Abstract
In all three kingdoms of life, nucleotides in ribosomal RNA (rRNA) are post-transcriptionally modified. One type of chemical modification is 2′-O-ribose methylation, which is, in eukaryotes and archaea, performed by box C/D small ribonucleoproteins (box C/D sRNPs in archaea) and box C/D small nucleolar ribonucleoproteins (box C/D snoRNPs in eukaryotes), respectively. Recently, the first structure of any catalytically active box C/D s(no)RNP determined by electron microscopy and single particle analysis surprisingly demonstrated that they are dimeric RNPs. Mutational analyses of the Nop5 protein interface suggested that di-sRNP formation is also required for the in vitro catalytic activity. We have now analyzed the functional relevance of the second interface, the sRNA interface, within the box C/D di-sRNP. Mutations in conserved sequence elements of the sRNA, which allow sRNP assembly but which severely interfere with the catalytic activity of box C/D sRNPs, prevent formation of the di-sRNP. In addition, we can observe the dimeric box C/D sRNP architecture with a different box C/D sRNP, suggesting that this architecture is conserved. Together, these results provide further support for the functional relevance of the di-sRNP architecture and also provide a structural explanation for the observed defects in catalysis of 2′-O-ribose methylation.
INTRODUCTION
Optimal mRNA translation by ribosomes requires the post-transcriptional modification of ribosomal RNAs (rRNAs). These chemical modifications include base methylation, pseudouridylation and 2′-O-ribose methylation of phylogenetically conserved and functionally critical regions of the ribosome. They are catalyzed either by protein-alone or by small (nucleolar) ribonucleoprotein [s(no)RNP] enzymes. The RNPs, classified as pseudouridylation box H/ACA s(no)RNPs or 2′-O-ribose methylation box C/D s(no)RNPs, utilize s(no)RNA-substrate RNA base pairing to direct the catalytic subunits of these RNPs to the target modification site. Besides rRNA, archaeal tRNAs and eukaryotic mRNAs and snRNAs are also substrates for box C/D and box H/ACA s(no)RNPs.
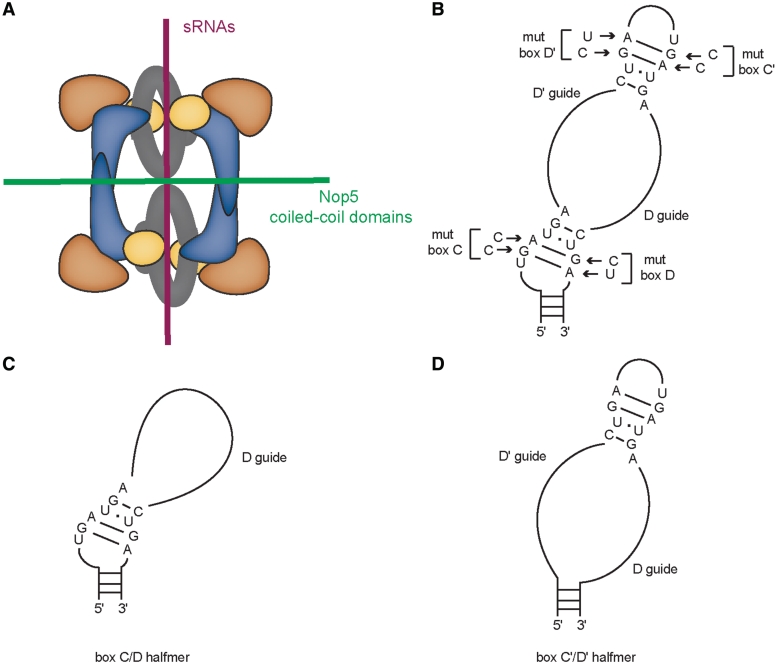
Box C/D s(no)RNPs are composed of small non-coding RNAs and conserved box C/D core proteins [reviewed in (1)]. The proteins include L7Ae, Nop5, and the methyltransferase, fibrillarin, in archaeal box C/D sRNPs (Figure 1A) and 15.5K/Snu13, Nop56 and Nop58, and fibrillarin/Nop1 in eukaryotic box C/D snoRNPs. The sRNA contains conserved sequence elements, boxes C, D, C′ and D′ with the consensus sequences RUGAUGA (boxes C and C′) and CUGA (boxes D and D′) that determine the name of this class of RNPs (Figure 1B). Boxes C and D and boxes C′ and D′ of the s(no)RNAs fold into k-turn and k-loop structures, respectively, that are recognized by the primary RNA-binding protein within box C/D s(no)RNPs, L7Ae or 15.5K (2–4). The k-turn (short for kink-turn) motif is a helix-bulge-helix motif, with an internal asymmetric 5+2nt loop (5–7). It contains two conserved G–A base pairs that are essential for archaeal L7Ae or mammalian 15.5K binding to the box C/D motif of box C/D s(no)RNAs (3,8,9). The internal C′/D′ elements in box C/D s(no)RNAs are predicted to fold into similar structures and are referred to as k-loops (4). While archaeal L7Ae can bind the k-loop formed by the C′/D′ motif, the eukaryotic homologs (15.5K in humans and Snu13 in yeast) cannot, suggesting differences between archaeal and eukaryotic s(no)RNP architecture (8,10,11).
Figure 1.
Schematic of archaeal box C/D sRNP and box C/D sRNA organization. (A) Di-sRNP model of box C/D sRNP architecture. The two independent interfaces that stabilize the archaeal di-sRNP are indicated: (i) the interface formed by dimerization of Nop5 coiled-coil domains and (ii) the interface formed by the sRNAs in the RNP. L7Ae—yellow, Nop5—blue, fibrillarin—orange. (B) Conserved sequence elements in the sRNA, boxes C, D, C′, and D′ were mutated as indicated. Halfmer sR8 sRNAs were generated that lack either the (C) C′/D′ or the (D) C/D motifs. Figure 1A and B are modified from ref. (14).
