Abstract
The transcription-repair coupling factor (TRCF, the product of the mfd gene) is a widely conserved bacterial protein that mediates transcription-coupled DNA repair. TRCF uses its ATP-dependent DNA translocase activity to remove transcription complexes stalled at sites of DNA damage, and stimulates repair by recruiting components of the nucleotide excision repair pathway to the site. A protein/protein interaction between TRCF and the β-subunit of RNA polymerase (RNAP) is essential for TRCF function. CarD (also called CdnL), an essential regulator of rRNA transcription in Mycobacterium tuberculosis, shares a homologous RNAP interacting domain with TRCF and also interacts with the RNAP β-subunit. We determined the 2.9-Å resolution X-ray crystal structure of the RNAP interacting domain of TRCF complexed with the RNAP-β1 domain, which harbors the TRCF interaction determinants. The structure reveals details of the TRCF/RNAP protein/protein interface, providing a basis for the design and interpretation of experiments probing TRCF, and by homology CarD, function and interactions with the RNAP.
INTRODUCTION
Transcribing RNA polymerase (RNAP) molecules stalled at sites of DNA damage elicit preferential repair of the DNA in a process called transcription-coupled repair (TCR) (1,2). Paradoxically, the stalled RNAP molecules are inhibitory to DNA repair in vitro (3), pointing to the role of additional factors mediating TCR in vivo. In bacteria, transcription-repair coupling factor (TRCF), the product of the mfd gene (4,5), was shown to be necessary and sufficient for TCR in vivo and in vitro. TRCF plays two key roles in mediating TCR: (i) relief of transcription-dependent inhibition of nucleotide excision repair (NER) by recognition and ATP-dependent removal of a stalled RNAP covering the damaged DNA and (ii) stimulation of DNA repair by recruitment of the Uvr(A)BC endonuclease (3,6,7)
TRCF is a large (130 kDa), evolutionarily conserved, multi-functional protein with a complex structure/function relationship. The 3.2-Å resolution X-ray crystal structure of Escherichia coli (Eco) TRCF comprises a compact arrangement of eight structured domains [D1a, D1b and D2–D7) linked by flexible linkers (8); Figure 1A]. These domains are arranged in functional modules that perform the various TRCF functions. Recruitment of the NER machinery through binding of the NER component UvrA is accomplished by the TRCF UvrB homology module (D1a, D1b and D2) (6,8–10). RNAP binding is mediated by the TRCF RNAP interacting domain [RID; (8)]. The ATP-dependent double-stranded DNA translocase activity is due to the translocation module [TD1 and TD2; (8)], which contains the seven signature sequence motifs of superfamily 2 helicases/ATPases (11). All of these activities are repressed in isolated TRCF through an interdomain interaction between D2 and the C-terminal D7 domain (12,13).
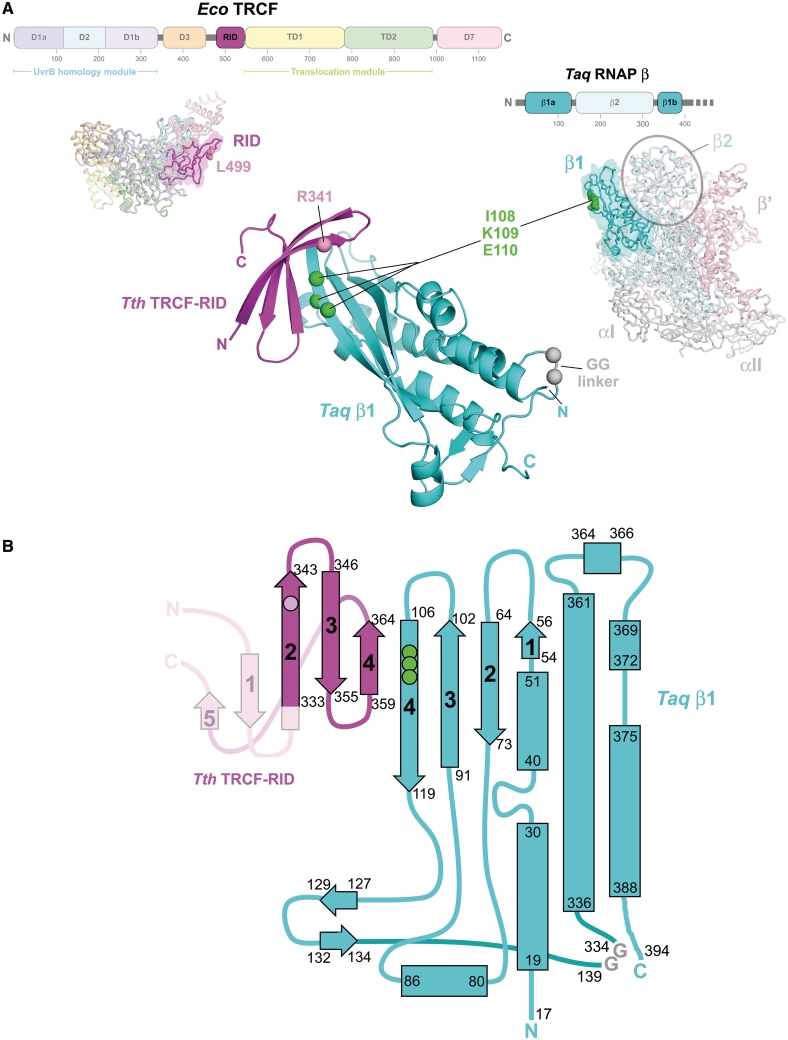
Figure 1.
The TRCF–RID/RNAP-β1 complex. (A) Structural context of the TRCF–RID and the RNAP-β1 domains. (Upper left) The horizontal bar represents the 1148-residue Eco TRCF primary sequence (every 100 residues are marked below the bar). Structural domains are represented as thick, colored bars; thin black bars represent linkers connecting the domains (8). The RID is highlighted and colored magenta. Below the bar, a side view of the structure of repressed Eco TRCF (8) is shown as an α-carbon backbone, with the domains color-coded as in the bar above. The RID (Eco TRCF479–545, colored magenta) is highlighted with a transparent molecular surface. L499 is shown as pink CPK atoms; an L499R substitution at this position disrupts the TRCF–RID/RNAP protein/protein interaction (8). (Upper right) The horizontal bar represents the N-terminal 400 residues of the Taq RNAP β-subunit primary sequence (every 100 residues are marked below the bar). Structural domains are represented as thick, colored bars, thin black bars represent linkers connecting the domains (25,26). The two sequence elements comprising the β1 domain (β1a, residues 17–139, and β1b, residues 334–395), which flank the β2 domain, are highlighted and colored cyan. Below the bar, the β-side view of the structure of Taq core RNAP (26) is shown as an α-carbon backbone and with the subunits color-coded as follows: αI, αII, gray; β, pale cyan, except the β1 domain is cyan; β′, pale pink. The β1 domain is also highlighted with a transparent molecular surface. The β2 domain is also denoted. β residues I108, K109, and I110 are shown as green CPK atoms; substitutions at corresponding positions in the Eco RNAP β-subunit (Eco RNAP-βI117,K118,E119) cause defects in the TRCF/RNAP protein/protein interaction (8,14). (Middle) The structure of the Tth TRCF–RID/ Taq RNAP-β1 complex, shown as a ribbon diagram (Tth TRCF–RID, magenta; Taq RNAP-β1, cyan). In the TRCF–RID, the α-carbon of R341 is shown as a pink sphere; Tth TRCF–RIDR341 corresponds to Eco TRCF–RIDL499, which has been shown to be involved in the TRCF–RID/RNAP protein/protein interface (8). In the RNAP-β1, the α-carbons of I108, K109 and E110 are shown as green spheres; Taq RNAP-β1I108,K109,E110 correspond to Eco RNAP-β1I117,K118,E119, which have been shown to be involved in the TRCF–RID/RNAP protein/protein interface (8,14). The –Gly–Gly– linker, introduced into the construct to connect β1a and β1b, is colored grey and shown as α-carbon spheres. (B) Topology diagram of the Tth TRCF–RID/ Taq RNAP-β1 complex; α-helices are shown as rectangles, β-strands as arrows. The start- and end-residues for each secondary structural element are shown. The β-strands involved in the intermolecular β-sheet formed in the complex are numbered for each protein. The TRCF–RID is colored magenta, but β-strand 1 and β-strand 5, disordered in all four heterodimers of the crystallographic asymmetric unit, are shown in pink. The location of the interface residue R341 is shown as a pink circle. The RNAP-β1 domain is colored cyan. The locations of the interface residues I108, K109 and E110 are shown as green circles.
