Abstract
Recent evidence points to homeotic proteins as actors in the crosstalk between development and DNA replication. The present work demonstrates that HOXC13, previously identified as a new member of human DNA replicative complexes, is a stable component of early replicating chromatin in living cells: it displays a slow nuclear dynamics due to its anchoring to the DNA minor groove via the arginine-5 residue of the homeodomain. HOXC13 binds in vivo to the lamin B2 origin in a cell-cycle-dependent manner consistent with origin function; the interaction maps with nucleotide precision within the replicative complex. HOXC13 displays in vitro affinity for other replicative complex proteins; it interacts also in vivo with the same proteins in a cell-cycle-dependent fashion. Chromatin-structure modifying treatments, disturbing origin function, reduce also HOXC13–origin interaction. The described interactions are not restricted to a single origin nor to a single homeotic protein (also HOXC10 binds the lamin B2 origin in vivo). Thus, HOX complexes probably contribute in a general, structure-dependent manner, to origin identification and assembly of replicative complexes thereon, in presence of specific chromatin configurations.
INTRODUCTION
The search for proteins participating in the regulation of human DNA replication recently led to the identification of homeotic proteins as members of the replication complexes (RCs) of several origins. The first indication came from a yeast mono-hybrid screen for human proteins with affinity for the lamin B2 origin (1). This study identified three proteins: HOXA13, HOXC10 and HOXC13, codified by the corresponding orthologs of the abdominal-B genes of Drosophila. The HOXC10 and HOXC13 proteins were shown to bind the same origin both in vivo (CAT assay) and in vitro. The HOXC10 protein is degraded early in mitosis by the ubiquitin pathway (2); mutations of two destruction boxes assure its permanence and delay the metaphase to anaphase transition. Fluorescent derivatives of HOXC13 co-localize only with early S replication foci (3) thanks to the homeodomain. Chromatin immuno-precipitation (ChIP) showed that HOXC13 binds the lamin B2 origin in asynchronous cultures but not in G0 cells, in agreement with the absence of a RC therein (4). The same behavior was observed also in the human origins close to the TOP1 and MCM4 genes. These studies suggest that the function-correlated interaction of HOXC13 with the RCs is not specific for one origin but may have a more general character in the origin functional cycle.
More recently, another abdominal-B ortholog, HOXD13 was found to interact in vivo with the lamin B2 origin and with the TOP1, MCM4, c-MYC and FMR1 origins (5); this was also confirmed for HOXA13 and HOXD11; HOXD13 stimulates pre-RC assembly in competition with geminin, an origin licensing inhibitor (6).
These data point to a direct intervention of homeotic proteins in origin regulation, with no mediation by transcription, previously considered as the only way through which HOX proteins act. A direct involvement in the regulation of origin activation of these proteins is not surprising, in light of their morphogenetic (and often proto-oncogenic) role (7), but raises questions on their actual role in DNA-replication regulation. Accordingly, we have explored in particular the spatial and temporal dynamics of the interaction of HOXC13 with the replication factories and origin sequence and of the possible interaction of this protein with other members of the RCs. Our observations stem from the combination of standard biochemical procedures and fluorescence techniques, the latter allowing to explore dynamics and interactions of proteins in living cells. We show here that HOXC13 is a rather stable component of chromatin, that it binds the origins at a precise moment of the cell cycle, specifically associating to DNA well within the pre-RC area, that the protein interacts with other members of the RC in coincidence with origin activation and that the interaction appears to be of general nature in the context of DNA replication regulation.
MATERIALS AND METHODS
Cell culture, transfection, synchronization and TSA treatment
U2OS, T98 G, NIH3T3 and HeLa cells (ATCC) were cultured, transfected and synchronized using standard procedures. For TSA treatment, asynchronously growing HeLa and T98 G cells were incubated or left untreated for 4 h with 100 ng/ml TSA in complete medium.
FRAP and FLIM acquisition
FRAP experiments were performed, according to the previously described ‘half-FRAP’ procedure (8), with an Olympus FluoView 1000-ASW-2.0 confocal laser scanning microscope, equipped with an incubator chamber set to 37°C and 5% CO2. The time-domain FLIM instrumental set up used was already described (9).
GST pull-down assay
[35S]-labelled proteins used for in vitro binding assays were produced using the TNT Reticulocyte Lysate System (Promega) according to the manufacturer’s instructions, by using the corresponding pcDNA3 and pIRES vectors as templates. The recombinant GST fusion proteins were produced and purified from BL21 bacteria transformed with the respective plasmids. The pull-down assay was performed as previously described (10).
In vivo DNA footprinting
Experiments were performed using a previously described procedure (11).
Time lapse imaging
Cells expressing E0GFP-Cdc6 and E0GFP-ORC2 (transiently with low expression profile, or stably) were imaged with the 488 nm laser line of a Leica TCS SP2 confocal microscope, equipped with an incubator chamber set to 37°C and 5% CO2 and a 40×/1.25 NA oil-immersion objective. To minimize photobleaching, images were acquired at low power (∼5 µW), using 1024 × 1024 pixels frame size, low zoom (3×) and pinhole set to 3AU. Four to five z-sections encompassing all nucleus thickness were imaged every 30 min for 16–20 h. The maximum Z-projection of each time point was used to build up the final movie.
Detailed protocols of cell culture, biochemical fractionation, chromatin and protein immuno-precipitation, GST pull-down assay, nascent DNA preparation, FRAP and FLIM data analysis are reported in Supplementary Data.
RESULTS
Spatial and temporal analysis of the localization of HOXC13 in cellular compartments
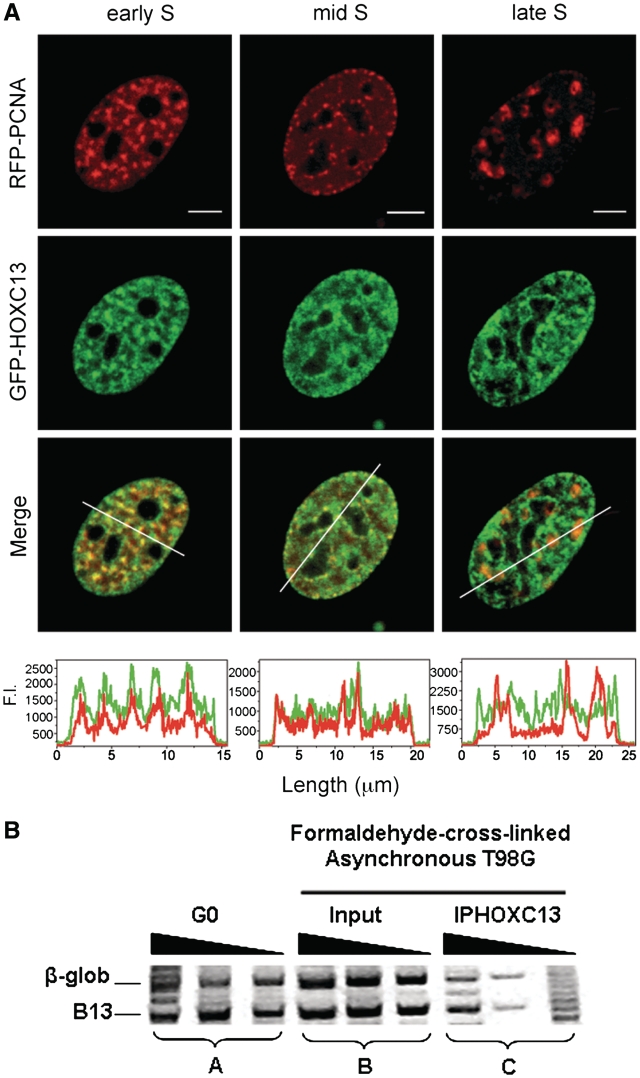
Our previous work showed that GFP-fusion variants of HOXC13 have a speckled-like nuclear distribution, very similar to that of their chromatin-bound endogenous counterpart. By pulsed-BrdUrd immunofluorescence we showed that EGFP-HOXC13 is distributed along the early replicating chromatin (3). We refined this observation by expressing GFP-HOXC13 together with RFP-PCNA, an in vivo marker of replication foci (12), in mouse NIH3T3 cells, which show more pronounced changes of the nuclear pattern of replication foci throughout the S phase. GFP-HOXC13 displayed a marked co-localization with early-S replication foci, but this was less evident in mid S, and was virtually completely absent in the late-S phase (Figure 1A). This observation was confirmed by time-lapse experiments throughout S in cells expressing both constructs (Supplementary Video 1). Another indication, supporting the presence of HOXC13 at early replicating chromatin only, came from a ChIP experiment showing that HOXC13 does not bind a late firing replication origin, namely the one close to the β-globin gene (Figure 1B).
Figure 1.
HOXC13 is present at early and not late replicating chromatin. (A) EGFP-HOXC13 was co-transfected with RFP-PCNA and their mutual localization was monitored in cells displaying three different replication-foci patterns (early, mid and late S replication foci). The intensity profile of green and red channels in a nuclear section (depicted as white bar across the nuclei of Merge images) is plotted below each Merge image. Scale bar: 5 µm. (B) The relative abundance of sequences corresponding to the β-globin origin and to the B13 non-origin control, was evaluated by probing the extracted DNA with sets of three serial 2-fold dilutions of the samples amplified using specific primers for the two sequences. Set A: control DNA extracted from T98 G cells synchronized in G0 phase; set B: DNA extracted from asynchronously growing formaldehyde cross-linked T98 G cells; set C: DNA extracted from asynchronously growing formaldehyde cross-linked T98 G cells after IP with anti-HOXC13 antibody.