L7Ae or 15.5K binding to box C/D s(no)RNAs allows recruitment of the remaining box C/D core protein components, Nop5 and fibrillarin, into the RNP (8,12,13). The recent structure of such a catalytically active archaeal box C/D sRNP that we determined by single particle electron microscopy suggests that this RNP is a di-sRNP, containing four sets of each core protein and two sRNAs (14). In this structure, two Nop5 dimerize via their coiled-coil domain, and two such Nop5 dimers are orchestrated into a single RNP by two sRNAs. Fibrillarin, the catalytic subunit in the sRNP, is positioned in the RNP via interaction with the N-terminal domain of Nop5. This di-sRNP model suggests that there are two major interfaces that stabilize the di-sRNP structure: the first interface is composed of the sRNA molecules, connecting the Nop5–fibrillarin heterotetramers, and the second interface is formed by the coiled-coil domain interactions between Nop5, resulting in two Nop5 dimers, one stabilizing either side of the di-sRNP (Figure 1A). For simplicity, we will refer to the sRNP containing two sRNAs and four sets of each core protein as the di-sRNP throughout this manuscript.
The dimeric structure for archaeal box C/D sRNPs was unexpected and raises the question of whether and how this specific structure contributes to the function of these RNPs. To resolve whether the di-sRNP structure is required for the function of archaeal box C/D sRNPs, we investigated whether enzymatic activity correlates with di-sRNP formation. We hypothesized that weakening the interaction at either of the two interfaces should (i) destabilize the di-sRNP and (ii) decrease the methylation activity of assembled sRNPs if the di-sRNP were indeed functionally relevant. We have previously shown that disruption of the Nop5 coiled-coil domain interface prevents di-sRNP formation and that this correlates with the catalytic activity of the respective box C/D sRNP (14). Here we extend these analyses to the sRNA interface and show that mutant sRNAs that assemble into sRNPs of reduced catalytic activity are impaired in di-sRNP formation. Furthermore, we provide evidence that another box C/D sRNA also assembles into the di-sRNP. Together these results indicate that the di-sRNP architecture is a conserved feature of archaeal box C/D sRNPs that is important for their function in catalyzing efficient 2′-O-ribose methylation of substrate RNAs.
MATERIALS AND METHODS
Production of sRNAs
We generated the wild-type (WT) and six mutant sR8 sRNAs by in vitro transcription. The construct used as template for the WT sR8 sRNA is described in (8) and the in vitro transcription was performed as in (14). Mutant sRNA genes were PCR amplified using an oligonucleotide representing the top strand of the gene as template and oligonucleotides described in Table 1. PCR products were cloned into pCR4-TOPO and verified by DNA sequencing. For in vitro transcription, the sRNA gene sequence was PCR amplified from plasmids with oligonucleotides in Table 1 and the PCR product was used as a template for in vitro transcription performed with the AmpliScribe T7-Flash Transcription Kit (Epicentre) according to the manufacturer’s instructions. Using the same strategy, the genes for the remaining Methanocaldococcus jannaschii sRNAs sR1-sR7 (http://lowelab.ucsc.edu/snoRNAdb/Archaea/Mja-align.htmlwere) were cloned into pCR4TOPO and amplified by PCR prior to RNA synthesis with oligonucleotides listed in Table 1.
Table 1.
Methanocaldococcus jannaschii sRNA gene sequences and oligonucleotides used for amplification
| sR8 RNA (WT) | |
| Gene sequence | AAA TCG CCA ATG ATG ACG ATT GGC TTT GCT GAG TCT GTG ATG AAC CGT ATG AGC A CT GAG GCG ATTT |
| sR8-T7-forward | CTA ATA CGA CTC ACT ATA GGC CAA ATC GCC AAT GAT GAC GAT TG |
| sR8.down | AAA TCG CCT CAG TGC TCA TAC GG |
| sR8 RNA C/D halfmer | |
| Gene sequence | AAA TCG CCA ATG ATG AAC CGT ATG AGC ACT GAG GCG ATT T |
| sR8-C/D-half-T7-forward.B | CTA ATA CGA CTC ACT ATA GGC CAA ATC G |
| sR8-C/D-half-reverse | AAA TCG CCT CAG TGC TCA TAC |
| sR8 RNA mut box C | |
| Gene sequence | AAA TCG CCA ATC CTG ACG ATT GGC TTT GCT GAG TCT GTG ATG AAC CGT ATG AGC A CT GAG GCG ATT T |
| sR8-C mut-T7-forward.B | CTA ATA CGA CTC ACT ATA GGC CAA ATC G |
| sR8.down | AAA TCG CCT CAG TGC TCA TAC GG |
| sR8 RNA mut box D | |
| Gene sequence | AAA TCG CCA ATG ATG ACG ATT GGC TTT GCT GAG TCT GTG ATG AAC CGT ATG AGC ACT CTG GCG ATT T |
| sR8-T7-forward | CTA ATA CGA CTC ACT ATA GGC CAA ATC GCC AAT GAT GAC GAT TG |
| sR8-Dmut-reverse | AAA TCG CCA GAG TGC TCA TAC GG |
| sR8 RNA C′/D′ halfmer | |
| Gene sequence | TCC TGG CG ATT GGC TTT GCT GAG TCT GTG ATG ACC GTA TGA GCA CTC CAG GA |