Critical among the TRCF functional modules is the RID, a Tudor-like domain that mediates protein/protein interactions between TRCF and RNAP that are essential for the RNAP release function (8,14) (Figure 1A). The interaction between the TRCF–RID and RNAP: (i) recruits TRCF to the site of the stalled RNAP; (ii) is thought to trigger the conformational changes associated with derepression of the TRCF activities (8) and (iii) provides an anchor for TRCF on RNAP, allowing the TRCF DNA-tracking translocation activity to exert forces on the DNA that are thought to cause collapse of the transcription bubble within the RNAP ternary elongation complex (TEC), resulting in RNAP and transcript release (15).
Taken together, biochemical, as well as yeast and bacterial two-hybrid analyses have identified an RNAP β-subunit segment, Eco β residues 19–142 (β19–142), as being sufficient for TRCF–RID interaction (8,14,16). Amino acid substitutions in both the TRCF–RID (Eco TRCFL499R) and in the RNAP β-subunit (Eco β I117A, K118A or E119A) disrupt the TRCF/RNAP protein/protein interaction and cause defects in the RNAP release activity of TRCF (8,14).
The TRCF–RID is homologous to a domain widely distributed in bacteria (17) and grouped as the CarD_TRCF protein family (Pfam: PF02559). CarD (also called CdnL) (18) is an essential regulator of rRNA transcription in Mycobacterium tuberculosis (Mtb) that is upregulated in the general stress response and plays a key role in persistence and pathogenesis (19). The CarD N-terminal domain (NTD) shares striking sequence homology with the TRCF–RID and interacts with the RNAP β-subunit in the same manner (18,19).
In this work, we determined a 2.9-Å resolution X-ray crystal structure of a complex between the Thermus thermophilus TRCF–RID and the T. aquaticus (Taq) RNAP β-subunit β1 domain (which harbors the TRCF–RID interaction determinant). The details of the protein/protein interface explain the effects of amino acid substitutions in both the TRCF–RID (8) and the RNAP-β1 domain (8,14) that cause defects in the protein/protein interaction, and guides the design of new substitutions tested herein to further elucidate the interaction. The structure also reveals a local conformational change in the β1 domain upon binding the TRCF–RID. This work provides a basis for the design and interpretation of experiments probing TRCF function and interactions with the RNAP, as well as the function and RNAP interactions of other members of the CarD_TRCF protein family.
MATERIALS AND METHODS
Construction of expression plasmids for Tth TRCF–RID and Taq RNAP-β1
The DNA encoding Tth HB27 TRCF–RID (residues 321–387) was amplified by the polymerase chain reaction (PCR) using primers that appended NdeI and HindIII sites at the 5′- and 3′-ends, respectively. The PCR-amplified DNA fragment was cleaved with NdeI and HindIII and cloned between the NdeI and HindIII sites of a pET28a-derived plasmid, creating pET28a TthHB27(His)6MfdRID. PCR was used to amplify and fuse the DNA encoding Taq β1a (Taq β17–139) and β1b (Taq β334–395). Primers were designed to introduce a –Gly–Gly– linker between the two β1 segments and NdeI and BamHI restriction endonuclease sites were appended to the 5′- and 3′-ends of the fused DNA fragment, respectively. The resulting DNA fragment was cleaved with NdeI and BamHI and cloned between the same sites of pET21a (Novagen), creating pET21a Taq β1. All DNA manipulations were confirmed by DNA sequencing.
Expression and purification of the TthTRCF–RID/Taq RNAP-β1 complex
The plasmids pET21a Taq β1 and pET28aTth HB27Mfd-RID were transformed simultaneously into Eco BL21 (DE3) cells (Novagen) and transformants were grown at 37°C in Luria–Bertani media supplemented with ampicillin (200 µg/ml) and kanamycin (50 µg/ml) to an A650 nm between 0.6 and 0.8. Subsequently, ampicillin (100 µg/ml) and isopropyl-β,d-thiogalactopyranoside (1 mM final concentration) were added to the culture. After incubation at 37°C for 3 h, the cells were harvested by centrifugation, resuspended in buffer A [20 mM Tris–HCl (pH 8.0 at 4°C), 200 mM NaCl, 5% (v/v) glycerol, 0.5 mM β-mercaptoethanol], lysed using a continuous-flow homogenizer (Avestin), and then centrifuged to remove insoluble debris. The clarified cell lysate was applied to a Ni2+-charged HiTrap column (GE Healthcare) equilibrated in buffer A + 5 mM imidazole. The column was washed with five column volumes (cv) of buffer A + 20 mM imidazole, 5 cv buffer A + 40 mM imidazole, 5 cv buffer A + 60 mM imidazole and finally 5 cv buffer A + 80 mM imidazole. Proteins bound to the column were eluted with buffer A + 250 mM imidazole. After overnight cleavage with PreScission protease (GE Healthcare) to remove the (His)6-tag and dialysis against buffer A + 5 mM imidazole, a subtractive Ni2+-chelating chromatographic step removed uncleaved (His)6TRCF–RID and the cleaved (His)6-tag. The sample was concentrated and applied to a Superdex 75 gel filtration column (GE Healthcare). Finally, the purified sample was concentrated to 10.6 mg/ml and exchanged into storage buffer (10 mM Tris–HCl [pH 8.0 at 4°C], 100 mM NaCl, 5 mM DTT). The purity of the complex was judged to be >95% as analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis and Coomassie blue staining (data not shown).