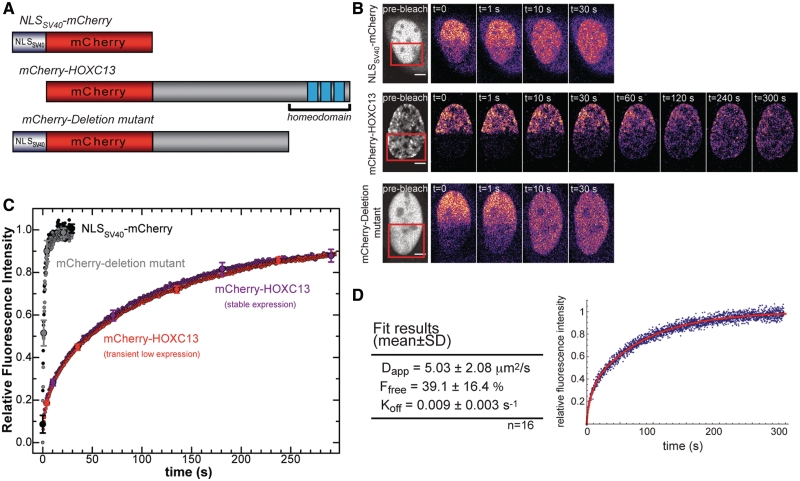
We further investigated HOXC13 dynamics within early replicating chromatin by FRAP (Fluorescence Recovery After Photobleaching). The mCherry-tagged constructs depicted in Figures 2A and 3A were expressed in U2OS cells and analyzed by the ‘half-FRAP’ procedure, an approach already used with several proteins, such as transcription factors and coactivators, structural, remodeling (8,13,14), and replication proteins (15–17). For each analyzed cell, the fluorescence of a half of the nucleus is bleached, and signal recovery is monitored in a subsequent time series (Figure 2B); the resulting normalized recovery of fluorescence intensity in the bleached area is plotted versus time (Figures 2C and 3C). wt mCherry-HOXC13 displayed the slowest average recovery curve of all constructs (Figure 2C). Its recovery halftime (t1/2) quantifies the rate at which molecules in the bleached area are replaced by molecules from the unbleached portion of the nucleus and was t1/2 = 35–40 s; within 300 s of post-bleach observation signal recovery reached a plateau corresponding to about 10% immobile fraction. These results did not depend on the choice of the mCherry fluorophore, as the GFP-HOXC13 construct displayed the same dynamics (data not shown). Such a slow diffusive behavior was reproducibly observed for both transient and stable expression of mCherry-HOXC13 in cells with low-expression profile. High-expression levels resulted in uncertain t1/2 estimates and dramatically increased the immobile fraction. On the contrary, as the protein concentration decreased, both parameters reached stable values in the same range of those obtained with stable protein expression (Supplementary Figure S1).
Figure 2.
FRAP analysis of wt mCherry-HOXC13. (A) Constructs used: (top, non-binding control) NLSSV40-mCherry, (middle) wt mCherry-HOXC13, and (bottom) mCherry-Deletion mutant of HOXC13 devoid of the homeodomain. (B) Representative FRAP experiments with the three constructs: about half of the nucleus (red square in the pre-bleach image) was bleached and time series were acquired until fluorescence recovery reached plateau. The pre-bleach image (grey look up table) is an average of 5–10 frames acquired before bleaching; the post-bleach images (colored look up table) are single frames corresponding to the indicated times. Scale bar: 5 µm. (C) Normalized fluorescence-intensity recovery versus time for the three analyzed constructs. The wt mCherry-HOXC13 construct was expressed either transiently (red dots) or stably (purple dots). Removal of the homeodomain makes the mCherry-Deletion mutant signal recovery (gray dots) very similar to that of the NLSSV40-mCherry non-binding control (black dots). Curves are averaged from data of all analyzed cells (NLSSV40-mCherry: n = 11, mCherry-Deletion mutant: n = 12, transient wt mCherry-HOXC13: n = 40, stable wt mCherry-HOXC13: n = 18). Standard errors values are reported on selected points. (D) The table reports the average fitting parameters obtained applying the reaction-diffusion model described in the text. Right: representative FRAP recovery curve for a single nucleus (blue dots) fitted using the model (red curve).
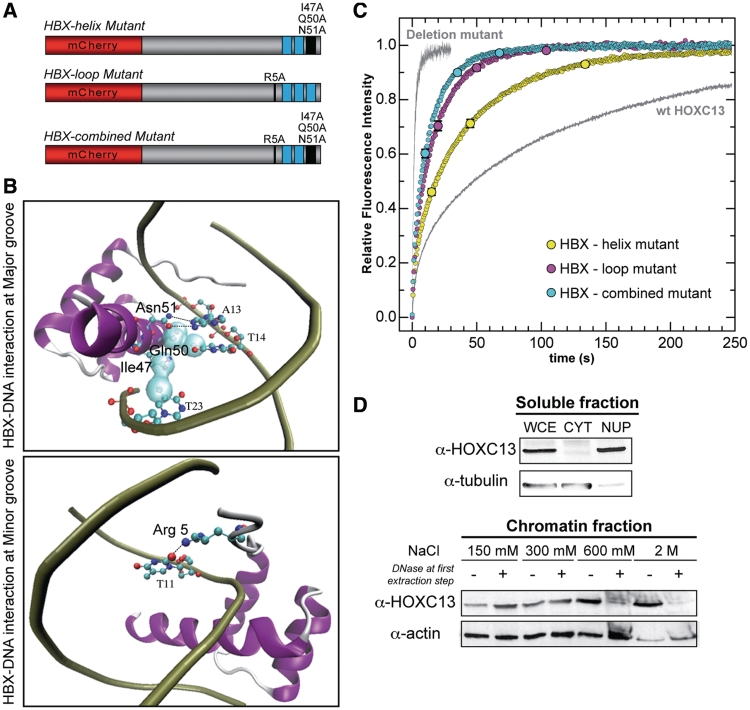
Figure 3.
FRAP analysis of three homeodomain (HBX) mutants of mCherry-HOXC13. (A) HBX mutant constructs: the HBX-helix mutant (top, three mutations in the third HBX recognition helix); the HBX-loop mutant (middle, single mutation at the HBX N-terminal loop); the HBX-combined mutant (bottom, combining both helix and loop mutations). (B) Main DNA–HBX interactions occurring at DNA major (top panel) and minor grooves (bottom panel). The co-crystal structure of the Drosophila engrailed homeodomain with DNA (20) (PDB no.: 1HDD) was taken as a reference and visualized using VMD software. Only interactions mediated by the mutated residues are highlighted in the two panels. Top panel: HBX-major groove interaction is mainly mediated by two hydrogen bonds (depicted with black dashed lines) between residue Asn51 and adenine-13 base and Van der Waals contacts (light cyan surface contour) between Ile47, adenine-13 and thymine-14 and between Gln50 and thymine-23. Bottom panel: HBX-minor groove interaction is mediated, among others, by a hydrogen bond (black dashed line) between Arg5 and thymine-11. (C) Normalized fluorescence-intensity recovery versus time of the three mutants (colored dot-curves). The mCherry-Deletion mutant and the wt mCherry-HOXC13 curves (same as Figure 2) are also reported in gray color, as a reference for 0 and 100% DNA-binding activity, respectively. All data represent mean recovery curves (±SE, reported at selected time points) obtained out of all analyzed cells (HBX-helix mutant: n = 38, HBX-loop mutant: n = 20, HBX-combined mutant: n = 37). (D) The soluble (top panel) and chromatin fractions (bottom panel) of biochemically fractionated asynchronous U2OS cells were investigated for the presence of endogenous HOXC13 by Western Blot (tubulin and actin are loading controls). All fractions were compared to the HOXC13 protein level detected in a whole-cell extract (WCE). CYT and NUP are cytoplasmic and nucleoplasmic fractions, respectively. The chromatin fractions are identified by the NaCl concentration used for the extraction. The first extraction step (150 mM) was performed in absence (–) or presence (+) of DNase I.
The slow dynamics of wt HOXC13 was compared to the recovery rate under the same bleaching conditions of the NLSSV40-mCherry construct (Figure 2). The latter was chosen as a control to monitor unspecific binding events: we obtained t1/2 ≈ 0.75 s, full recovery in ∼6.5 s [similarly to what previously reported for NLSSV40-GFP (18)]. In order to investigate whether this difference was caused by the anchoring of the homeodomain to the nuclear structure, we measured FRAP for an mCherry-HOXC13 mutant devoid of the homeodomain (3). We found that upon deletion of the homeodomain and insertion of NLSSV40 to rescue protein nuclear localization, the dynamics was very similar to that of the NLSSV40-mCherry control protein (t1/2 ≈ 1 s, full recovery in ∼10.5 s). Hence, the peculiar slow nuclear mobility of wt HOXC13 relies on the DNA-binding and chromatin-interaction properties of its homeodomain.
