene expression, cell cycle or transport. In addition to explore how mRNA specificity is achieved, we provide direct evidence that each yeast 4E-BP can interact with specific PUF proteins. As PUF proteins bind RNAs with sequence specificity, this could present one mechanism by which the yeast 4E-BPs target individual mRNAs. By analogy with higher eukaryotes, it is likely that these complexes contain additional components that remain to be determined and that other 3′-UTR-binding protein/4E-BP combinations also await identification. These studies, therefore, provide the first evidence that the yeast 4E-BPs regulate translation of hundreds of mRNAs and that by further study it will be possible to uncover molecular details in yeast that inform studies in higher eukaryotes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biotechnology and Biological Research Council (UK) (grants G17520, BBD000106 and BBG012571). Funding for open access charge: BBSRC grant BBG012571.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Microarray facility at the University of Manchester (especially A. Hayes and L. Wardleworth) for providing technical support for Affymetrix arrays. A. Gerber (Zurich) and D. Herschlag (Stanford) for providing the PUF-TAP strains. M. Stark (Dundee) for plasmid pRSK. A. Blomberg (Göteborg) for supplying the MS Excel macro and D. Burgess (Manchester) for macro adaptation and growth assay development. We especially thank those who supplied us antibodies to complete our target verification analyses including M. Swanson (Florida), Sandra Wolin (Yale), D. Wolf (Stuttgart), T. Kokubo (Yokohama City) and James McNew (Rice) whose reagents were used for immunoblots in Figure 2.
REFERENCES
- 1.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 3.Mothe-Satney I, Yang D, Fadden P, Haystead TA, Lawrence JC., Jr Multiple mechanisms control phosphorylation of PHAS-I in five (S/T)P sites that govern translational repression. Mol. Cell. Biol. 2000;20:3558–3567. doi: 10.1128/mcb.20.10.3558-3567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson B, Wickens M, Kimble J. Translational control in development. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor: CHSL Press; 2007. pp. 507–544. [Google Scholar]
- 6.Altmann M, Schmitz N, Berset C, Trachsel H. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 1997;16:1114–1121. doi: 10.1093/emboj/16.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosentino GP, Schmelzle T, Haghighat A, Helliwell SB, Hall MN, Sonenberg N. Eap1p, a novel eukaryotic translation initiation factor 4E-associated protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:4604–4613. doi: 10.1128/mcb.20.13.4604-4613.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Cruz J, Iost I, Kressler D, Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mascarenhas C, Edwards-Ingram LC, Zeef L, Shenton D, Ashe MP, Grant CM. Gcn4 Is Required for the Response to Peroxide Stress in the Yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2008;19:2995–3007. doi: 10.1091/mbc.E07-11-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibrahimo S, Holmes LE, Ashe MP. Regulation of translation initiation by the yeast eIF4E binding proteins is required for the pseudohyphal response. Yeast. 2006;23:1075–1088. doi: 10.1002/yea.1415. [DOI] [PubMed] [Google Scholar]
- 11.Meier KD, Deloche O, Kajiwara K, Funato K, Riezman H. Sphingoid base is required for translation initiation during heat stress in Saccharomyces cerevisiae. Mol. Biol. Cell. 2006;17:1164–1175. doi: 10.1091/mbc.E05-11-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deloche O, de la Cruz J, Kressler D, Doere M, Linder P. A membrane transport defect leads to a rapid attenuation of translation initiation in Saccharomyces cerevisiae. Mol. Cell. 2004;13:357–366. doi: 10.1016/s1097-2765(04)00008-5. [DOI] [PubMed] [Google Scholar]
- 13.Polymenis M, Schmidt EV. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danaie P, Altmann M, Hall MN, Trachsel H, Helliwell SB. CLN3 expression is sufficient to restore G1-to-S-phase progression in Saccharomyces cerevisiae mutants defective in translation initiation factor eIF4E. Biochem. J. 1999;340:135–141. [PMC free article] [PubMed] [Google Scholar]
- 15.Sezen B, Seedorf M, Schiebel E. The SESA network links duplication of the yeast centrosome with the protein translation machinery. Genes Dev. 2009;23:1559–1570. doi: 10.1101/gad.524209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast genetics: A Cold Spring Harbor Laboratory Course Manual. 1997 edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 18.Warringer J, Blomberg A. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast. 2003;20:53–67. doi: 10.1002/yea.931. [DOI] [PubMed] [Google Scholar]
- 19.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 21.Smirnova JB, Selley JN, Sanchez-Cabo F, Carroll K, Eddy AA, McCarthy JE, Hubbard SJ, Pavitt GD, Grant CM, Ashe MP. Global gene expression profiling reveals widespread yet distinctive translational responses to different eukaryotic translation initiation factor 2B-targeting stress pathways. Mol. Cell. Biol. 2005;25:9340–9349. doi: 10.1128/MCB.25.21.9340-9349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shenton D, Smirnova JB, Selley JN, Carroll K, Hubbard SJ, Pavitt GD, Ashe MP, Grant CM. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J. Biol. Chem. 2006;281:29011–29021. doi: 10.1074/jbc.M601545200. [DOI] [PubMed] [Google Scholar]