Crystallization and structure determination
Crystals were grown by hanging-drop vapor diffusion by mixing 2 µl protein solution (10–12 mg/ml in storage buffer) with 1 µl crystallization solution (0.1 M Tris–HCl [pH 7.5 at 22°C], 1.6 M di-potassium ammonium phosphate), and incubating over a well containing crystallization solution. Crystals (10–20 µm octahedra) grew in ∼1 week. The crystals were prepared for cryo-crystallography by stepwise transfer into 0.1 M Tris–HCl [pH 7.5 at 22°C], 100 mM NaCl, 7 M ammonium formate, then frozen in liquid nitrogen. X-ray diffraction data sets (Table 1) were collected at the Advanced Photon Source (Argonne National Laboratory) NE-CAT 24-ID-E beamline, using an MD-2 microdiffractometer with a 20 µm aperture. Because of the small size of the crystals, the data quality degraded before a full data set from a single crystal could be collected, but partial data sets were collected from nine separate crystals. The crystals appeared to belong to space group P43212, but subsequent analysis indicated the crystals were hemihedrally twinned and belonged to space group P43. One data set was chosen as a reference (crystal a, Table 1). The additional data sets were processed in two orientations (h, k, l and −h, k, −l), and were then combined, one at a time, and kept or discarded, depending on whether the overall data improved, resulting in the final combined data set from partial data sets collected from three separate crystals (Table 1).
Table 1.
Data collection and refinement statistics (molecular replacement)
| a | b | c | Comb (a,b,c) | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P43a | P43a | P43a | P43a |
| Cell dimensions | ||||
| a/b, c (Å) | 106.55, 122.23 | 106.19, 120.76 | 106.32, 120.74 | 106.59, 122.30 |
| Resolution (Å) | 30.0−2.90 (3.00−2.90) | 30.0−2.90 (3.00−2.90) | 30.0−2.90 (3.00−2.90) | 30.0−2.90 (3.00−2.90) |
| Rsym or Rmerge | 0.063 (0.340) | 0.063 (0.423) | 0.064 (0.521) | 0.088 (0.501) |
| I/σI | 9.98 (1.35) | 10.0 (1.31) | 11.2 (1.30) | 14.1 (1.80) |
| Completeness (%) | 58.4 (60.0) | 56.9 (58.3) | 46.3 (47.6) | 91.3 (92.4) |
| Redundancy | 1.2 (1.1) | 1.3 (1.2) | 1.2 (1.2) | 2.2(2.1) |
| Refinement | ||||
| Resolution (Å) | 30.0−2.90 | |||
| No. reflections | 25 538 | |||
| Rwork/Rfreeb | 0.228/0.250 | |||
| No. atoms | 6 868 | |||
| Protein | 6 863 | |||
| Ligand/ion | 5(1 PO4− ion) | |||
| Water | 0 | |||
| B-factors | ||||
| Protein | 52.02 | |||
| Ligand/ion | 53.78 | |||
| Rmsd | ||||
| Bond lengths (Å) | 0.008 | |||
| Bond angles (°) | 1.236 | |||
| Ramachandran plotc | ||||
| Favored (% residues) | 90.7 | |||
| Allowed (% residues) | 9.3 | |||
| Disallowed (% residues) | 0 | |||
Using the structure of the Taq RNAP-β1 (Taq RNAP-β[17–195 and 334–395]) as a search model, a molecular replacement solution containing four copies in the asymmetric unit was obtained using CNS (20). Additional molecular replacement searches to locate the TRCF–RID, using a homology model of the Tth TRCF–RID based on the Eco TRCF–RID (8) as a search model, using either CNS or BRUTEPTF (21) were unsuccessful. Nevertheless, maps phased from the Taq RNAP-β1 molecular replacement solution alone, combined with density modification using CNS, revealed clear electron density for the TRCF–RIDs (Supplementary Figure S1A). After iterative rounds of building and minimization to 2.9 Å, the final model was refined to an R/Rfree of 0.228/0.250 using Refmac5 (22). Initial rounds of refinement incorporated tight non-crystallographic symmetry (NCS) restraints (with two NCS groups, the TRCF–RID and RNAP-β1). Final rounds kept loose NCS restraints and also utilized TLS refinement (with two TLS groups, the TRCF–RID and RNAP-β1).
Bacterial two-hybrid assays
Plasmids pAC λCI-Eco β, pAC λCI-Eco TRCF, pAC λCI-Tth β and pAC λCI-Tth TRCF encode residues 1–236 of the bacteriophage λCI protein fused to residues 19–142 of the Eco RNAP β-subunit, residues 472–603 of Eco TRCF, residues 10–133 of the Tth RNAP β-subunit or residues 314–444 of Tth TRCF, respectively, under the control of the IPTG-inducible lacUV5 promoter. Plasmids pBR α-Eco β, pBRα-Eco TRCF, pBRα-Tth β and pBRα-Tth TRCF encode residues 1–248 of the Eco RNAP α-subunit fused to residues 19–142 of the Eco RNAP β-subunit, residues 472–603 of Eco TRCF, residues 10–133 of the Tth RNAP β-subunit or residues 314–444 of Tth TRCF, respectively, under the control of tandem lpp and IPTG-inducible lacUV5 promoters (8). Substitutions Tth RNAP-β1Q99R, Tth TRCF–RIDR341L, Eco TRCF–RIDL499R, Eco RNAP-β1 R101Q and R101 E, were introduced into the appropriate plasmid by PCR. Plasmid pBRα encodes wild-type α and plasmid pACλCI encodes the bacteriophage λCI protein (23).
FW102 OL262 reporter strain cells were transformed with the indicated plasmids. Individual transformants were selected and grown in LB supplemented with carbenicillin (100 µg/ml), kanamycin (50 µg/ml), chloramphenicol (25 µg/ml), and the indicated concentration of IPTG. β-galactosidase assays were performed as described earlier (24) using microtitre plates and a microtitre plate reader. β-Galactosidase activity (reported as Miller Units) was calculated as described earlier (24).
RESULTS
Crystallization and structure determination of TRCF–RID/RNAP-β1
The TRCF/RNAP protein/protein interaction was discovered and characterized studying the Eco system (8,10,14). Nevertheless, biochemical experiments suggested that the Eco TRCF–RID/RNAP-β1 complex was relatively unstable and might not be suitable for structural studies. For instance, while we could detect and study the Eco TRCF–RID/RNAP-β1 interaction using the highly sensitive bacterial two-hybrid system, we did not observe a stable interaction in vitro using typical biochemical methods, such as affinity isolations (8). However, in other experiments, we observed evidence for robust interaction between Tth TRCF and Tth RNAP (L.F. Westblade, B.T. Chait and S.A. Darst, unpublished data), suggesting that the Thermus TRCF–RID/RNAP-β1 interaction is suitably stable for X-ray crystallographic studies. The two protein domains are well conserved; the Tth TRCF–RID is 40% identical in sequence to the Eco TRCF–RID and the Thermus RNAP-β1 domain is 50% identical to Eco RNAP-β1 (Figure 2A). We chose to crystallize the complex of only the protein domains required for the TRCF/RNAP protein/protein interaction (i.e. the Tth TRCF–RID and Taq RNAP-β1 domains) to increase the chances of obtaining crystals of the complex that diffract to high resolution.