| sR8-C′/D′-half-T7-forward.B | CTA ATA CGA CTC ACT ATA GGC CTC CTG G |
| sR8-C′/D′-half-reverse | TCC TGG AGT GCT CAT ACG GTT C |
| sR8 RNA mut box C′ | |
| Gene sequence | AAA TCG CCA ATG ATG ACG ATT GGC TTT GCT GAG TCT GTC CTG AAC CGT ATG AGC ACT GAG GCG ATT T |
| sR8-T7-forward | CTA ATA CGA CTC ACT ATA GGC CAA ATC GCC AAT GAT GAC GAT TG |
| sR8.down | AAA TCG CCT CAG TGC TCA TAC GG |
| sR8 RNA mut box D′ | |
| Gene sequence | AAA TCG CCA ATG ATG ACG ATT GGC TTT GCT CTG TCT GTG ATG AAC CGT ATG AGC ACT GAG GCG ATT T |
| sR8-T7-forward | CTA ATA CGA CTC ACT ATA GGC CAA ATC GCC AAT GAT GAC GAT TG |
| sR8.down | AAA TCG CCT CAG TGC TCA TAC GG |
| sR1 RNA | |
| Gene sequence | TGG CAG ATG ATG ACG TTT ATC CCC GTC TGA GTT ATG ATG AGT AGC AAG CCG GCT GAT GCC A |
| sR1-T7-forward | CTA ATA CGA CTC ACT ATA GGC CTG GCA GAT GAT GAC GTT TAT C |
| sR1-reverse | TGG CAT CAG CCG GCT TGC TAC |
| sR2 RNA | |
| Gene sequence | TGG CGG ATG ATG AAC GGA GTA GCT GCT GAG CTA TGA TGA TTG ATG GGC GAA CTG ACG CCA |
| sR2-T7-forward | CTA ATA CGA CTC ACT ATA GGC CTG GCG GAT GAT GAA CGG AGT AG |
| sR2-reverse | TGG CGT CAG TTC GCC CAT CAA TC |
| sR3 RNA | |
| Gene sequence | ATG GCA ATG ATG AAA AGA GGG TTA GCT GAA CTG TGA TGA TAC TTA CCC GAA CTG AGC CAT |
| sR3-T7-forward.B | CTA ATA CGA CTC ACT ATA GGC CAT GGC AAT G |
| sR3-reverse | ATG GCT CAG TTC GGG TAA GTA TC |
| sR5 RNA | |
| Gene sequence | TAA TTC CTC GAT GAT GAG CAA TAA AAA GCT GAC TTA ATA TGA TGA ACC TTT CGG GGT ATC TGA GAG GAA TTA |
| sR5-T7-forward.B | CTA ATA CGA CTC ACT ATA GGC CTA ATT CC |
| sR5-reverse | TAA TTC CTC TCA GAT ACC C |
| sR6 RNA | |
| Gene sequence | AAA CTG GCG ATG ATG ACA ATT TCG CTA TCT GAT TCT GTG ATG ACT ACT CCC GCA GCT GAG CCA GTT T |
| sR6-T7-forward | CTA ATA CGA CTC ACT ATA GGC CAA ACT GGC GAT GAT GAC AAT TT |
| sR6-reverse | AAA CTG GCT CAG CTG CGG GAG TA |
| sR7 RNA | |
| Gene sequence | TTT TAT GGG GAT GAT GAT ACA TCG ATG TGC TGA ATA TTG ATG ATG AAC GCG CCC TTC TCT GAC CTT TAA AA |
| sR7-T7-forward.B | CTA ATA CGA CTC ACT ATA GGC CTT TTA TGG |
| sR7-reverse | TTT TAA AGG TCA GAG AAG G |
The T7 promoter sequence is highlighted in italics
sRNP assembly and purification
sRNPs were assembled and purified by glycerol gradient centrifugation as described previously (14). Briefly, 10.5 µM of sRNA and 13 µM of each of the M. jannaschii box C/D proteins L7Ae, Nop5 (amino acids 1–367) and N-terminally FLAG-tagged fibrillarin were mixed and RNPs assembled at 75°C for 8 min in 20 mM HEPES pH 7.0, 500 mM NaCl, 1.5 mM MgCl2 and 10% glycerol. Assembly reactions were slowly cooled to room temperature and used for further analyses. For RNP purification on glycerol gradients, 250 µl of assembled sRNP was loaded on a 10–25% glycerol gradient (20 mM HEPES pH 7.0, 300 mM NaCl, 1.5 mM MgCl2) and centrifuged at 35 000 rpm in a SW41 rotor for 18 h at 4°C. Six-hundred microliter fractions were harvested and used for further analyses. Immunoprecipitations were performed as described in (14).
Native gel electrophoresis
An amount of 5–10 µl of assembled sRNPs was loaded on native polyacrylamide gels composed of a separating gel (6 or 8% polyacrylamide, 90 mM Tris–borate pH 9, 1.5 mM MgCl2) and a stacking gel (3.5% polyacrylamide, 50 mM Tris–HCl pH 6.8, 2.5% sucrose, 1.5 mM MgCl2). Complexes were resolved by electrophoresis in 90 mM Tris–borate buffer pH 9.0 in the presence of 1.5 mM MgCl2 at 4°C for 2 h at 150 V followed by 3 h at 300 V. For analysis of gradient fractions by native gel electrophoresis, 50 µl of each fraction was loaded. Resolved complexes were analyzed by silver staining.
Electron microscopy of the M. jannaschii sR6 sRNP
sR6 sRNP from peak glycerol gradient fractions were buffer exchanged into 20 mM HEPES pH 7.0, 500 mM NaCl, 1.5 mM MgCl2, 6% trehalose. An amount of 5 µl was applied to a glow discharged holey carbon EM grid coated with one layer of thin continuous carbon film. After 2 min incubation, the EM grid was stained in four consecutive drops of 2% uranyl formate, and an additional thin layer of carbon film was applied to EM grids prior to air-drying. EM grids were analyzed in a Tecnai T12 electron microscope with a LaB6 filament operated at 120 keV. EM images were recorded at a nominal magnification of 30 000 with a Gatan 1k × 1k CCD camera. In total, 2207 particles were boxed, low-pass filtered and subjected to reference-free alignment, followed by multivariate statistical analysis and classification. Representative class averages were chosen as references in subsequent iterative reference-based alignments, multivariate statistical analyses and classifications until class averages were stable. Image processing was performed with IMAGIC-5.