We analyzed the dynamics of chromatin binding by HOXC13 by applying to the FRAP data a simplified diffusion-reaction mathematical model (see Supplementary Data). The model describes the 2D redistribution of unbleached proteins within an area resembling a half-bleached nucleus, assuming that it is governed both by free diffusion and by chromatin-binding events, as demonstrated for other proteins (13,19). We applied this model to the experiments which best fitted the model geometry (a total of 16 out of 40 cells, selection criteria are reported in Supplementary data, together with the summary of the results on all cells, which does not change the picture reported here). We estimated three parameters (Figure 2D): Dapp (apparent diffusion coefficient), Ffree (fraction of unbound protein) and Koff (dissociation rate). We obtained Dapp = 5.03 ± 2.08 µm2/s, a value consistent with other D values calculated by FRAP for transcription factors with similar molecular weight [the diffusion coefficient is called ‘apparent’ to highlight the fact that its value may underestimate the real nuclear D, since it may include both unspecific/transient chromatin interaction and free nucleoplasmic diffusion by the protein (13)]. The obtained mean Ffree reveals that HOXC13 is in equilibrium between the chromatin-bound (≈60%) and the freely diffusing/transiently interacting forms (≈40%), similarly to most nuclear factors investigated by FRAP (8). A relatively low value for Koff (0.009 ± 0.003 s−1) was found that corresponds to a mean residence time on chromatin of 110 ± 40 s.
We further investigated HOXC13 nuclear mobility by characterizing its dependence on the homeodomain–DNA affinity. To this end we mutated to Ala the homeodomain residues which create crucial contacts with DNA (20,21). Figure 3A (top construct) schematically shows our ‘HBX-helix mutant’, obtained by mutating three residues of the third helix of the homeobox (Ile47, Gln50, Asn51, numbers correspond to the classical homeobox numeration). This is also known as ‘recognition helix’: it inserts into the DNA major groove, where these residues provide a contact with DNA bases (Figure 3B, top panel). Mutation of these to Ala causes abolition of DNA-binding capacity for residues 50 and 51 (21). This HBX-helix mutant was investigated by FRAP with the same protocol used for wt HOXC13: the observed nuclear mobility of this mutant was increased more than twice (t1/2 ≈17.5 s, 98% recovery in ∼160 s) with respect to the wt protein (Figure 3C). In the ‘HBX-loop mutant’ (Figure 3A, middle construct), Arg5 was mutated to Ala; Arg5 belongs to the N-terminal loop of the homeodomain and makes contact with the DNA minor groove (Figure 3B, bottom panel). Crystal structure shows that among all DNA–homeobox contacts, Arg5 forms the strongest hydrogen bond by interacting with a thymine base of the minor groove (20,22). Arg5 mutated to Ala causes the total abolition of DNA-binding capacity in vitro (21). Our FRAP data revealed that this mutant displays more than 4-fold increased mobility with respect to the wt protein (t1/2 ≈ 9 s, full recovery in ∼100 s). We also combined the three mutations of HBX-helix with the HBX-loop one to get the ‘HBX-combined mutant’ (Figure 3A, bottom construct), which yielded a 5-fold mobility increase (t1/2 ≈ 7 s and full recovery in ∼80 s). Thus, DNA-binding ability is responsible for the slow nuclear dynamics of the protein, and its progressive impairment causes an up to 5-fold increased mobility as well as the loss of immobile fraction in the observed time range.
Based on our data a new scenario emerges for the role of HOXC13 that veers significantly from the commonly accepted dynamic vision of nuclear architecture and compartmentalization (23). In fact, a large amount of HOXC13 is actually bound to chromatin very stably, with a mean residence time of the order of minutes. This makes HOXC13 different from most nuclear proteins, especially transcription factors, and supports the idea that, once the protein reaches its binding sites, it forms stable complexes and dissociates very slowly from chromatin.
The tight association of HOXC13 to chromatin was confirmed by biochemical fractionation analysis. We separated the cytoplasmic fraction of asynchronous U2OS cells (24,25) from intact nuclei by low-speed centrifugation; the nuclear envelope was then broken and the nucleoplasm separated by centrifugation (26). The residual chromatin was sequentially extracted with increasing NaCl concentration (150 mM to 2 M), performing the first extraction step either without or with DNase I. We then determined the presence of endogenous HOXC13 in the various fractions in comparison with a whole cell extract. Figure 3D shows that almost no HOXC13 was found in the cytoplasm, while both nucleoplasm and chromatin fractions contained a significant amount. These data are consistent with the fact that both endogenous and recombinant HOXC13 are almost exclusively localized in the nucleus (3), where the protein is in equilibrium between the chromatin-bound and nucleoplasm forms as shown by FRAP. The bulk of HOXC13 could be extracted from chromatin mostly at fractions ≥300 mM NaCl, thus confirming that this protein is indeed tightly bound to the nuclear structure. DNase treatment changed significantly the extraction profile: most of the protein was eluted at 150–300 mM NaCl, while the 600 mM and 2 M fractions were almost empty, confirming that HOXC13 is stably bound to the nuclear structure through its interaction with DNA. Similar experiments performed in U2OS cells expressing the fluorolabeled construct of HOXC13 wt and the Deletion mutant, confirmed that the presence of the homeodomain is essential for chromatin binding (Supplementary Figure S2).
Thus, the interaction of HOXC13 with chromatin in nuclear regions corresponding to replication factories (Figure 1A) is very tight and is mediated by the homeodomain. If the interaction were related to origin function, considering its absence in G0 (3), we would expect cell-cycle-related variations of the presence of the protein on the origin, in the area occupied by the RCs.
Spatial and temporal analysis of the localization of HOXC13 in origin DNA
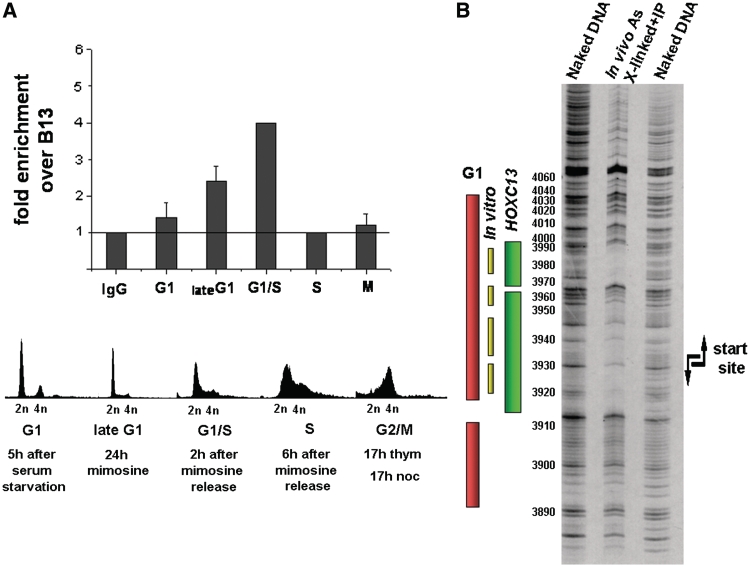
T98 G cells were synchronized in G1 by release from serum starvation, in late G1-G1/S-S with mimosine and in G2/M with thymidine plus nocodazole. Cells in different phases were investigated for the presence of HOXC13 on the origin by ChIP and measure of the enrichment of the precipitated DNA in the lamin B2 origin sequence. Figure 4A shows that the protein appears on the origin region at the beginning of G1 and reaches a peak value at the G1/S transition. As the cells enter S, HOXC13 leaves the origin and returns to it only at the next G1. These observations agree with the data of Figure 1 and provide new insight on HOXC13 origin binding: not only does the protein bind specifically early-firing origins (lamin B2 among them), but it also leaves them in the course of S, when the early replicating origins go out of the foci.
Figure 4.
Spatial and temporal analysis of the localization of HOXC13 in origin DNA. (A) Cell-cycle-dependent association of HOXC13 with human lamin B2 replication origin. Quantification of cross-linked lamin B2 origin DNA immuno-precipitated by ChIP from synchronized cells using HOXC13 antibody. Fold enrichments of lamin B2 origin region over non-origin control region (B13) are reported for each analyzed phase. The line indicates the threshold enrichment level obtained by using rabbit IgG antibody as negative control. Cell-cycle phase was determined by FACS. (B) Analysis of HOXC13-lamin B2 origin interaction in vivo. In vivo cross-linked and sonicated chromatin was immuno-precipitated with HOXC13 antibody and the resulting DNA-protein complexes were subjected to footprint analysis followed by ligation-mediated PCR (LM-PCR). Bars indicate the area covered by pre-RC in G1 phase in vivo (pink) and in vitro (yellow) and the area covered by HOXC13 in asynchronous HeLa cells (green).
Our data demonstrate the presence of HOXC13 at a given moment of the cell cycle in the neighborhood of the origin, in particular on chromatin fragments of a size ranging between 200 and 1000 bp. But we must still establish whether the interaction of this molecule in the lamin B2 and other origin areas involves nearby promoters (like the TIMM promoter for lamin B2) rather than directly the RC-covered sequence. Since the RC assembled on the lamin B2 origin covers a DNA length ≤100 bp, we investigated whether the HOXC13–DNA interaction occurs within or merely close to the RC area (4,27,28). Accordingly, the position of HOXC13 was explored in more detail by submitting the cross-linked, immuno-purified DNA to dimethyl sulfate (DMS) degradation followed by ligation-mediated PCR (LM–PCR) of the produced fragments. A clear footprint was observed (Figure 4B) on the lower strand within nt 3910 and 3990, that is, within the area protected by the pre-RC in G1. This area includes also the sequence recognized in vitro by HOXC13 (1). The large protection observed in Figure 4B is certainly not due exclusively to this molecule: the formaldehyde treatment causes also protein–protein cross-links and the footprint is most probably due to the covalent linking of several RC members, including HOXC13. Also, the quantitative variations in the course of G1 may in part reflect a greater exposure of HOXC13 epitopes as the RC evolves rather than an actual increase in amount of bound protein.