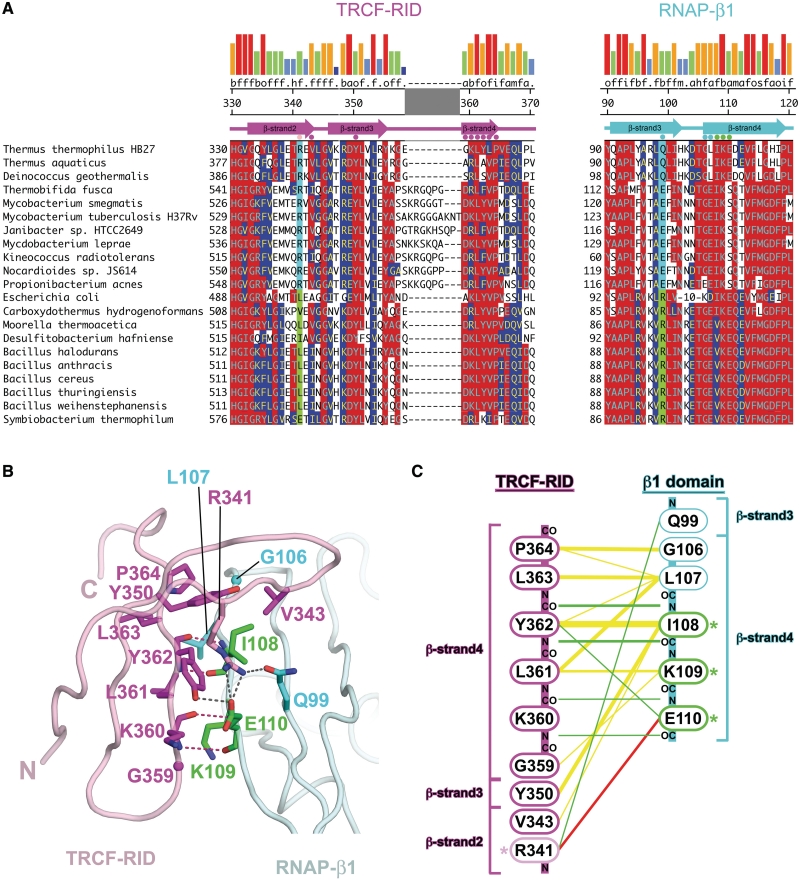
Figure 2.
Sequence and structural features of the TRCF–RID/RNAP-β1 interface. (A) Sequence conservation in the TRCF–RID/RNAP-β1 interface. Sequences are shown in one-letter amino acid code and identified by species at the left. The numbers at the beginning of each line indicate amino acid positions relative to the start of each protein sequence. Residues involved in direct interprotein contacts in the TRCF–RID/RNAP-β1 interface are denoted by colored dots directly above the sequences. The locations of β-strands are denoted schematically above the sequences. The numbers above indicate the amino acid position in Tth TRCF and RNAP-β. Positions in the alignment that share >50% identity with the consensus are indicated by red shading, while positions that share >50% homology are indicated by blue shading. Homology groups are defined as: a, acidic (DE); b, basic (HKR); f, aliphatic (AGILV); m, amide (NQ); o, aromatic (FWY), h, hydroxyl (ST); i, imino (P); s, sulfur (CM). Shown at the very top is the consensus homology, and the strength of the homology shown in a histogram (tall red bar, 100% homologous, short blue bar, <20% homologous). The positions correlated between the TRCF–RID and RNAP-β1 are highlighted in cyan/green). If the position corresponding to Tth TRCF–RIDR341 is R, then the position corresponding to Tth RNAP-β1Q99 is Q or E (cyan). On the other hand, if the TRCF–RID residue is I, L, V, or E, then the RNAP-β1 residue is R (green). (B) Molecular structure of the TRCF–RID/RNAP-β1 interface. The TRCF–RID is shown as a pale pink backbone worm. Residues involved in direct contacts with RNAP-β1 are shown in stick format (nitrogen atoms, blue; oxygen, red; carbon, magenta, except R341 is pink). The RNAP-β1 is shown as a pale cyan backbone worm. Residues involved in direct contacts with the TRCF–RID are shown in stick format (nitrogen atoms, blue; oxygen, red; carbon, cyan, except I108, K109 and E110 are green). Polar contacts are shown as dashed lines (backbone–backbone hydrogen bonds, red; side-chain polar contacts, grey). (C) Schematic diagram illustrating TRCF–RID/RNAP-β1 intermolecular contacts: yellow lines, van der Waals contacts (<4 Å); green lines, hydrogen bonds (<3.2 Å); red lines, salt bridges/hydrogen bonds. Note that the TRCF–RID is shown with the C-terminus at the top, while the RNAP-β1 is shown with the N-terminus at the top.
While the RNAP-β1 domain is a well-folded structural domain with a distinct hydrophobic core, a complication for our strategy was that the β1 structural domain is not contiguous in the RNAP β-subunit sequence. In Thermus RNAP, the β1 domain comprises β residues 17–139 (β1a) and 334–395 [β1b; (25,26)]. The RNAP mutants that disrupt the TRCF/RNAP protein/protein interaction (Eco β117–119/Tth β108–110) (8,14) lie in the larger β1a, but this segment alone is unlikely to form a stable, well-folded domain suitable for structural studies. Fortunately, within the RNAP structure, the C-terminus of β1a (residue 139) and the N-terminus of β1b (residue 334) are only 5.4 Å apart (Cα–Cα distance), allowing them to be connected by a –Gly–Gly– linker. We therefore sub-cloned the Tth TRCF–RID (Tth TRCF321–387) into a pET28a-based co-expression cassette (27) along with the –Gly–Gly– linked segments of the Taq RNAP-β1 domain (Taq RNAP-β[17–130]–GG–[334–395]), or expressed the two proteins simultaneously from separate plasmids. The Taq and Tth RNAP-β1 domains are 93% identical/98% homologous over 185 residues, and all of the β1 residues that interact with the TRCF–RID are identical between Taq and Tth (Figure 2A).
Upon co-expression, both the Tth TRCF–RID and the –Gly–Gly– linked segments of the Taq RNAP-β1 domain assembled into a stable, soluble complex that was purified and crystallized as small (15–20 µm width) octahedra. SDS–PAGE and mass-spectrometric analyses confirmed that the crystals contained both proteins. Subsequent optimization procedures yielded slightly larger crystals (25 µm width). Despite the small size of the crystals, X-ray diffraction to ∼5 Å-resolution was observed at various synchrotron sources, where the smallest achievable beamsize was ∼50 µm in diameter. Diffraction data <3 Å-resolution was collected at a microdiffractometer beamline, using a collimated 20 µm diameter beam. Individual crystals succumbed to radiation damage before complete data sets could be collected, but a data set to 2.9-Å resolution was obtained by combining partial data sets from three separate crystals (Table 1).
The hemihedrally twinned crystals belong to the space group P43. The structure was solved by molecular replacement using the Taq RNAP-β1 fragment as a search model. Four solutions were eventually found, consistent with four copies of the heterodimeric complex in the asymmetric unit. Additional molecular replacement searches including a homology model of the Tth TRCF–RID (20,28), and even ‘brute-force’ phased translation searches (21) failed to position the TRCF–RID. Nevertheless, electron density maps calculated from the RNAP-β1 molecular replacement solution alone showed good density corresponding to the TRCF–RID (Supplementary Figure S1A). An atomic model of the TRCF–RID/RNAP-β1 complex was built and refined to a twinned R/Rfree of 0.228/0.250 at 2.9-Å resolution (Table 1 and Figure 1A).