RESULTS
Previous studies of the role of conserved sRNA sequences in M. jannaschii sRNP assembly focused on six mutant sRNAs (8): four sRNAs that each contain mutations in conserved GA dinucleotides in the asymmetric bulges of the k-turn or k-loop motifs in the sR8 sRNA (Figure 1B) and two constructs that each generate an sR8 sRNA halfmer, one that contains only the box C/D motif and the corresponding D guide sequence (C/D halfmer; Figure 1C) and one that contains only the box C′/D′ motif and both guide sequences (C′/D′ halfmer; Figure 1D). Biochemical studies of these sRNAs have shown that binding of L7Ae to the box C/D and box C′/D′ motifs can occur independently and that abrogation of L7Ae binding by mutating the tandem sheared GA base pairs of one motif does not interfere with binding of L7Ae to the other motif (8). Furthermore, Nop5 and fibrillarin can still be recruited into these RNPs, most likely through the L7Ae bound to the intact k-turn or k-loop (8). Surprisingly however, the RNPs were found to be strongly diminished in their catalytic activity as compared to sRNPs assembled with the WT sR8 sRNA (8).
In light of the newly proposed di-sRNP model, we re-examined the role of the sRNA interfaces in di-sRNP assembly using the previously described six mutant sRNAs, all of which would be predicted to disrupt the sRNA interface in the dimeric box C/D sRNP. Here we test the hypothesis that the previously described diminished catalytic activity of these mutant sRNPs is due to inefficient di-sRNP formation because the sRNA interface is compromised in these mutant sRNPs. We thus determine whether there is a structural explanation for the previously observed biochemical results.
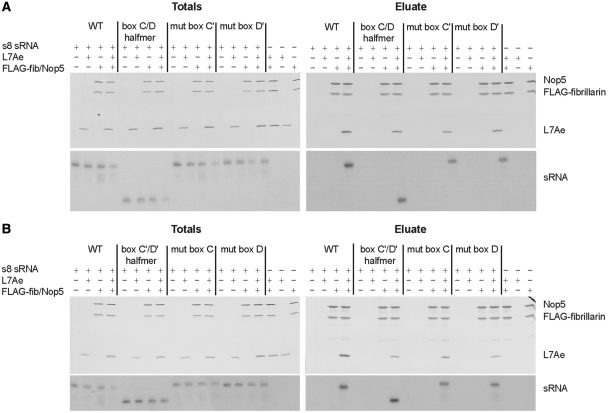
To analyze the RNPs assembled with mutant sRNAs, we initially confirmed that all mutant sRNAs can indeed assemble as expected with all common box C/D proteins under our experimental conditions. Mutant sRNAs were assembled into box C/D sRNPs where the fibrillarin protein was tagged with the FLAG epitope. Immunoprecipitations with anti-FLAG antibody were analyzed for the presence of the protein components on SDS–PAGE by silver staining and for the sRNA component by northern blotting (Figure 2). We found efficient immunoprecipitation of all core proteins and each of the six mutant sRNAs in reactions containing all sRNP components but not in control reactions lacking one sRNP component. This indicated that all mutant sRNAs could still assemble into an RNP. These results are consistent with previous observations by Tran et al. where it was shown that all mutant sRNAs assemble into RNPs by electrophoretic gel mobility shift assays (8). However, neither our assay nor theirs provides information on the size of the assembled RNPs and the stoichiometry of their components, and hence it remains unresolved whether the assembled sRNPs consisting of mutant sRNAs form di-sRNPs similar in architecture to WT sRNPs.
Figure 2.
Mutant sR8 sRNAs assemble efficiently with all core box C/D proteins into RNPs. WT and mutant sR8 sRNAs [C/D halfmer, mut box C′, and mut box D′ in (A) and box C′/D′ halfmer, mut box C, and mut box D in (B)] were incubated with all box C/D proteins and subjected to immunoprecipitation using anti-FLAG antibodies. Proteins in assembly reactions (Totals) and eluates from beads after immunoprecipitation were separated by SDS–PAGE and visualized by silver staining. RNAs were extracted, separated by gel electrophoresis, and visualized by northern blotting.
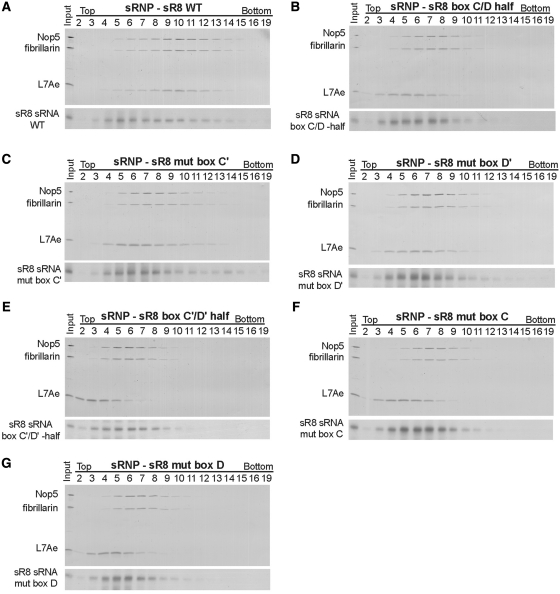
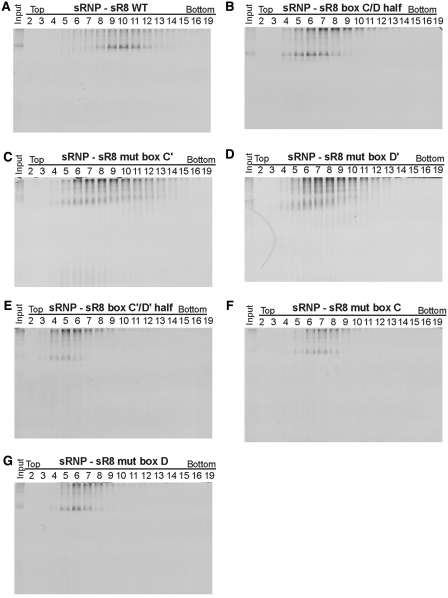
To investigate the mutant RNPs in more detail and to obtain information on their sizes, we subjected them to glycerol gradient centrifugation. Di-sRNPs assembled with unmutated sR8 sRNA migrated around peak fraction 10 (Figure 3A) (14). In contrast, peak fractions of the components of sRNPs assembled with any of the sRNAs containing mutations in conserved sequence elements, or with halfmer RNAs, were shifted towards the top of the gradient, suggesting smaller complexes different from the di-sRNP (Figure 3B–G). These results could reflect either instability of the initially assembled di-sRNPs during the extensive time required for glycerol gradient centrifugation or a decrease in the efficiency of di-sRNP assembly.