We can conclude that HOXC13 assembles in G1 together with the other pre-RC molecules on the origin and leaves it after firing. This suggests that it must closely interact with other members of the pre-RC.
Interactions of HOXC13 with other RC proteins: in vitro studies
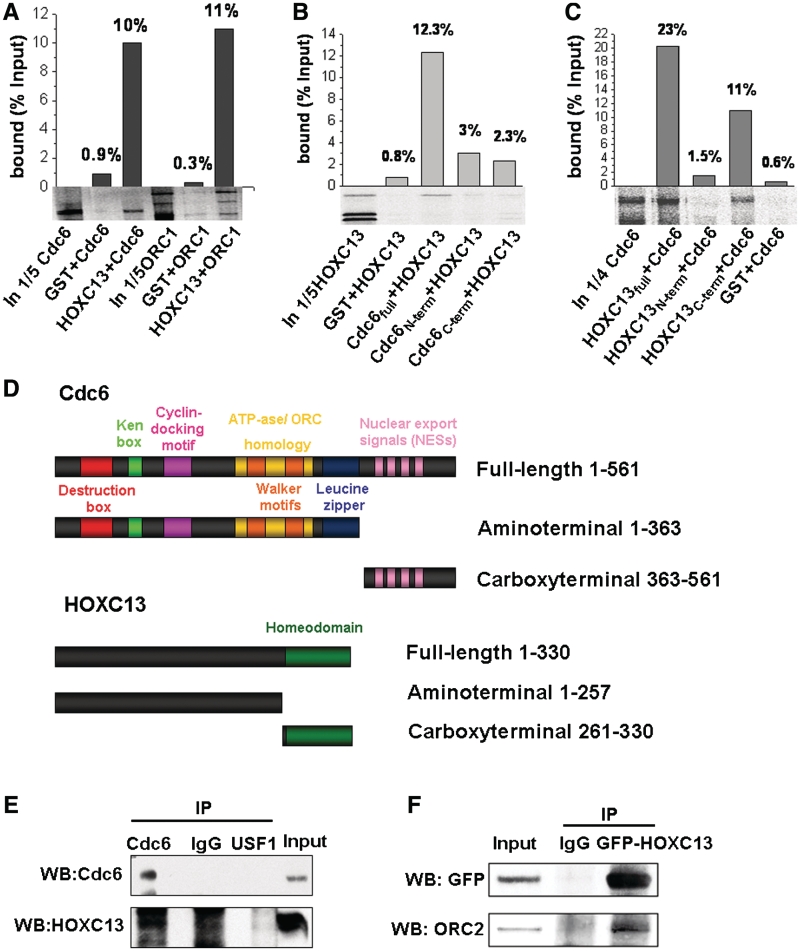
Affinity of HOXC13 for other members of the pre-RC was investigated by GST pull-down and by co-immuno-precipitation. In vitro translated [35S]-labeled Cdc6, ORC1, MCM2 and MCM3 were challenged for their ability to bind a GST-HOXC13 fusion protein. Figure 5A shows that Cdc6 and ORC1 were specifically retained on GST-HOXC13 agarose beads but not on GST beads; differently, MCM2 and MCM3 were not retained (data not shown). A more detailed analysis of the interacting moieties of HOXC13 and Cdc6 was carried out, as reported in Figures 5B, C. Two Cdc6 fragments, one from the N-terminal to aa 363 and the other from aa 364 to the C terminal (Figure 5D), were immobilized on the beads and challenged with full-length [35S]-HOXC13. Neither fragment interacted significantly with HOXC13 (Figure 5B) indicating the central portion of the protein in its integrity as the likely interacting moiety. Conversely, the absence from the beads of the fragment of HOXC13 from the N-terminal to aa 260 and the presence instead of the fragment from aa 261 to the C-terminal, indicate clearly the latter, containing the homeodomain, as the site of interaction with Cdc6 (Figure 5C).
Figure 5.
HOXC13 binds Cdc6, ORC1 and ORC2 in vitro. (A) GST or GST-HOXC13, immobilized on agarose beads were incubated with in vitro translated [35S]-labeled Cdc6 or ORC1. Proteins retained by extensively washed beads were loaded onto a 10% acrylamide-SDS gel. The input lanes contain a fraction of the radiolabeled proteins prior to binding (IN). Graphs show the amount of bound radiolabeled proteins as % of the input. (B) [35S]-labeled HOXC13 was incubated with GST, GST-Cdc6 full length, GST-Cdc6 aminoterminal or GST-Cdc6 carboxyterminal and processed as described in A. (C) [35S]-labeled Cdc6 was incubated with GST-HOXC13 full length, GST-HOXC13 aminoterminal, GST-HOXC13 carboxyterminal or GST, and processed as described in A. (D) Cdc6 and HOXC13 constructs used in the assay. (E) Co-immuno-precipitation experiments performed with lysates from asynchronous crosslinked T98 G cells (Input) using the indicated antibodies for immuno-precipitation and blottings. (F) Immunodetection of endogenous ORC2 after co-immuno-precipitation with exogenous GFP-tagged HOXC13 in transiently-transfected asynchronous U2OS cells (Input).
Co-immuno-precipitations with an anti-Cdc6 antibody confirmed the affinity of HOXC13 for Cdc6 (Figure 6 E). Experiments with an antibody against GFP-HOXC13 demonstrated also an affinity of the homeotic protein for ORC2. Importantly, this interaction was detected only when the fraction of HOXC13 tightly bound to chromatin was extracted (Figure 6 F).
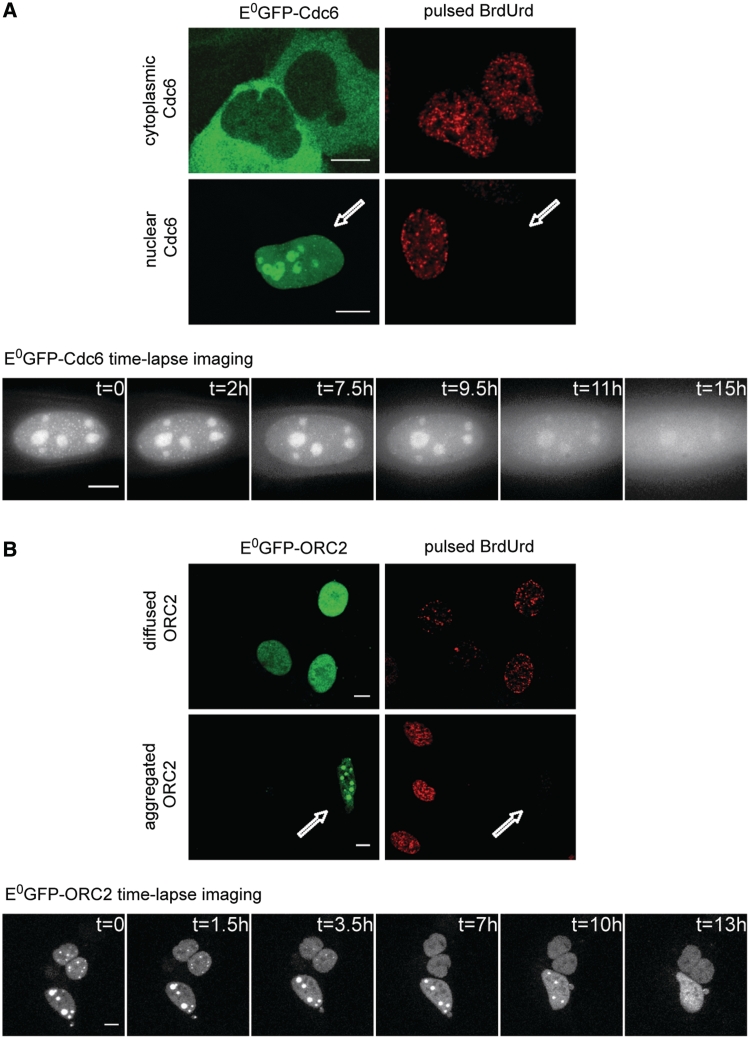
Figure 6.
Dynamics of E0GFP-Cdc6 and E0GFP-ORC2 localization at the G1/S transition. (A) Two different localizations were found for E0GFP-Cdc6: cytoplasmic and nuclear. Pulsed BrdUrd immunofluorescence (top color-panel) shows that cells displaying cytoplasmic E0GFP-Cdc6 localization are positively stained with BrdUrd, while cells displaying the nuclear localization (white arrow) do not incorporate BrdUrd. Time-lapse imaging (bottom gray-scale panel) shows that nuclear E0GFP-Cdc6 is gradually exported to cytoplasm within 15 h (representative time for cells with low-expression profile of the construct). (B) E0GFP-ORC2 displayed different subnuclear distributions. Pulsed BrdUrd immunofluorescence (top color-panel) shows that cells having a diffused nuclear E0GFP-ORC2 localization are positively stained with BrdUrd, while cells displaying a nuclear focal structure for E0GFP-ORC2 do not incorporate BrdUrd (white arrow). Time-lapse imaging (bottom gray-scale panel) reveals that the big E0GFP-ORC2 nuclear foci are first disassembled into smaller ones, until a final nuclear diffused localization is reached within 13 h. Scale bar: 10 µm.