Overall structure
The TRCF–RID/RNAP-β1 structure reveals the expected 1:1 heterodimer (Figure 1). The four crystallographically independent heterodimers are all very similar in structure; the maximum root-mean-square-deviation (rmsd) in α-carbon positions when comparing 190 positions within well-defined regions of the structure (excluding flexible loops) was 0.41 Å.
The two proteins engage in a complex involving interfaces containing the residues found previously to be important for the protein/protein interaction. This includes Tth TRCF–RIDR341 [corresponding to Eco TRCF–RIDL499; (8)] and Taq RNAP-β1I108/K109/E110 [corresponding to Eco RNAP-β1I117/K118/E119; (8,14)] (Figures 1, 2A and B). Formation of the complex results in the burial of a modest 555 Å2 of otherwise exposed surface area.
The structural core of the RNAP-β1 domain can be described as a four-stranded antiparallel β-sheet buttressed on one face by α-helices (Figure 1). The TRCF–RID comprises a Tudor-like fold, a highly bent, five-stranded antiparallel β-sheet that folds into a barrel-like roll (8). The C-terminal β-strand of the β1 domain (β1 β-strand 4, Figure 1B) and the penultimate TRCF–RID β-strand (TRCF–RID β-strand 4, Figure 1B), are exposed at the edge of the individual domain structures (Figure 1). In the TRCF–RID/RNAP-β1 complex, these two edge-exposed β-strands interact to form an antiparallel, intermolecular β-sheet that extends across the two separate proteins (Figure 1).
The Tth TRCF–RID in the TRCF–RID/RNAP-β1 complex is very similar in structure to the Eco TRCF–RID, except that the N-terminal and C-terminal β-strands (TRCF–RID β-strand 1 and β-strand 5, Figures 1B and 3A) are disordered in all four crystallographically independent copies of the Tth TRCF–RID. Thus, the extended, intermolecular β-sheet in the TRCF–RID/RNAP-β1 complex comprises seven β-strands, rather than the expected nine (Figures 1B, 3A). Analysis of the TRCF–RID/RNAP-β1 structure indicates that the intermolecular crystal packing is incompatible with the presence of the TRCF–RID β-strand 1 and β-strand 5 (Supplementary Figure S2), suggesting that crystal packing forces induced partial unfolding of the TRCF–RID. This may explain the small size of the crystals. Superimposition of the ordered portion of the Tth TRCF–RID with the Eco TRCF–RID yields an rmsd of 1.15 Å over 32 α-carbon positions (Figure 3A).
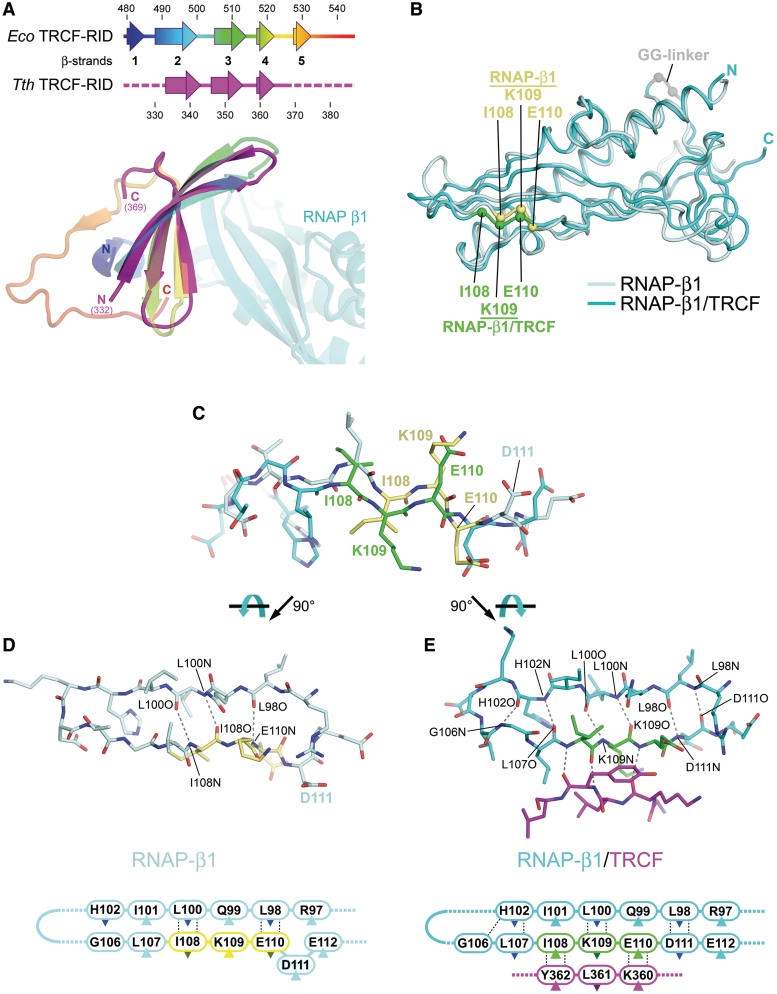
Figure 3.
Structural comparisons. (A) Comparison of the Eco TRCF–RID (residues 479–545 from the Eco TRCF structure; (8), colored as a ramp from blue (N-terminus) to red (C-terminus), and the Tth TRCF–RID (colored magenta) from the TRCF–RID/RNAP-β1 complex. The secondary structure of the two proteins is shown in schematic form on top. Below, the superimposed TRCF–RID structures are shown as ribbon diagrams, color-coded as in the schematic. The Taq RNAP-β1 complexed with Tth TRCF–RID is also shown. (B) Comparison of the Taq RNAP-β1 domains from the Taq core RNAP (pale cyan) (26), and from the TRCF–RID/RNAP-β1 complex (cyan). The superimposed structures are shown as backbone worms. The α-carbons of the TRCF–RID interface residues I108, K109, and E110 are shown as spheres (RNAP, yellow; complex with TRCF–RID, green), illustrating the register shift. (C) Side view of RNAP-β1 residues 102–112 from the RNAP structure (pale cyan, but with I108/K109/E110 colored yellow) and from the TRCF–RID/RNAP-β1 structure (cyan, but with I108/K109/E110 colored green), illustrating the register shift. The structures were aligned over the entire β1 domain. (D) Top view of RNAP-β1 β-strand 3 and β-strand 4 from the RNAP structure. Backbone-backbone hydrogen bonding interactions are shown as dashed grey lines. Below is the same, shown as a schematic. The dark-colored triangles denote side chains that point away from the viewer, down into the plane of the page. The light-colored triangles denote side chains that point up towards the viewer, out of the plane of the page. (E) Top view of RNAP-β1 β-strand 3 and β-strand 4 from the TRCF–RID/RNAP-β1 structure. Also shown are TRCF–RID360–362 (magenta). Backbone-backbone hydrogen-bonding interactions are shown as dashed grey lines. Below is the same, shown as a schematic.