Figure 3.
Intact C/D and C′/D′ motifs are both required for stable di-sRNP assembly. WT sR8 sRNA (A), sR8 sRNAs lacking an intact C′/D′ motif [box C/D half (B), mut box C′ (C) and mut box D′ (D)], and sR8 sRNAs lacking an intact C/D motif [box C′/D′ half (E), mut box C (F) and mut box D (G)] were incubated with L7Ae, Nop5, and fibrillarin and assembled RNPs were separated on 10–25% glycerol gradients. Proteins and RNA in harvested fractions were separated by denaturing polyacrylamide gel electrophoresis and visualized by silver staining and northern blotting, respectively.
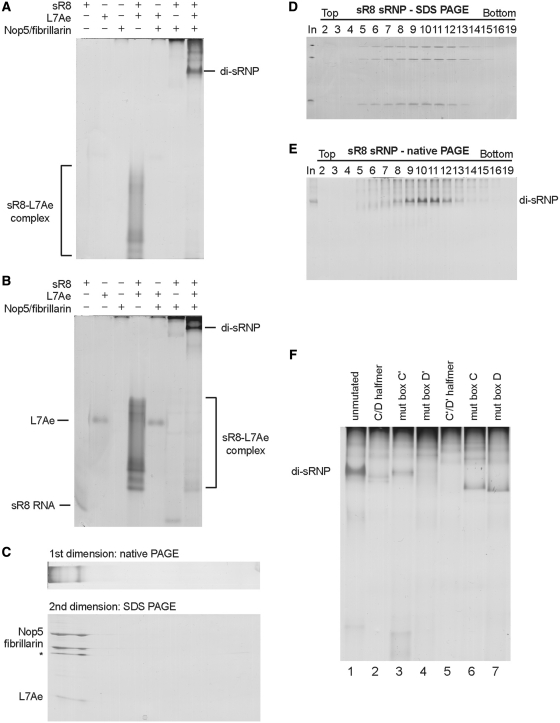
To distinguish between these possibilities, we utilized a native gel electrophoretic assay (native PAGE) to follow di-sRNP assembly. In contrast to the previously used electrophoretic gel mobility shift assay, which only reports differences in the mobility of labeled sRNAs, we visualize the biological complexes by silver staining. Since both RNAs and proteins are stained with this method, our results are not selectively biased towards RNA-containing complexes but reflect detection of the most abundant macromolecules. Analysis of fully assembled sR8 box C/D sRNP by this assay demonstrates that the RNP migrates as a predominant band in both 6 and 8% native polyacrylamide gels (Figure 4A and B). This band is not observed with sRNP components individually or in different combinations with each other, indicating that this band is specific to the fully assembled sRNP. While the sR8 sRNA and L7Ae enter native gels by themselves, the Nop5–fibrillarin complex does not do so very efficiently, suggesting that the net charge in its native form prevents migration towards the anode. Subsequent SDS–PAGE analysis in the second dimension after 6% native PAGE confirms that the RNP complex observed in native gels contains all three box C/D core proteins (Figure 4C). To provide evidence that the major band that we detect corresponds to the di-sRNP, we purified sRNPs by glycerol gradient centrifugation and analyzed fractions by both SDS–PAGE and native PAGE, followed by detection by silver staining (Figure 4D and E). As observed previously (14), the di-sRNP migrates in peak fractions 10+11 (Figure 4D). Importantly, when the glycerol gradient fractions are analyzed by native PAGE, a single band is observed in the peak fractions (Figure 4E). This band also contains the sRNA which was detected by northern blotting after native PAGE (data not shown). This demonstrates that the predominant band observed in native gels after silver staining corresponds to the di-sRNP. Thus, the native gel electrophoretic assay in combination with silver staining is a valid method to assess box C/D di-sRNP assembly quickly and accurately.
Figure 4.
Native gel electrophoresis (PAGE) can be used to follow di-sRNP assembly and demonstrates that the sRNA mutants do not assemble efficiently into di-sRNPs. Native gel electrophoresis of sRNP components individually and in different combinations with each other were separated on 6% in (A) or 8% in (B) native polyacrylamide gels. Proteins and RNA were visualized by silver staining. (C) The sRNP complex observed on native gels contains all sRNP core proteins. Assembled sR8 sRNP was separated by 6% native PAGE initially in the first dimension and then the entire lane was loaded onto a SDS–PAGE gel to separate individual protein components. sRNP components and complexes were visualized by silver staining. Incomplete denaturation of the sRNA–L7Ae complex results in a fourth band after SDS–PAGE which is indicated by an asterisk. (D) and (E) sR8 sRNPs were separated on 10–25% glycerol gradients and harvested fractions were analyzed by both SDS–PAGE and 6% native PAGE. The complex observed on native gels coincides with the migration of the di-sRNP on glycerol gradients, indicating it corresponds to the di-sRNP. (F) sRNPs were assembled with either WT or mutant sR8 sRNAs as indicated and separated on 6% native polyacrylamide gels followed by silver staining.
Having established that native PAGE and silver staining can be used to study di-sRNP assembly, we asked whether the mutant sRNPs form di-sRNPs using this method (Figure 4F). Box C/D sRNPs reconstituted with WT sRNA, which are di-sRNPs, were abundant and easily detected on native gels (Figure 4F, lane 1). In contrast, mutant sRNAs did not assemble into this complex (C/D half, mut box D′, C′/D′ half, mut box C, mut box D in Figure 4F, lanes 2, 4, 5, 6 and 7) or very inefficiently (mut box C′ in Figure 4F, lane 3). sR8 mut box C, sR8 mut box D, and sR8 C/D halfmer sRNAs formed complexes with the core box C/D proteins that migrated faster than the di-sRNP on native gels, indicating a different composition of these RNPs when compared to the di-sRNP.