In summary, HOXC13 displayed a significant in vitro affinity with three members of the RCs, namely ORC1, ORC2 and Cdc6. Additionally we showed that this interaction is mediated by the HOXC13 homeodomain, at least for Cdc6.
Interaction of HOXC13 with other RC proteins: in vivo studies
We inquired whether the above-described protein–protein interactions occur also in vivo, in the nuclear compartments and during specific cell-cycle intervals.
We preliminarily performed an in vivo study of the cell-cycle dynamics of the intracellular distribution of the analyzed proteins in U2OS cells. HOXC13 has an exclusively nuclear localization (Figures 1A and 3D) and does not undergo changes of intracellular distribution throughout the cell cycle (data not shown). We found instead significant cell-cycle-related changes of localization for the other probed proteins (Figure 6). We observed two different localizations (nuclear and cytoplasmic) for E0GFP-Cdc6: cells in S (i.e. positively stained with BrdUrd: see Figure 6A, top) displayed a cytoplasmic E0-Cdc6 localization, in agreement with what reported for its endogenous counterpart (10,29,30). Time-lapse imaging of E0-Cdc6 (Figure 6A, bottom) showed that nuclear localization precedes the cytoplasmic one, as nuclear Cdc6 was gradually excluded to cytoplasm within 16–20 h observation, a change certainly related to the transition from G1 to S. Hence, the G1 phase is the only moment of the cell cycle when E0-Cdc6 is present in the nucleus for a possible interaction with HOXC13.
The localization of E0GFP-ORC2 showed a more pronounced cell-cycle modulation. BrdUrd immuno-fluorescence revealed that in S cells E0GFP-ORC2 is homogeneously diffused in the nucleus, whereas it displays a focal distribution outside S (Figure 6B, top). Further analysis showed that this focal organization relies on the association with heterochromatin (Supplementary Figure S3), similarly to endogenous ORC2 (31–33). Time-lapse analysis showed that this focal structure is gradually lost within 13 h: few big foci are disassembled into smaller ones, which then dissolve until a final homogeneous nucleoplasmic distribution is reached (Figure 6B, bottom and Supplementary Video 2). Thus, similarly to endogenous ORC2, the focal structure corresponds to the G1 distribution and precedes the homogenization of nuclear distribution typical of S.
The ORC1 protein (pulled down by HOXC13 in vitro, Figure 5A) was also tested for the in vivo interaction, but it was not possible to obtain significant observations, since the E0GFP construct displayed peculiar localization that hampered a clear evaluation of the results (Supplementary Figure S4).
We then moved to the analysis of the possible interaction of HOXC13 with ORC2 and Cdc6. To this purpose, we utilized Fluorescence Resonance Energy Transfer (FRET) to detect only short-range interactions (≤10 nm). FRET was monitored as reduction of the donor fluorescence lifetime in the presence of a close acceptor, as can be achieved on a pixel-by-pixel basis by Fluorescence Lifetime Imaging Microscopy (FLIM) (34). FLIM measurements were performed by using E0GFP (35) and mCherry (36) as donor and acceptor, respectively. This optimized FRET pair shows negligible donor/acceptor cross-talk when compared to other FRET pairs and features a rather long, mono-exponential donor-fluorescence decay when excited at 403 nm (τ = 3.01 ns in vivo) (9). This allowed us to minimize the spectral bleed-through and to simplify fitting procedures in the FLIM analysis.
E0GFP fusions of the Cdc6 and ORC2 proteins were expressed together with mCherry-HOXC13. We also tested an E0GFP-MCM3 fusion construct, although MCM3 did not interact in vitro with HOXC13, nor did fluorolabeled MCM3 undergo significant cell-cycle related changes of localization (data not shown).
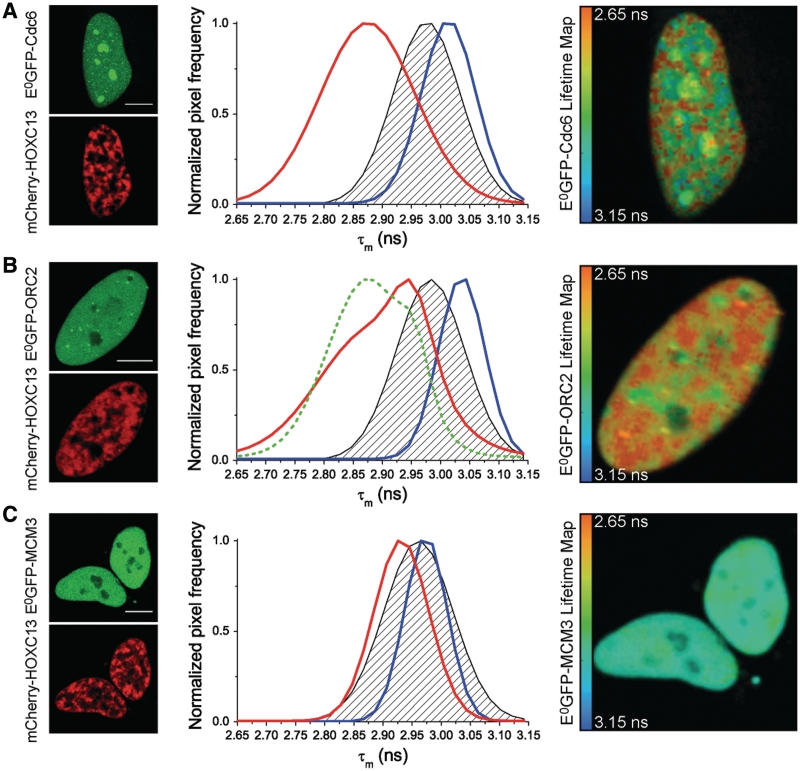
Figure 7 reports on the left side of each panel three sets of representative images in which fluorolabeled HOXC13 was co-expressed with E0GFP fusions of Cdc6 (A), ORC2 (B) and MCM3 (C). To obtain lifetime values, E0GFP was excited with a pulsed 403 nm laser and fluorescence decay was recorded and fitted in all nuclear pixels. This yielded the distribution of the frequency of nuclear pixels versus mean donor lifetime (τm). Cumulative τm distribution curves were obtained based on all analyzed cells expressing either E0GFP-Cdc6, or E0GFP-ORC2, or E0GFP-MCM3 together with mCherry-HOXC13 (red curves in the graphs of Figure 7A, B and C, respectively). In the same graphs, the blue curve is the τm distribution of cells expressing the respective E0GFP-RC protein alone; the black dashed area represents the negative control, i.e. an estimate of unspecific FRET signal between donor and acceptor when they are not actually interacting.
Figure 7.
FLIM analysis of mCherry-HOXC13 interactions with E0GFP-fusions of Cdc6 (A), ORC2 (B), MCM3 (C) in living cells. Left side of each panel: representative fluorescence-intensity images of the cells used in the FLIM study. Cells expressed both donor E0GFP (green top image) fused in turn to Cdc6 (A), ORC2 (B), MCM3 (C), and acceptor mCherry (red bottom image) fused to HOXC13. In the middle graphs of each panel: plot of the frequency of nuclear pixels versus the mean donor lifetime (τm) calculated for each pixel. The blue curve represents the τm distribution of cells expressing the respective E0GFP-fusion of the probed protein alone. The black dashed area represents the negative control: this was obtained either transfecting E0GFP-NLSSV40 with mCherry-HOXC13 or E0GFP-pre-RC proteins with untagged mCherry (the average of the two controls is presented here and in Table 1). The red curve shows τm distribution for cells co-expressing the respective E0GFP-fusion of the probed protein with mCherry-HOXC13. All reported curves are cumulative sum-distribution data relative to all analyzed cells (Table 1); the pixel frequency is normalized to the peak of each distribution curve. Positive in vivo interaction, is detected by FRET as a significant shift of the red curve versus lower τm values when compared to the dashed area of the negative control, this is evident for the case of Cdc6 (A) and ORC2 (B). On the right side of each panel: representative donor-lifetime maps in the presence of mCherry-HOXC13. Scale bar: 10 µm.
The graphs show that, when expressed alone, all E0GFP-RC proteins display a similar average lifetime in the nucleus (from 2.97 to 3.03 ns, see also Table 1, first row): these values are very close to those reported for untagged E0GFP (9). Conversely, when co-expressed with mCherry-HOXC13, E0GFP-Cdc6 and E0GFP-ORC2, but not E0GFP-MCM3, displayed a significant τm reduction when compared either to the donor alone, or to the negative control curves (Figure 7 and Table 1, second and third row). Thus, Cdc6 and ORC2 do interact with HOXC13 in living cells, in agreement with what observed in vitro (Figure 5).
Table 1.