Although the overall structure of the RNAP-β1 domain in the TRCF–RID/RNAP-β1 complex is similar to the RNAP-β1 domain in the context of RNAP (rmsd of 0.97 Å over 114 well-defined α-carbon positions; Figure 3B), a significant, local conformational change was observed (Figure 3B–E). The conformational change entails a ‘register shift’ of β1 β-strand 4 with respect to β-strand 3 (Figure 3B–E). In all available bacterial RNAP structures, the register of the β1 domain β-sheet is such that L98 pairs with E110 (L98:E110) and L100:I108 (Figure 3D and E, left). In the β-side view of the RNAP (looking down on the β-subunit from outside the active site channel, the perspective seen in Figure 3D and E), the side-chains of I108 and E110 point down away from the viewer, while the side-chain of K109 points up towards the viewer (Figure 3D and E, left). On the other hand, in the complex with the TRCF–RID, the register is such that L98:D111 and L100:K109. The side-chains of I108 and E110 now point up towards the viewer, while K109 points down away from the viewer (Figure 3D and E, right). The overall register shift involves residues 103–111. In the RNAP without TRCF, E110 or D111 is ‘pinched out’ of β1 β-strand 4, while in the complex with TRCF, these residues are incorporated into the β-strand (Figure 3D and E). The shift in β1 β-strand 4 is accommodated in the flexible loop connecting β-strand 3 and β-strand 4 (residues 103–105). Since this is an unusual conformational change and the resolution of our analysis (2.9 Å) is modest, we performed test refinements in which the RNAP-β1 β-strand 4 register was modeled as in the available RNAP structures. The results of these tests confirmed that the RNAP-β1 β-strand 4 register shift was not the result of a mistracing (Supplementary Data).
Although this conformational change is very localized, it must play an important role in the TRCF–RID interaction with the RNAP β1 domain, since the residues involved in the shift and reorientation are exactly the residues of RNAP that make critical interactions with the TRCF–RID, corresponding to Thermus I108, K109 and E110. We presume that the RNAP-β1 fluctuates normally between these two conformational states, and that the TRCF–RID binds to and stabilizes the state observed in the TRCF–RID/RNAP-β1 structure (Figure 3D and E, right).
TRCF–RID/RNAP-β1 interface
The predominant interaction in the TRCF–RID/RNAP-β1 interface occurs across the extended, intermolecular β-sheet and involves van der Waals as well as polypeptide backbone hydrogen bonding between RNAP-β1 βstrand4 and TRCF–RID βstrand4 (Figures 1 and 2). On the β1 domain, this interface extends from Taq RNAP-β1 G106 to E110, and includes the three RNAP residues shown to be important for the TRCF–RID/RNAP protein/protein interaction, corresponding to Thermus I108/K109/E110 (8,14). On the TRCF–RID, this interface extends from Tth TRCF–RID G359 to P364. TRCF–RID residues 361–364 comprise a well-conserved motif, f-o-f-P, where ‘f’ stands for hydrophobic aliphatic residue (I, L or V), and ‘o’ stands for aromatic (Y or F) (Figure 2A) (8). TRCF–RID β-strand 2 and β-strand 3 extend past the end of TRCF–RID β-strand 4 and arch over the top of the RNAP-β1 domain, affording additional interactions. Y350 (from TRCF–RID β-strand 3) is highly conserved as an aromatic residue (Figure 2A), and makes van der Waals contacts with RNAP-βI108. Finally, TRCF–RIDR341 (from TRCF–RID β-strand 2) makes polar contacts with RNAP-β E110 as well as with Q99 (Figure 2). Interestingly, Tth TRCF–RIDR341 corresponds to Eco TRCF–RIDL499, which is critical for the TRCF/RNAP protein/protein interaction (8), but is poorly conserved, as is one of the residues it interacts with, RNAP-βQ99 (Figure 2A). Examination of the sequence alignment, however, reveals a strong correlation between the identity of these two residues (corresponding to Tth TRCF–RIDR341 and Tth RNAP-βQ99) across phyla (Figure 2A, columns highlighted in cyan and green). Figure 2A shows a limited set of 21 sequences, but the rules denoted below were determined from a more extensive alignment containing more than 100 sequences from actinobacteria, cyanobacteria, deinococcus-thermus, firmicutes, fusobacteria, planctomycetes, proteobacteria and spirochaetes:
If the TRCF–RID has an R at this position, then the RNAP-β1 has a Q, E or D at this position. So, for example, Tth TRCF–RID341 is R, Tth RNAP-β99 is Q. All of the Deinococcus–Thermus and virtually all of the Actinobacteria have R in the TRCF–RID (a handful have Q, one has K, one has E). R does not occur at this position in any other phylum examined.
If the TRCF–RID has a hydrophobic residue (either F, I, L, M or V) or E or H at this position, then the RNAP-β1 has an R or K (i.e. Eco TRCF–RIDL499, Eco RNAP-β1R101).
This suggests that conceptually, the TRCF–RID/RNAP-β1 interface can be thought of as bipartite. A central, relatively conserved set of contacts across the intermolecular β-sheet, primarily between TRCF–RID β-strand 4 and RNAP-β1 β-strand 4 is relatively conserved across all phyla. On the other hand, a peripheral interaction between residues corresponding to Tth TRCF–RIDR341 and Tth RNAP-β1Q99 may occur in a phylum-specific manner.
This hypothesis makes four predictions for the results of single amino acid substitutions in the Thermus or Eco proteins:
The substitution Tth TRCF–RIDR341L should disrupt the interaction with Tth RNAP-β1 but improve the interaction with Eco RNAP-β1.
The substitution Eco TRCF–RIDL499R should disrupt the interaction with Eco RNAP-β1 but improve the interaction with Tth RNAP-β1.
The substitution Tth RNAP-β1Q99R should disrupt the interaction with Tth TRCF–RID but improve the interaction with Eco TRCF–RID.
The substitution Eco RNAP-β1 R101Q or R101E should disrupt the interaction with Eco TRCF–RID but improve the interaction with Tth TRCF–RID.
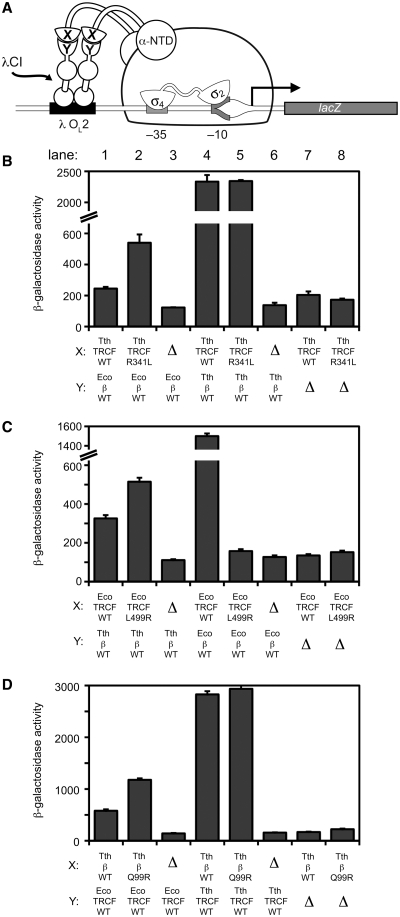
We tested these predictions using the bacterial two-hybrid assay with the individual protein domains (TRCF–RID and RNAP-β1; Figure 4A). The bacterial two-hybrid assay previously established the minimal domains required for the TRCF/RNAP protein/protein interaction, and demonstrated that single amino acid substitutions in either the TRCF–RID or the RNAP-β1 domains disrupt the interaction (8). The results show that predictions 1–3 are fulfilled (Figure 4B–D, Supplementary Figure S3B and C), and prediction 4 is partially fulfilled (Supplementary Figure S3D):
-
Prediction (1)
The substitution Tth TRCF–RIDR341L improves the interaction with the heterologous Eco RNAP-β1 more than 2-fold (Figure 4B, compare lanes 1 and 2). At the conditions of this experiment (50 µM IPTG), there is no apparent defect in the interaction of Tth TRCF–RIDR341L with Tth RNAP-β1 (Figure 4B, lanes 4 and 5), but an ∼2-fold defect is revealed when no IPTG is used (Supplementary Figure S3B, lanes 1 and 2), where leaky expression results in very low concentrations of the binding partners.