Analysis of glycerol gradient fractions of mutant sRNPs by native PAGE revealed that the observed RNPs were indeed smaller than the di-sRNP assembled with the WT sR8 RNA (Figure 5A–G). As also observed in Figure 3, the mutant sRNPs sedimented in peak fractions closer to the top of the gradient, indicating a smaller size. In addition, in every case, the mobility of the predominant species from the peak fractions on native PAGE was different from that of the di-sRNP (compare Figure 5A with Figure 5B–G). These complexes observed by native PAGE did not correspond to core proteins alone (compare with Figure 4A and B; data not shown). The low abundance of some of the mutant sRNPs could indicate that the assembled RNPs were very heterogeneous and that the individual complexes were below the limits of detection, or that these complexes were extremely unstable. Complex instability and heterogeneity of mutant sRNPs was also supported by the glycerol gradient sedimentation profiles on denaturing PAGE, which revealed that not all sRNP components completely co-migrated with each other (Figure 3). Alternatively, we cannot exclude that mutant sRNPs were not of a sufficient negative charge to migrate towards the anode due to a high pI, which is determined by the exact RNA and protein composition of the RNP. Nevertheless, taken together, the results demonstrate that both an intact C/D and an intact C′/D′ motif are essential for di-sRNP assembly.
Figure 5.
Native PAGE analysis of mutant sR8 RNPs after glycerol gradient centrifugation reveals heterogeneous RNP complexes. sRNPs were assembled with wild type (A) or mutant sR8 sRNAs as indicated (B–G) and all core box C/D proteins and separated by centrifugation in 10–25% glycerol gradients. Harvested fractions were analyzed by electrophoresis on 6% native gels and silver staining.
To visualize sRNPs assembled with mutant sRNAs directly, we analyzed them by electron microscopy (data not shown). Consistent with our biochemical results, particles observed by electron microscopy were smaller than the di-sRNP assembled with WT sRNA but also showed a substantial degree of heterogeneity.
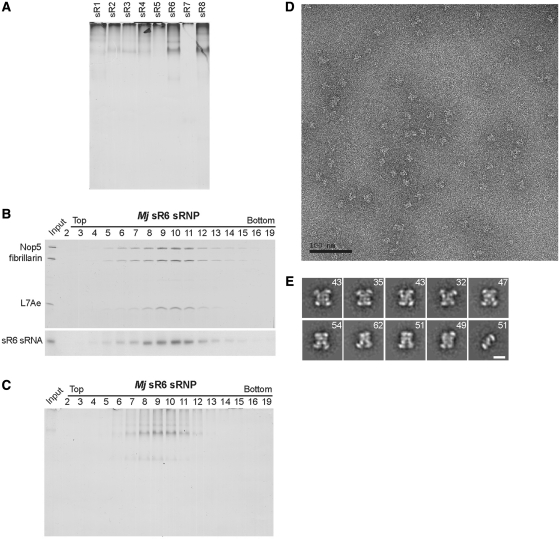
Bioinformatic analysis aimed at identifying archaeal box C/D sRNAs predicts that there are eight different box C/D sRNPs in Methanocaldococcus jannaschii (an alignment can be found at http://lowelab.ucsc.edu/snoRNAdb/Archaea/Mja-align.html, 15). To date, most biochemical studies with M. jannaschii methylation guide sRNPs, as well as our structural studies, have been performed with sRNPs assembled with the sR8 sRNA (8,14,16–19). The only other M. jannaschii box C/D sRNP that had been previously assembled in vitro and shown to be enzymatically active is the sR6 sRNP (17). To test whether the di-sRNP architecture is conserved among M. jannaschii box C/D sRNPs, we attempted to reconstitute all predicted M. jannaschii box C/D sRNPs in vitro. Native gel electrophoresis of reconstituted sRNPs with all bioinformatically predicted M. jannaschii box C/D sRNAs suggested that, besides sR8, the sR6 sRNA efficiently assembled into RNPs with similar electrophoretic mobility as the sR8 sRNP (Figure 6A). In contrast, the remaining box C/D sRNAs assembled only very inefficiently into box C/D sRNPs. The low abundant complexes observed with sR2-sR4 sRNAs migrated similarly to the sR8 sRNP in native gels, indicating that they may also be di-sRNPs.
Figure 6.
The M. jannaschii sR6 sRNP also assembles into a di-sRNP. (A) Box C/D sRNPs with different predicted M. jannaschii box C/D sRNAs assemble with different efficiency. All predicted M. jannaschii sRNAs (sR1–sR8) were in vitro transcribed and assembled with the M. jannaschii core box C/D proteins in vitro. sRNP assembly was analyzed by native gel electrophoresis and silver staining. (B) The sR6 sRNP was reconstituted and purified on 10–25% glycerol gradients. Indicated fractions were analyzed by SDS–PAGE and silver staining for the presence of box C/D sRNP protein components and by northern blotting for the presence of the sR6 sRNA. (C) Fractions from (B) were also analyzed by electrophoresis on 6% native gels and subsequent silver staining. (D) Electron micrograph of negatively stained sR6 sRNP particles from the peak gradient fraction. (F) Experimental class averages of the sR6 sRNP. The number of images averaged in each class is indicated. The last class average most likely corresponds to contaminating free Nop5–fibrillarin heterotetrameric complex. Scale bar is 10 nm.