Mean lifetime values (mean±SEM) obtained with FLIM analysis
| Cdc6 |
ORC2 |
MCM3 |
||||
|---|---|---|---|---|---|---|
| τm (ns) | n | τm (ns) | n | τm (ns) | n | |
| E0GFP-replicative complex protein alone | 3.00 ± 0.04 | 49 | 3.03 ± 0.04 | 51 | 2.97 ± 0.04 | 46 |
| Negative control | 2.97 ± 0.04 | 85 | 2.97 ± 0.04 | 116 | 2.95 ± 0.04 | 76 |
| E0GFP-replicative complex protein+mCherry-HOXC13 | 2.86 ± 0.04 | 106 | 2.86 ± 0.06 | 64 | 2.91 ± 0.05 | 36 |
A lifetime value was calculated for all nuclear pixels of the analyzed cells. In this way we generated donor-lifetime maps at sub-cellular level (right side in Figure 7A, B and C). These τm maps are superimposed onto the corresponding donor-intensity images (green channel on the left side of Figure 7). Pseudocolors represent lifetime values: as indicated in the calibration bar, red-shifted colors correspond to lower lifetime values and indicate FRET occurrence. Only Cdc6 and ORC2 (Figure 7A and B) display a red-shifted lifetime map in presence of HOXC13, while the lifetime map of MCM3 co-expressed with HOXC13 (Figure 7C) retains a blue-shifted color similarly to the maps of the donor alone (Supplementary Figure S5). Notably, the red areas in Cdc6 and ORC2 lifetime maps are super-imposable to the areas of HOXC13 localization (red channel on the left side of Figure 7). Thus, the interaction of both Cdc6 and ORC2 with HOXC13 occurs in the same nuclear regions where the homeodomain binds to chromatin (Figures 2 and 3). It must be mentioned that, particularly for the case of Cdc6, some reduction of donor lifetime could be detected in nucleoli (green/yellow color of these subcellular structures in the lifetime maps of Figure 7A and Supplementary Figure S5A). This does not significantly affect the final lifetime distribution, however, since these areas are relatively small and excluded from HOXC13 characteristic nuclear compartments (data not shown).
Moreover, by relating lifetime measurements (Figure 7) with the cell-cycle-dependent localization of Cdc6 and ORC2 (Figure 6), we were able to temporally define their interaction with HOXC13. The interaction of HOXC13 with Cdc6 is restricted to G1, the only phase when these proteins co-localize in the nucleus. As for the interaction with ORC2, the lifetime distribution of E0GFP-ORC2 co-expressed with mCherry-HOXC13 (Figure 7B, red curve) displays two distinct peaks, one close to the negative control (∼2.95 ns), the other one shifted towards lower-lifetime values (∼2.82 ns). We argue that this stems from the different behavior in different moments of the cell cycle that are simultaneously present when the interaction is investigated in asynchronous cells. In order to verify this, cells were synchronized at the G1/S border. Resulting FLIM data are shown as a green, dashed curve in the same graph (Figure 7B). Upon aphidicolin block, an inversion of the amplitude of the two lifetime peaks was observed, as a consequence of an increase of the cell population in which ORC2 interacts with HOXC13. Thus, the maximum of direct interaction between these two proteins occurs in G1, before the start of S. Considering that the homeotic protein reaches its peak on origin DNA only at the end of G1 it is not surprising that its interaction with ORC2 and Cdc6 turns out to be maximized at this cell-cycle stage.
Thus, HOXC13 interacts in vivo with both Cdc6 and ORC2 when bound to the origin in G1, within the pre-RCs, and this interaction is not restricted to one or few origins, but appears to have a general significance in cell-cycle transactions.
Dispensability of HOXC13 and presence of other homeotic proteins on the lamin B2 origin
The general character of the interaction of HOXC13 with pre-RC proteins raises two questions: does HOXC13 play an essential role for origin function? And, in view of the multiplicity of homeotic proteins, is the role of HOXC13 in this process unique, or is it shared by other related molecules?
We studied the effect of HOXC13 depletion in both T98 G and U2OS cells by either transient or stable shRNA transfection. By FACS analysis of the cell-cycle profiles and of BrdUrd incorporation in asynchronous or synchronized cultures, we found that HOXC13 depletion did not affect either cell-cycle progression, or S phase entry (Supplementary Figure S6). This lack of effect is not surprising in light of the observation that in mice the mutation of HOXC13 alleles is compatible with life and only causes a defect in hair morphogenesis (37); actually, HOX-gene knock-out often gives viable progenies (38). This is suggestive of a redundant availability of other HOX proteins that can surrogate structurally and functionally the missing one.
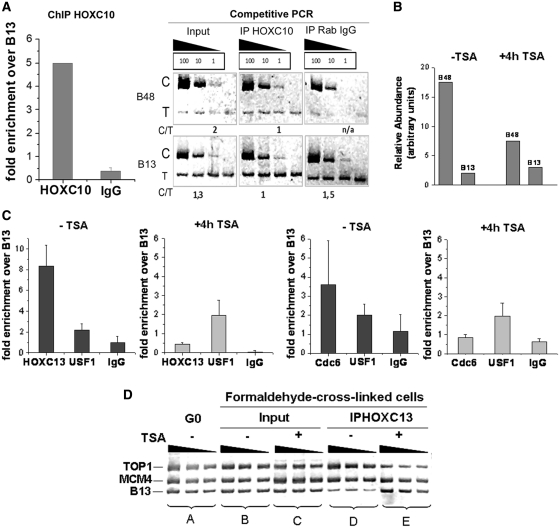
We investigated by ChIP the possible presence of other HOX proteins on the lamin B2 origin: we tested HOXC10 that, like HOXC13 and HOXA13, was initially identified by mono-hybrid screen for binding the origin sequence (1). We found a significant enrichment of the origin sequence in the HOXC10 immuno-precipitate (Figure 8A). These data agree with the recent observation that also HOXA13 binds lamin B2 origin in vivo (together with two other abdominal-B orthologs, HOXD11 and HOXD13) (5).
Figure 8.
(A) Binding of HOXC10 to the lamin B2 origin in asynchronous T98 G cells. Chromatin was immuno-precipitated with HOXC10 antibody or rabbit IgG as control. DNA was then analyzed by competitive PCR for the abundance of origin (B48) over non-origin sequence (B13) (Supplementary Data). C: competitor. T: target DNA. C/T: ratio of the net intensities. Histograms report the relative enrichment of the origin sequence over the non-origin one in the HOXC10 IP and its control, as derived from the analysis of the PCR products shown alongside. (B) TSA inhibition of the lamin B2 origin activity. The abundance of nascent DNA for origin (B48) and non-origin (B13) regions was measured by competitive PCR analysis in cells treated or not with TSA. (C) Comparison of ChIP performed without and with TSA treatment, using HOXC13 antibody (left) or Cdc6 antibody (right). For each ChIP experiment USF1 and rabbit IgG were used as positive and negative control, respectively. (D) Association of HOXC13 with TOP1 and MCM4 origins in vivo. PCR was performed on genomic DNA isolated from quiescent cells (G0), on chromatin fragments from cross-linked asynchronously growing cells with and without TSA treatment (Input) and on chromatin fragments isolated after the immuno-precipitation with HOXC13 antibody with and without TSA treatment (IP HOXC13), in serial 2-fold dilution.
These data fit with the contention that HOX proteins operate, as a rule, by binding DNA as part of a multi-protein complex, typically associating to other HOX proteins and specific co-factors, like Pbx or Meis1 (39,40). The association in these multi-molecular complexes confers the sequence-specificity for the interaction that is absent in isolated HOX proteins. In these complexes, one missing HOX protein may be surrogated by a similar one, structurally fitting the complex and ensuring its function.
Thus, HOX proteins display an affinity in vivo for origins and for other RC proteins, probably in the context of a complex with each other. We may wonder whether these complexes specify or contribute to the specification of origins, considering that in metazoan genomes no clear common motifs were yet discovered that confer this function. This hypothesis suffers, nonetheless, from the lack of consensus sequence also for the motifs bound by different homeotic complexes. This raises the possibility that, rather than sheer sequence, particular local chromatin configurations may actually provide the appropriate conditions for RC assembly.
Effects of disruption of origin chromatin
In order to investigate the impact of the disruption of chromatin organization at the origin sequence, we induced histone hyperacetylation by treating cells with trichostatin A (TSA), an inhibitor of histone deacetylase. The disturbance caused by TSA is reflected in the functionality of the lamin B2 origin: Figure 8B shows that TSA reduces origin activity, as previously reported (41). This reduced functionality is paralleled by the reduced ability of the origin area to bind other RC members: as shown in Figure 8C, also Cdc6 and its partner HOXC13 cannot bind the origin as a consequence of TSA treatment.
The TSA-induced disturbance in HOXC13 binding is observed also for the TOP1 and MCM4 origins. Figure 8D reports a comparison of the PCR products of set B (non-immuno-precipitated chromatin of cycling cells) and set C (the same, in presence of TSA) and shows that the abundance of the three sequences (including a non-origin control) is not significantly changed; conversely, in the samples immuno-precipitated with anti-HOXC13 antibody, the ones obtained from TSA-treated cells (set E) lose the enrichment in origin sequences present in the non-treated ones (set D).
Thus, an appropriate chromatin organization of the origin area appears to be required for the formation of an active RC. Notably, chromatin disruption does not affect other different protein–DNA interactions even if occurring in close proximity: the binding of the USF transcription factor to the nearby promoter for the TIMM gene (28) is unaffected by chromatin disruption (Figure 8C).
In conclusion, the structural features of chromatin at the origins appear to play an important role in origin function rather independently from the detailed sequence features.
DISCUSSION
The data reported in Figures 1–3 demonstrate that HOXC13 is present only in the nucleus and, at least in interphase, over half of it is bound to early replicating chromatin via the homeodomain, with a mean residence time of the order of minutes. Thus, HOXC13 can be considered a stably chromatin-bound protein, with properties intermediate between the fastly exchanging transcription factors (8) and the almost immobile structural proteins like histones (42), cohesin (14), CENP at centromeres (43) or PCNA at replication foci (16). HOXC13 distribution correlates well spatially and temporally with the sites (replication foci) where origins are collected in G1: the affinity for at least three different origins [(3) and Figure 8D] lead to infer that HOXC13 is stably bound to at least a good fraction of the origins, specifically the early replicating ones, to which all the three tested origins belong and that represent anyhow the majority. This hypothesis is further confirmed by our observations that HOXC13 does not co-localize with late S replication foci (Figure 1A), nor does it bind a late firing replication origin, namely the one close to the β-globin gene (Figure 1B).