- 23.Mohammad-Qureshi SS, Haddad R, Hemingway EJ, Richardson JP, Pavitt GD. Critical contacts between the eukaryotic initiation factor 2B (eIF2B) catalytic domain and both eIF2beta and -2gamma mediate guanine nucleotide exchange. Mol. Cell. Biol. 2007;27:5225–5234. doi: 10.1128/MCB.00495-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jager S, Strayle J, Heinemeyer W, Wolf DH. Cic1, an adaptor protein specifically linking the 26S proteasome to its substrate, the SCF component Cdc4. EMBO J. 2001;20:4423–4431. doi: 10.1093/emboj/20.16.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pannone BK, Kim SD, Noe DA, Wolin SL. Multiple functional interactions between components of the Lsm2-Lsm8 complex, U6 snRNA, and the yeast La protein. Genetics. 2001;158:187–196. doi: 10.1093/genetics/158.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahata S, Kasahara K, Kawaichi M, Kokubo T. Autonomous function of the amino-terminal inhibitory domain of TAF1 in transcriptional regulation. Mol. Cell. Biol. 2004;24:3089–3099. doi: 10.1128/MCB.24.8.3089-3099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Komen JS, Bai X, Scott BL, McNew JA. An intramolecular t-SNARE complex functions in vivo without the syntaxin NH2-terminal regulatory domain. J. Cell Biol. 2006;172:295–307. doi: 10.1083/jcb.200507138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson JT, Paddy MR, Swanson MS. PUB1 is a major nuclear and cytoplasmic polyadenylated RNA-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:6102–6113. doi: 10.1128/mcb.13.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preiss T, Baron-Benhamou J, Ansorge W, Hentze MW. Homodirectional changes in transcriptome composition and mRNA translation induced by rapamycin and heat shock. Nat. Struct. Biol. 2003;10:1039–1047. doi: 10.1038/nsb1015. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev. Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- 31.Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl Acad. Sci. USA. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paquin N, Menade M, Poirier G, Donato D, Drouet E, Chartrand P. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol. Cell. 2007;26:795–809. doi: 10.1016/j.molcel.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 33.Deng Y, Singer RH, Gu W. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev. 2008;22:1037–1050. doi: 10.1101/gad.1611308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park YU, Hur H, Ka M, Kim J. Identification of translational regulation target genes during filamentous growth in Saccharomyces cerevisiae: regulatory role of Caf20 and Dhh1. Eukaryot. Cell. 2006;5:2120–2127. doi: 10.1128/EC.00121-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Regenberg B, Holmberg S, Olsen LD, Kielland-Brandt MC. Dip5p mediates high-affinity and high-capacity transport of L-glutamate and L-aspartate in Saccharomyces cerevisiae. Curr. Genet. 1998;33:171–177. doi: 10.1007/s002940050324. [DOI] [PubMed] [Google Scholar]
- 37.Duttagupta R, Tian B, Wilusz CJ, Khounh DT, Soteropoulos P, Ouyang M, Dougherty JP, Peltz SW. Global analysis of Pub1p targets reveals a coordinate control of gene expression through modulation of binding and stability. Mol. Cell. Biol. 2005;25:5499–5513. doi: 10.1128/MCB.25.13.5499-5513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inada M, Guthrie C. Identification of Lhp1p-associated RNAs by microarray analysis in Saccharomyces cerevisiae reveals association with coding and noncoding RNAs. Proc. Natl Acad. Sci. USA. 2004;101:434–439. doi: 10.1073/pnas.0307425100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 41.Mee CJ, Pym EC, Moffat KG, Baines RA. Regulation of neuronal excitability through pumilio-dependent control of a sodium channel gene. J. Neurosci. 2004;24:8695–8703. doi: 10.1523/JNEUROSCI.2282-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–289. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- 43.Edwards TA, Wilkinson BD, Wharton RP, Aggarwal AK. Model of the brain tumor-Pumilio translation repressor complex. Genes Dev. 2003;17:2508–2513. doi: 10.1101/gad.1119403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu W, Deng Y, Zenklusen D, Singer RH. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 2004;18:1452–1465. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackson JS, Jr, Houshmandi SS, Lopez Leban F, Olivas WM. Recruitment of the Puf3 protein to its mRNA target for regulation of mRNA decay in yeast. RNA. 2004;10:1625–1636. doi: 10.1261/rna.7270204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houshmandi SS, Olivas WM. Yeast Puf3 mutants reveal the complexity of Puf-RNA binding and identify a loop required for regulation of mRNA decay. RNA. 2005;11:1655–1666. doi: 10.1261/rna.2168505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ulbricht RJ, Olivas WM. Puf1p acts in combination with other yeast Puf proteins to control mRNA stability. RNA. 2008;14:246–262. doi: 10.1261/rna.847408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hook BA, Goldstrohm AC, Seay DJ, Wickens M. Two yeast PUF proteins negatively regulate a single mRNA. J. Biol. Chem. 2007;282:15430–15438. doi: 10.1074/jbc.M611253200. [DOI] [PubMed] [Google Scholar]
- 49.Chritton JJ, Wickens M. Translational repression by PUF proteins in vitro. RNA. 2010;16:1217–1225. doi: 10.1261/rna.2070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.