Since the sR6 sRNPs efficiently assembled in vitro, we investigated their architecture in more detail. To do that, we purified the M. jannaschii sR6 sRNP by glycerol gradient centrifugation and analyzed harvested fractions by both SDS–PAGE (Figure 6B) and native PAGE (Figure 6C). sRNP components as well as the assembled RNP migrate in peak fractions 9–11, which is similar to what was observed with the sR8 sRNP (14), supporting that the sR6 sRNP is of similar size as the sR8 di-sRNP. We next subjected sR6 sRNPs from the peak gradient fraction to electron microscopic studies. Electron micrographs of negatively stained sR6 sRNP particles (Figure 6D) and class averages (Figure 6E) showed that sR6 sRNP particles have similar dimensions and structural features as the previously analyzed sR8 sRNP particles (14). These results are consistent with a di-sRNP architecture for sR6 sRNPs, suggesting that the di-sRNP structure is not restricted to the M. jannaschii sR8 sRNP but rather that it is conserved among different M. jannaschii methylation guide sRNPs.
DISCUSSION
Recently, we determined the first structure of a fully assembled and catalytically active archaeal box C/D sRNP (14). It suggested that these RNPs are dimeric RNPs, contradicting a long-standing monomeric model for box C/D sRNP architecture (14,20). However, questions regarding the functional relevance and conservation of this di-sRNP structure remained unresolved. Here we have analyzed the sRNAs containing mutations in conserved sequence elements, correlating di-sRNP assembly with efficient methylation activity. We also provide evidence that the di-sRNP architecture is conserved among archaeal box C/D sRNPs from the same species.
Our results demonstrate that di-sRNP formation depends on two native k-turn/k-loop motifs within a single box C/D sRNA. While abrogation of one of these motifs continues to allow association of all core box C/D proteins with the sRNA to form an RNP, these RNPs are of different composition and smaller than the di-sRNPs assembled with the WT sRNA. However, the copy number of the sRNA and protein components in these mutant sRNPs remains unknown. Hence, mutant sRNPs could still contain two mutant sRNAs but less than four sets of the core proteins, resulting in architecture different from that of the di-sRNP. These results are expected based on the di-sRNP model that predicts that impeding L7Ae binding to one of the motifs would destroy the sRNA interface, resulting in sRNPs that are about half the size of the di-sRNP. In contrast, this change in size of RNPs assembled on mutant sRNAs is not reconcilable with the conventional mono-sRNP model because in this model the sRNA interface is not independent from the Nop5 coiled-coil domain interface.
Previous studies of the exact same sRNA mutants used in this study found that the assembled RNPs are diminished in their catalytic activity compared to sRNPs assembled with the WT sR8 sRNA (8). Most importantly, we observe that the mutant M. jannaschii sR8 sRNAs do not assemble into di-sRNPs, suggesting di-sRNP assembly is important for efficient enzymatic activity (Table 2). The consequences of mutations in the conserved sequence elements of box C/D sRNAs on catalytic activity are not specific to the M. jannaschii sR8 mutant sRNA but have also been observed with box C/D or box C′/D′ mutant sRNAs from other archaeal species (9,21–23).
Table 2.
The ability of mutant sRNAs to assemble into di-sRNPs correlates with efficient methylation activity
| sR8 RNA mutant | Enzymatic activity (as percentage of WT sR8 sRNP activity for D/D′ targets; from ref. 8) | RNP formation [pulldowns in Figure 2 and (8)] | di-sRNP formation (Figures 3–5) |
|---|---|---|---|
| C/D halfmer | 34%/n.a. | + | ↓ |
| C′/D′ halfmer | n.a./0% | + | ↓ |
| mut box C | 25%/5% | + | ↓ |
| mut box D | 2%/58% | + | ↓ |
| mut box C′ | 0%/45% | + | ↓ |
| mut box D′ | 27%/5% | + | ↓ |
Note that the term ‘di-sRNP’ refers to an RNP containing two sRNAs and four sets of each core protein. Methylation activity results are taken from ref. (8).
Previous interpretations of the diminished enzymatic activity of sRNA mutant box C/D sRNPs in light of the conventional mono-sRNP model proposed that mutation of one of the conserved motifs resulted in an asymmetric sRNP lacking one L7Ae. This was further proposed to interfere with crosstalk between the C/D and C′/D′ motifs (8,9). In contrast, our results here provide an alternative explanation for the impaired enzymatic activity of these sRNPs and suggest that it is caused by lack of or by inefficient di-sRNP assembly. The inability of the 6 sRNA mutants to assemble into the di-sRNP correlates with a decrease in their 2′-O-ribose methylation activity (Table 2). We propose that the reduced catalytic activity of these mutant box C/D sRNPs is caused by altered positions or conformations of the guide sequences in these mutant RNPs. The predicted greater flexibility of the guide sequences expected in mutant sRNPs may decrease the likelihood for the guide:substrate RNA duplex to be correctly positioned with respect to the active site of fibrillarin. Consistent with this interpretation, chemical probing studies of box C/D or box C′/D′ mutant sRNPs showed altered protection patterns in their guide sequences (24).
Analyses of all predicted M. jannaschii sRNAs for RNP assembly revealed that they assemble with different efficiencies into sRNPs, with sR6 and sR8 sRNAs being the most efficient. The different behavior of different M. jannaschii sRNAs in RNP assembly is somewhat surprising considering that all predicted sRNAs contain the conserved sequence elements and only differ in the sequences of their guide regions and in the sequences and lengths of their terminal stems (an alignment of all M. jannaschii sRNAs can be found at http://lowelab.ucsc.edu/snoRNAdb/Archaea/Mja-align.html). While at this point it is unknown whether these predicted M. jannaschii sRNAs are bona fide box C/D sRNAs in vivo, the in vitro results indicate that additional regions beyond the conserved motifs (boxes C, D, C′, and D′) may play a role in box C/D sRNP assembly, probably by mediating RNA-protein interactions or by stabilizing particular RNA structures. Higher resolution structures of box C/D sRNPs will be needed to test this hypothesis.
Gagnon et al. previously showed that in vitro reconstituted M. jannaschii sR6 sRNPs are catalytically active in guiding the 2′-O-ribose methylation of substrate RNAs (17). Results of native PAGE, glycerol gradient centrifugation and EM analyses suggest that the sR6 sRNP is also a di-sRNP. Importantly, 2D image analysis further indicates that the RNP architecture previously observed in the sR8 sRNP is maintained in the sR6 sRNP (14). The assembly of the M. jannaschii sR6 sRNA into di-sRNPs and the fact that all core proteins and important sRNA elements are conserved in other archaeal species makes it very likely that the di-sRNP structure is a general feature of methylation guide sRNPs.