The HOXC13 protein is bound to origin chromatin, at least for the lamin B2 origin, at a precise site within the pre-RC (Figure 4B) at specific moments of the cell cycle (Figure 4A), and interacts in a time-dependent way with canonical members of the pre-RC (Figures 6 and 7). Interaction with the origin occurs within the area protected by the pre-RC in G1, very close to the start sites of leading strand synthesis and to the binding sites of ORC1, ORC2, Cdc6, topoisomerase (topo) I and topo II (27,44). The protein is absent from the origin in M and appears on it at the beginning of G1, interacts with ORC2 in this phase to reach a peak at G1/S, that is the moment when the interaction with Cdc6 can occur. As synthesis starts, interaction of HOXC13 with the origin and the other RC members fades, in parallel with the transition from this large pre-RC to a smaller and differently organized post-RC (27). We can surmise that the protein, although slowly exchanging with nucleoplasm, gradually moves from origin to non-origin DNA chromatin throughout S, consistently with the observation that mid S and late S replication foci are excluded from HOXC13 nuclear compartments (Figure 1A and Supplementary Video 1). Even if the data on the precise site and time of interaction of HOXC13 with the origin were here referred to the lamin B2 case, we stress that the interaction with ORC2 and Cdc6 is a global feature of the cell.
The interaction of HOXC13 with the origins, in all probability, occurs in the context of a multi-protein homeotic effector, containing different homeotic proteins. This conclusion is suggested by the observation that several homeotic proteins, namely HOXC13, HOXA13, HOXC10, HOXD11 and HOXD13 bind replication origins in vivo [Figure 8A and (1,5)]; moreover, HOXC13 and HOXD13 are both involved in protein–protein interactions with the pre-RC, and share an affinity for Cdc6, while the fact that ORC2 is found to interact with HOXC13 but not with HOXD13 could depend on the different experimental procedures used in the investigation [Figures 5–7 and (5)]. In this perspective, we should also expect that, besides HOXC13, other homeotic proteins might display a stable interaction with chromatin. Depletion of one of these proteins is compatible with the continuation of the cell cycle, as evident for both HOXC13 and HOXD13 (5) proteins: probably, thanks to the redundancy of homeotic protein structures, a missing protein may be surrogated in the complex by an analogous one, at least insofar as a relatively ‘generic’ function (marking of a large fraction of replication origins) is required, rather than the regulation of a specific gene. In this view, it was not surprising to find that depletion of HOXC13 does not alter cell cycle progression or S phase entry (Supplementary Figure S6). The delay in initiation of DNA synthesis in HEK293 cells reported upon HOXD13 depletion (5) could be the result of the synergistic silencing of several HOX members when using siRNA targeted to highly conserved protein sequences like that of the homeodomain; or it could be due to a possible higher level of HOXD13 in HEK293 cells when compared to the levels of HOXC13 in U2OS and T98 G cells.
A connection between the morphogenetic function of homeotic proteins and the regulation of DNA replication could be postulated a priori. Indeed both in the case of development and cancer a key role of HOX proteins in modulating cell proliferation was amply demonstrated [reviewed by (7,45)]. Thus, which specific role can we ascribe to the binding of HOXC13 to a large fraction of replication origins, in the context of the pre-RC assembly and operation? It is tempting to hypothesize that homeotic complexes could contribute to the specification of origins in the genome, considering the lack of sequence consensus in metazoan origins, apart from a relative frequency of asymmetric A/T stretches (46). On the other hand, also the motifs bound by homeotic complexes, when considered for their role of transcription (positive or negative) effectors, do not display any obvious consensus. This raises the possibility that, rather than sheer sequence, particular local topological constraints and/or specific chromatin configurations may actually provide the appropriate conditions for the assembly of the RCs. This apparent paradox stems from the observation that interaction of the origins with HOXC13 occurs in the absence of sequence conservation. In fact, TOP1 and MCM4 origins do not contain any motif similar to the one bound by HOXC13 on the lamin B2 origin in vivo or in vitro. DNA topology and chromatin structure are closely interrelated, affect each other and depend also on relatively long-range effects of DNA sequence, such as the tendency to form unusual DNA structures (triple strands, Hoogsteen structures) and to bend the duplex. The lamin B2 origin sequence is characterized by an intrinsic tendency to form such unusual structures (47) and contains a bent DNA sequence between nt 3923 and 3928. Furthermore, topological status is clearly critical for origin recognition by ORC in yeast and Drosophila origins (48,49), while topo I and II bind precise sites in the lamin B2 pre-RC area in precise moments of the cell cycle, topo I being essential for synthesis initiation (44,50). In this context, it is particularly important to observe that both topos precisely recognize in vitro the same origin nucleotides cleaved in vivo. Finally, disruption of chromatin structure caused by TSA sharply reduces origin function and disturbs the binding of RC members like HOXC13 and Cdc6, whereas it does not affect the binding of USF to the TIMM promoter, that is located only 50-bp away from the right border of the RC, and that, contrary to what happens with RC members, remains bound throughout the cell cycle (Figure 8C).
With these considerations, how can we rationalize the riddle of origin specification and of the role of homeotic proteins in origin function? A possible working hypothesis requires looking at the problem in a network, rather than strictly deterministic, logical frame: a combination of local chromatin organization, loose but not nil sequence specificity of the topos and of a homeotic complex, plus other yet-to-be-identified proteins and elements (presence of asymmetric AT-rich stretches? Presence of nearby sites for binding transcription factors, like the E-box present in the close TIMM13 promoter, to which USF is bound as well as, possibly, cMYC? Presence of sequences causing bends in DNA? Closeness to a chromosome scaffold attachment site?) could synergize in creating a chromosomal region fit for assembly of the RC. In this view, we might hypothesize that constitutively open-chromatin sequences may be bound preferentially by topos, assuring a local negatively supercoiled status, or by other proteins recognizing yet to be defined features of the sequence, and by the homeoprotein complex that, thanks to the interaction with the pre-RC members, facilitates the assembly of this complex on the origin.
Homeotic proteins have a preference for AT-rich sequences (51) and our FRAP data indicate that most of the HOXC13 stabilization onto DNA comes from the anchoring of the homeodomain (Figures 2 and 3). Notably, not all mutations that did show to completely abolish DNA binding in vitro (21) had the same effect on our in vivo FRAP analysis. In particular, our data rather indicate that most of the stabilization on chromatin depends on the interaction of an Arg residue of the homeodomain with the DNA minor groove. Interestingly, N-terminal Arg residues were recently indicated as the only residues necessary to confer specificity in the DNA binding by HOX and other proteins (52); indeed, minor groove anchoring by Arg residues seems to constitute the basis for a new DNA recognition mechanism used by many families of DNA-binding proteins (53). Such recognition property may provide the relatively loose sequence specificity contribution of HOX complexes that could lead to precise origin specification, in combination with other partial but converging specificities. In this context, it is worth mentioning that, among pre-RC proteins, HOXC13 shares some similarity with Schizosaccharomyces pombe ORC4 (54) which was shown to bind to the minor groove of AT-rich sequences and thus contribute to origin specification.
We should like to propose this network/combinatorial approach as a possible guide for solving the puzzle of metazoan origin selection: according to this view, the property of replication origin might emerge from a limited number of combinations of the structural features mentioned above, in an appropriate cellular context, in which certain essential proteins are available.
It appears certainly stimulating to discover a connection between an element of the DNA replication regulation and an actor of development and differentiation (7): actually, the HOXC13 protein may be only the first identified of possibly several go-betweens of the two processes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Associazione Italiana per la Ricerca sul Cancro and Istituto Toscano Tumori (to A.F.); Fondazione Monte del Paschi di Siena. Funding for open access charge: Scuola Normale Superiore, Pisa, Italy.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Dr Cardoso and Prof. Dr Leonhardt for kindly making the RFP-PCNA construct available. This work is dedicated to the memory of Prof. Arturo Falaschi, who supervised the entire course of this research and contributed at most in interpreting the data and in writing this article.