-
Prediction (2)
The substitution Eco TRCF–RIDL499R improves the interaction with the heterologous Tth RNAP-β1 (Figure 4C, lanes 1 and 2), and causes a dramatic defect (∼10-fold) in binding to Eco RNAP-β1 (Figure 4C, lanes 4 and 5), as observed previously earlier (8).
-
Prediction (3)
The substitution Tth RNAP-β1Q99R improves the interaction with the heterologous Eco TRCF–RID ∼2-fold (Figure 4D, lanes 1 and 2). At the conditions of this experiment (50 µM IPTG), there is no apparent defect in the interaction of Tth RNAP-β1Q99R with Tth TRCF–RID (Figure 4B, lanes 4 and 5), but an ∼2-fold defect is revealed when no IPTG is used (Supplementary Figure S3C, lanes 1 and 2).
-
Prediction (4)
The substitutions Eco RNAP-β1 R101Q or R101E cause dramatic (∼5-fold) defects in binding to the Eco TRCF–RID (Supplementary Figure S3D, lanes 5–7), but under no conditions did these substitutions improve the interaction with the heterologous Tth TRCF–RID (Supplementary Figure S3D, lanes 1–3).
Figure 4.
Bacterial two-hybrid analysis of the interaction between heterologous TRCF–RID and RNAP-β1 proteins. (A) Bacterial two-hybrid assay (23,45) used to study the interaction between the TRCF–RID and RNAP-β1 domains. Cartoon depicts how the interaction between protein domain X (fused to the N terminal domain of the RNAP α-subunit, α-NTD) and protein domain Y (fused to the bacteriophage λ CI protein) activates transcription from test promoter placOL262, which bears the λ operator OL2 centered 62-bp upstream of the start site of the lac core promoter. In reporter strain FW102 OL262, test promoter placOL262 is located on an F′ episome and drives the expression of a linked lacZ transcriptional fusion (8,45). (B–D). Results of β-galactosidase assays performed with FW102 OL262 cells containing two compatible plasmids, one encoding either α (Δ) or the indicated α fusion protein, and the other encoding λCI (Δ) or the indicated λCI fusion protein. The plasmids directed the synthesis of α, λCI, or the fusion proteins under the control of isopropyl-β-d-thiogalactoside (IPTG)-inducible promoters and the cells were grown in the presence of either 50 µM IPTG (B, D) or 20 µM IPTG (C). Plotted on the graphs are the mean and SEM of four (B), six (C) or 12 (D) independent measurements.
DISCUSSION
The specific protein/protein interaction between TRCF and RNAP is key to TRCF function and regulation in bacterial TCR. TRCF does not recognize DNA damage per se, it recognizes sites of DNA damage by proxy through the protein/protein interaction with elongating RNAP stalled at sites of damage. Furthermore, the TRCF/RNAP protein/protein interaction is thought to trigger the conformational changes in TRCF that are necessary for derepression of key TRCF activities, UvrA binding and ATP-dependent DNA translocase activity (8,12,13). Finally, the TRCF/RNAP protein/protein interaction provides an anchor point for TRCF on RNAP, allowing the TRCF DNA-tracking translocation activity to exert forces on the DNA that are ultimately responsible for releasing the RNAP from the DNA template and RNA transcript, enabling NER (15). In this work, the structural basis for the TRCF/RNAP protein/protein interaction has been elucidated with the 2.9-Å resolution X-ray crystal structure of a complex between the interacting domains (the TRCF–RID and the RNAP-β1 domain) necessary and sufficient for the protein/protein interaction.
The structural details of the complex explain many aspects of previous mutagenesis data for both binding partners. An amino acid substitution was identified within the Eco TRCF–RID, L499R, that abolished an activity of TRCF essential for the displacement of stalled elongating RNAP, but did not abolish DNA binding, ATP binding or ATP-hydrolysis (8). The simplest interpretation of these results was that the L499R substitution disrupted the protein/protein interaction between the TRCF–RID and the RNAP. This interpretation was supported by previous bacterial two-hybrid experiments (8) as well as in this work (Figure 4C, compare lanes 4 and 5). In the structure of the Tth TRCF–RID/Taq RNAP-β1 complex, Tth TRCF–RIDR341 (corresponding to Eco TRCF–RIDL499; Figure 2A) forms a hydrogen-bond with RNAP-β1Q99 as well as salt-bridge/hydrogen-bond interactions with RNAP-β1E110 (Figure 2B and C). Although the residue at this position of the TRCF–RID (corresponding to Tth TRCF–RID341) is not conserved across species, its identity is correlated with the RNAP-β1 residue corresponding to Tth RNAP-β199 (Figure 2A).
Amino acid substitutions were identified in three consecutive residues of RNAP-β1 that abrogated the RNAP displacement function of TRCF. The effect of these substitutions was interpreted to mean that these three residues participate in the TRCF/RNAP protein/protein interaction (14). This interpretation was supported by bacterial two-hybrid experiments (8). In the structure of the Tth TRCF–RID/Taq RNAP-β1 complex, these three RNAP-β1 residues are central to the protein/protein interface (Figure 2B and C). In addition to participating in backbone hydrogen bonding that mediates the formation of the intermolecular β-sheet (Figures 2B and C), the side chain of I108 makes extensive van der Waals contacts with TRCF–RID Y350 and Y362, the side chain of K109 makes van der Waals contacts with TRCF–RID residues (primarily TRCF–RIDL361), and the side chain of E110 participates in polar interactions, primarily with TRCF–RIDR341 (Figure 2B and C). Thus, qualitatively, the finding that substitutions at these three positions of the RNAP-β1 domain cause defects in the TRCF/RNAP protein/protein interaction (8,14) is explained by the structure.
Smith and Savery (14) found that substitutions in Eco TRCF corresponding to Tth TRCF–RID I108A and K109A had only mild effects on the TRCF/RNAP protein/protein interaction, while the E110A substitution had a more severe effect. Quantitatively, these results are more complicated to reconcile with the structure. Given the extensive participation of RNAP-β1I108 in van der Waals contacts with TRCF (Figure 2B and C), one might expect the I108A substitution to have a more severe defect. Moreover, it is difficult to see from the structure why lysine is so relatively well conserved at position 109. Also, despite the apparent importance of RNAP-β1E110, it is relatively poorly conserved (Figure 2A). These observations may be reconciled, however, by the finding that the RNAP-β1 undergoes a local conformational change in the complex with the TRCF–RID (Figure 3B–E). The conformational change involves a register shift of RNAP-β1 β-strand 4 and includes I108-E110. Thus, the effects of amino acid substitutions in this region of RNAP-β1 may affect the direct interaction with the TRCF–RID, but can also influence the interaction through its effect on the conformational equilibrium of RNAP-β1 β-strand 4. An amino acid substitution in RNAP-β1 that favored the conformational state of RNAP-β1 seen in the RNAP structures (Figure 3D and E, left) over the state seen in the complex with TRCF (Figure 3D, right) would introduce an apparent defect in the interaction with TRCF by reducing the population of RNAP competent to bind TRCF, even if that residue did not directly contact TRCF. In this case, it could be misleading to interpret the results of mutagenesis studies on the TRCF/RNAP protein/protein interaction by considering only the final TRCF–RID/RNAP-β1 structure.