Collectively, the results presented here obtained with mutant sRNAs and the previously published studies with Nop5 containing mutations in the coiled-coil domain (14) demonstrate that both di-sRNP interfaces (Figure 1A) are important for di-sRNP formation. Furthermore, the conservation of the di-sRNP architecture in other archaeal box C/D sRNPs from the same species, as well as the correlation between structure and function, both underline the importance of the formation of the di-sRNP structure for efficient methylation activity of the box C/D sRNP, and hence the relevance for their function. These results provide an explanation for previous biochemical results and, more importantly, provide a framework for future studies deciphering the mechanism of box C/D sRNP action.
FUNDING
National Institute of Health (grant GM52581 to S.J.B); Boehringer Ingelheim Fonds PhD scholarship (to F.B.). Funding for open access charge: National Institutes of Health R01.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Keith Gagnon and Stuart Maxwell for providing the template and oligonucleotides to clone M. jannaschii sR4 and sR6 sRNAs.
REFERENCES
- 1.Reichow SL, Hamma T, Ferre-D'Amare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhn JF, Tran EJ, Maxwell ES. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res. 2002;30:931–941. doi: 10.1093/nar/30.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Luhrmann R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 4.Nolivos S, Carpousis AJ, Clouet-d'Orval B. The K-loop, a general feature of the Pyrococcus C/D guide RNAs, is an RNA structural motif related to the K-turn. Nucleic Acids Res. 2005;33:6507–6514. doi: 10.1093/nar/gki962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 6.Moore T, Zhang Y, Fenley MO, Li H. Molecular basis of box C/D RNA-protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004;12:807–818. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 7.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran EJ, Zhang X, Maxwell ES. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C'/D' RNPs. EMBO J. 2003;22:3930–3940. doi: 10.1093/emboj/cdg368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rashid R, Aittaleb M, Chen Q, Spiegel K, Demeler B, Li H. Functional requirement for symmetric assembly of archaeal box C/D small ribonucleoprotein particles. J. Mol. Biol. 2003;333:295–306. doi: 10.1016/j.jmb.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Szewczak LB, DeGregorio SJ, Strobel SA, Steitz JA. Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 2002;9:1095–1107. doi: 10.1016/s1074-5521(02)00239-9. [DOI] [PubMed] [Google Scholar]
- 11.Szewczak LB, Gabrielsen JS, Degregorio SJ, Strobel SA, Steitz JA. Molecular basis for RNA kink-turn recognition by the h15.5K small RNP protein. RNA. 2005;11:1407–1419. doi: 10.1261/rna.2830905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Omer AD, Ziesche S, Ebhardt H, Dennis PP. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl Acad. Sci. USA. 2002;99:5289–5294. doi: 10.1073/pnas.082101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watkins NJ, Dickmanns A, Luhrmann R. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 2002;22:8342–8352. doi: 10.1128/MCB.22.23.8342-8352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bleichert F, Gagnon KT, Brown BA, 2nd, Maxwell ES, Leschziner AE, Unger VM, Baserga SJ. A dimeric structure for archaeal box C/D small ribonucleoproteins. Science. 2009;325:1384–1387. doi: 10.1126/science.1176099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Omer AD, Lowe TM, Russell AG, Ebhardt H, Eddy SR, Dennis PP. Homologs of small nucleolar RNAs in Archaea. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- 16.Tran E, Zhang X, Lackey L, Maxwell ES. Conserved spacing between the box C/D and C'/D' RNPs of the archaeal box C/D sRNP complex is required for efficient 2′-O-methylation of target RNAs. RNA. 2005;11:285–293. doi: 10.1261/rna.7223405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gagnon KT, Zhang X, Agris PF, Maxwell ES. Assembly of the archaeal box C/D sRNP can occur via alternative pathways and requires temperature-facilitated sRNA remodeling. J. Mol. Biol. 2006;362:1025–1042. doi: 10.1016/j.jmb.2006.07.091. [DOI] [PubMed] [Google Scholar]
- 18.Appel CD, Maxwell ES. Structural features of the guide:target RNA duplex required for archaeal box C/D sRNA-guided nucleotide 2′-O-methylation. RNA. 2007;13:899–911. doi: 10.1261/rna.517307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gagnon KT, Zhang X, Qu G, Biswas S, Suryadi J, Brown BA, 2nd, Maxwell ES. Signature amino acids enable the archaeal L7Ae box C/D RNP core protein to recognize and bind the K-loop RNA motif. RNA. 2010;16:79–90. doi: 10.1261/rna.1692310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aittaleb M, Rashid R, Chen Q, Palmer JR, Daniels CJ, Li H. Structure and function of archaeal box C/D sRNP core proteins. Nat. Struct. Biol. 2003;10:256–263. doi: 10.1038/nsb905. [DOI] [PubMed] [Google Scholar]
- 21.Omer AD, Zago M, Chang A, Dennis PP. Probing the structure and function of an archaeal C/D-box methylation guide sRNA. RNA. 2006;12:1708–1720. doi: 10.1261/rna.31506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bortolin ML, Bachellerie JP, Clouet-d'Orval B. In vitro RNP assembly and methylation guide activity of an unusual box C/D RNA, cis-acting archaeal pre-tRNA(Trp) Nucleic Acids Res. 2003;31:6524–6535. doi: 10.1093/nar/gkg860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardin JW, Batey RT. The bipartite architecture of the sRNA in an archaeal box C/D complex is a primary determinant of specificity. Nucleic Acids Res. 2006;34:5039–5051. doi: 10.1093/nar/gkl644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh SK, Gurha P, Gupta R. Dynamic guide-target interactions contribute to sequential 2′-O-methylation by a unique archaeal dual guide box C/D sRNP. RNA. 2008;14:1411–1423. doi: 10.1261/rna.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]