REFERENCES
- 1.de Stanchina E, Gabellini D, Norio P, Giacca M, Peverali FA, Riva S, Falaschi A, Biamonti G. Selection of homeotic proteins for binding to a human DNA replication origin. J. Mol. Biol. 2000;299:667–680. doi: 10.1006/jmbi.2000.3782. [DOI] [PubMed] [Google Scholar]
- 2.Gabellini D, Colaluca IN, Vodermaier HC, Biamonti G, Giacca M, Falaschi A, Riva S, Peverali FA. Early mitotic degradation of the homeoprotein HOXC10 is potentially linked to cell cycle progression. EMBO J. 2003;22:3715–3724. doi: 10.1093/emboj/cdg340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Comelli L, Marchetti L, Arosio D, Riva S, Abdurashidova G, Beltram F, Falaschi A. The homeotic protein HOXC13 is a member of human DNA replication complexes. Cell Cycle. 2009;8:454–459. doi: 10.4161/cc.8.3.7649. [DOI] [PubMed] [Google Scholar]
- 4.Abdurashidova G, Riva S, Biamonti G, Giacca M, Falaschi A. Cell cycle modulation of protein-DNA interactions at a human replication origin. EMBO J. 1998;17:2961–2969. doi: 10.1093/emboj/17.10.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salsi V, Ferrari S, Ferraresi R, Cossarizza A, Grande A, Zappavigna V. HOXD13 binds DNA replication origins to promote origin licensing and is inhibited by geminin. Mol. Cell. Biol. 2009;29:5775–5788. doi: 10.1128/MCB.00509-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 7.Falaschi A, Abdurashidova G, Biamonti G. DNA replication, development and cancer: a homeotic connection? Crit. Rev. Biochem. Mol. Biol. 2010;45:14–22. doi: 10.3109/10409230903365608. [DOI] [PubMed] [Google Scholar]
- 8.Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albertazzi L, Arosio D, Marchetti L, Ricci F, Beltram F. Quantitative FRET analysis with the EGFP-mCherry fluorescent protein pair. Photochem. Photobiol. 2009;85:287–297. doi: 10.1111/j.1751-1097.2008.00435.x. [DOI] [PubMed] [Google Scholar]
- 10.Paolinelli R, Mendoza-Maldonado R, Cereseto A, Giacca M. Acetylation by GCN5 regulates CDC6 phosphorylation in the S phase of the cell cycle. Nat. Struct. Mol. Biol. 2009;16:412–420. doi: 10.1038/nsmb.1583. [DOI] [PubMed] [Google Scholar]
- 11.Kang SH, Vieira K, Bungert J. Combining chromatin immunoprecipitation and DNA footprinting: a novel method to analyze protein-DNA interactions in vivo. Nucleic Acids Res. 2002;30:e44. doi: 10.1093/nar/30.10.e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso MC. Dynamics of DNA replication factories in living cells. J. Cell. Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beaudouin J, Mora-Bermudez F, Klee T, Daigle N, Ellenberg J. Dissecting the contribution of diffusion and interactions to the mobility of nuclear proteins. Biophys. J. 2006;90:1878–1894. doi: 10.1529/biophysj.105.071241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr. Biol. 2006;16:1571–1578. doi: 10.1016/j.cub.2006.06.068. [DOI] [PubMed] [Google Scholar]
- 15.Essers J, Theil AF, Baldeyron C, van Cappellen WA, Houtsmuller AB, Kanaar R, Vermeulen W. Nuclear dynamics of PCNA in DNA replication and repair. Mol. Cell. Biol. 2005;25:9350–9359. doi: 10.1128/MCB.25.21.9350-9359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sporbert A, Domaing P, Leonhardt H, Cardoso MC. PCNA acts as a stationary loading platform for transiently interacting Okazaki fragment maturation proteins. Nucleic Acids Res. 2005;33:3521–3528. doi: 10.1093/nar/gki665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schermelleh L, Haemmer A, Spada F, Rosing N, Meilinger D, Rothbauer U, Cardoso MC, Leonhardt H. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 2007;35:4301–4312. doi: 10.1093/nar/gkm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xouri G, Squire A, Dimaki M, Geverts B, Verveer PJ, Taraviras S, Nishitani H, Houtsmuller AB, Bastiaens PI, Lygerou Z. Cdt1 associates dynamically with chromatin throughout G1 and recruits Geminin onto chromatin. EMBO J. 2007;26:1303–1314. doi: 10.1038/sj.emboj.7601597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sprague BL, Pego RL, Stavreva DA, McNally JG. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 2004;86:3473–3495. doi: 10.1529/biophysj.103.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kissinger CR, Liu BS, Martin-Blanco E, Kornberg TB, Pabo CO. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- 21.Ades SE, Sauer RT. Specificity of minor-groove and major-groove interactions in a homeodomain-DNA complex. Biochemistry. 1995;34:14601–14608. doi: 10.1021/bi00044a040. [DOI] [PubMed] [Google Scholar]
- 22.Fraenkel E, Rould MA, Chambers KA, Pabo CO. Engrailed homeodomain-DNA complex at 2.2 A resolution: a detailed view of the interface and comparison with other engrailed structures. J. Mol. Biol. 1998;284:351–361. doi: 10.1006/jmbi.1998.2147. [DOI] [PubMed] [Google Scholar]
- 23.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreitz S, Ritzi M, Baack M, Knippers R. The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J. Biol. Chem. 2001;276:6337–6342. doi: 10.1074/jbc.M009473200. [DOI] [PubMed] [Google Scholar]
- 27.Abdurashidova G, Danailov MB, Ochem A, Triolo G, Djeliova V, Radulescu S, Vindigni A, Riva S, Falaschi A. Localization of proteins bound to a replication origin of human DNA along the cell cycle. EMBO J. 2003;22:4294–4303. doi: 10.1093/emboj/cdg404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dimitrova DS, Giacca M, Demarchi F, Biamonti G, Riva S, Falaschi A. In vivo protein-DNA interactions at human DNA replication origin. Proc. Natl Acad. Sci. USA. 1996;93:1498–1503. doi: 10.1073/pnas.93.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delmolino LM, Saha P, Dutta A. Multiple mechanisms regulate subcellular localization of human CDC6. J. Biol. Chem. 2001;276:26947–26954. doi: 10.1074/jbc.M101870200. [DOI] [PubMed] [Google Scholar]
- 30.Fujita M, Ishimi Y, Nakamura H, Kiyono T, Tsurumi T. Nuclear organization of DNA replication initiation proteins in mammalian cells. J. Biol. Chem. 2002;277:10354–10361. doi: 10.1074/jbc.M111398200. [DOI] [PubMed] [Google Scholar]
- 31.Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 2004;23:2651–2663. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Craig JM, Earle E, Canham P, Wong LH, Anderson M, Choo KH. Analysis of mammalian proteins involved in chromatin modification reveals new metaphase centromeric proteins and distinct chromosomal distribution patterns. Hum. Mol. Genet. 2003;12:3109–3121. doi: 10.1093/hmg/ddg330. [DOI] [PubMed] [Google Scholar]
- 33.Auth T, Kunkel E, Grummt F. Interaction between HP1alpha and replication proteins in mammalian cells. Exp. Cell Res. 2006;312:3349–3359. doi: 10.1016/j.yexcr.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 34.Wallrabe H, Periasamy A. Imaging protein molecules using FRET and FLIM microscopy. Curr Opin. Biotechnol. 2005;16:19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Arosio D, Garau G, Ricci F, Marchetti L, Bizzarri R, Nifosi R, Beltram F. Spectroscopic and structural study of proton and halide ion cooperative binding to gfp. Biophys. J. 2007;93:232–244. doi: 10.1529/biophysj.106.102319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 37.Godwin AR, Capecchi MR. Hoxc13 mutant mice lack external hair. Genes Dev. 1998;12:11–20. doi: 10.1101/gad.12.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wellik D. Hox genes and vertebrate axial pattern. In: Pourquie O, editor. HOX Genes. Oxford, UK: AP Elsevier; 2009. pp. 257–264. [DOI] [PubMed] [Google Scholar]
- 39.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 40.Shen WF, Rozenfeld S, Kwong A, Kom ves LG, Lawrence HJ, Largman C. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol. Cell. Biol. 1999;19:3051–3061. doi: 10.1128/mcb.19.4.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kemp MG, Ghosh M, Liu G, Leffak M. The histone deacetylase inhibitor trichostatin A alters the pattern of DNA replication origin activity in human cells. Nucleic Acids Res. 2005;33:325–336. doi: 10.1093/nar/gki177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell. Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell. Biol. 2008;180:1101–1114. doi: 10.1083/jcb.200710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abdurashidova G, Radulescu S, Sandoval O, Zahariev S, Danailov MB, Demidovich A, Santamaria L, Biamonti G, Riva S, Falaschi A. Functional interactions of DNA topoisomerases with a human replication origin. EMBO J. 2007;26:998–1009. doi: 10.1038/sj.emboj.7601578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Del Bene F, Wittbrodt J. Cell cycle control by homeobox genes in development and disease. Semin. Cell Dev. Biol. 2005;16:449–460. doi: 10.1016/j.semcdb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Karnani N, Taylor CM, Malhotra A, Dutta A. Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol. Biol. Cell. 2010;21:393–404. doi: 10.1091/mbc.E09-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kusic J, Kojic S, Divac A, Stefanovic D. Noncanonical DNA elements in the lamin B2 origin of DNA replication. J. Biol. Chem. 2005;280:9848–9854. doi: 10.1074/jbc.M408310200. [DOI] [PubMed] [Google Scholar]
- 48.Mitkova AV, Biswas-Fiss EE, Biswas SB. Modulation of DNA synthesis in Saccharomyces cerevisiae nuclear extract by DNA polymerases and the origin recognition complex. J. Biol. Chem. 2005;280:6285–6292. doi: 10.1074/jbc.M410129200. [DOI] [PubMed] [Google Scholar]
- 49.Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 2004;23:897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Falaschi A, Abdurashidova G, Sandoval O, Radulescu S, Biamonti G, Riva S. Molecular and structural transactions at human DNA replication origins. Cell Cycle. 2007;6:1705–1712. doi: 10.4161/cc.6.14.4495. [DOI] [PubMed] [Google Scholar]
- 51.Billeter M. Homeodomain-type DNA recognition. Prog. Biophys. Mol. Biol. 1996;66:211–225. doi: 10.1016/s0079-6107(97)00006-0. [DOI] [PubMed] [Google Scholar]
- 52.Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kong D, DePamphilis ML. Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit. Mol. Cell. Biol. 2001;21:8095–8103. doi: 10.1128/MCB.21.23.8095-8103.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.