The observed conformational change in RNAP-β1 raises another, more interesting issue; whether the conformational state of RNAP-β1 could be controlled allosterically as a consequence of the functional state of the RNAP. Could the TRCF-binding conformation of RNAP-β1 be favored in stalled elongating RNAPs, marking it as a target for TRCF function? A priori, marking stalled elongating RNAPs is not necessary if the rate of TRCF-mediated RNAP release is much slower than elongation. In general, this appears to be the case; TRCF-mediated RNAP release occurs on a time scale of minutes (8,14,16), while nucleotide addition by RNAP can occur on a time scale of milliseconds (29). As a rule, then, TRCF would not be kinetically competent to disrupt actively elongating RNAPs in vivo unless they were stalled.
Several crystal structures of elongating Tth RNAP are available. In one, the elongation complex was in the post-translocated state, but transcription elongation was stalled by nucleotide deprivation (30). In principal, such an RNAP elongation complex stalled by nucleotide deprivation would be subject to TRCF release (16), but the conformational state of RNAP-β1 in both crystallographically independent complexes is essentially identical to that observed in all other bacterial RNAP structures (to date, more than 30 crystallographically independent complexes), where RNAP-β1 is not in the TRCF-binding conformation. Taken together, consideration of kinetic parameters and available structural evidence suggests that the conformational change in the RNAP-β1 domain does not serve as an allosteric signal for stalled RNAPs, but such a scenario cannot be completely ruled out. There may be situations in vivo, such as at regulatory transcriptional pause sites (31), where the kinetics of TRCF-mediated RNAP release could compete with RNAP elongation, and TRCF-mediated release of these paused elongation complexes might be disadvantageous for the cells. From a structural viewpoint, the conformational change in RNAP-β1 could conceivably occur in an elongation intermediate that has not been trapped in any crystal structures. The TRCF–RID/RNAP-β1 structure presented here provides a basis for designing experiments to address this question.
In addition to the previous mutagenesis studies of (14), we tested additional amino acid substitutions at two positions (corresponding to Tth TRCF–RID341 and Thermus RNAP-β199) that are not conserved but appear to be correlated with each other in a phylum-specific manner (Figure 2A). As a rule, the cross-species interactions of the wt Eco and Tth proteins were relatively weak compared to the correct binding partners (Figure 4B–D, compare lane 1 and 4). In three cases (Tth TRCF–RIDR341L, Figure 4B; Eco TRCF–RIDL499R, Figure 4C; Tth RNAP-β1Q99R; Figure 4D), the predicted mutations at the correlated positions improved the cross-species interactions (Figure 4B–D, compare lane 1 and 2). In one case (Eco RNAP-β1 R101Q or R101 E), the predicted mutation had little effect (Supplementary Figure S3). In this case, the effects of this substitution in the Eco RNAP-β1 on the TRCF–RID/RNAP-β1 interaction may be complicated by possible effects of the substitution on the RNAP-β1 conformational change that occurs upon TRCF–RID binding.
Using the apo-TRCF crystal structure, a model of the RNAP TEC, and additional constraints, a preliminary model for the TRCF/TEC assembly was constructed (8). The model was preliminary since it was concluded that the conformation of TRCF observed in the crystal structure may not to correspond to the active conformation (8). Indeed, it is now established that the conformation of the apo-TRCF observed in the crystal structure corresponds to a repressed state in which the UvrA binding determinants are occluded, ATPase activity is very low, and DNA translocase activity is essentially nonexistent (12,13). Derepression of these TRCF activities is expected to be associated with profound conformational rearrangements of the TRCF domains with respect to each other. It is thus not surprising that the orientation of the TRCF–RID with respect to the RNAP-β1 domain in the TRCF–RID/RNAP-β1 crystal structure is quite different from the previous TRCF/TEC model (Supplementary Figure S4). Because the conformation of the derepressed TRCF associated with the TEC is expected to be very different from the repressed conformation of the TRCF crystal structure, it is not fruitful to update the TRCF/TEC model by superimposing the RID in the repressed TRCF crystal structure onto the RID from the TRCF–RID/RNAP-β1 structure.
CarD has been identified as an essential Mtb protein that is induced by DNA damage and starvation, and controls rRNA transcription through a direct interaction with the RNAP (19). CarD is a widely conserved, two-domain protein (17) with an NTD with striking sequence similarity to the TRCF–RID (19), and a C-terminal domain of unknown structure. Moreover, the TRCF–RID-like CarD-NTD is sufficient for RNAP interaction, which, like the TRCF–RID, is targeted to the RNAP-β1 domain (19). Thus, the Tth TRCF-RID/Taq RNAP-β1 structure serves as an excellent model for understanding the CarD/RNAP protein/protein interaction.
A host of accessory factors directly interact with the RNAP to modulate every step of the transcription cycle. As the structural delineation of accessory factor/RNAP interactions progresses, a handful of RNAP structural features have emerged as regulatory ‘hot spots’, such as the β-flap [NusA (32); σ factors (33–35); bacteriophage T4 AsiA (36); T4 gp33 (37); bacteriophage λ Q (38)], the secondary channel [Gre-factors, (39–41); DksA (42,43)], and the β′ clamp helices [σ (34,35); RfaH (44)]. The interaction of the RNAP-β1 domain with the TRCF–RID detailed here, along with the analagous CarD–RID interaction, point to the RNAP-β1 domain as another hot spot for RNAP regulation.
The TRCF–RID/RNAP-β1 crystal structure presented here reveals the structural details of the TRCF/RNAP protein/protein interaction, which is key for TRCF function and regulation. Structural analysis also reveals a local conformational change in the RNAP-β1 in the TRCF–RID-bound state. The effects of amino acid substitutions in RNAP-β1 on this conformational change must be taken into account when interpreting the results of protein interaction assays. This structure provides a basis for the design and interpretation of experiments probing TRCF and CarD function and interactions with the RNAP.
ACCESSION NUMBERS
Structure coordinates and structure factors have been deposited in the Protein Data Bank under ID code 3MLQ.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NE-CAT beam lines of the Advanced Photon Source (APS), supported by (award RR-15301) from the National Center for Research Resources at the National Institutes of Health; Use of the APS is supported by the United States Department of Energy, Office of Basic Energy Sciences, under (contract No. W-31-109-ENG-38); National Institutes of Health (RR00862 and RR022220, to B.T. Chait); (GM073829, to S.A.D.). Funding for open access charge: Laboratory funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank M. Glickman, K. Rajashankar, N. Savery and C. Stallings for helpful discussion and advice on the manuscript, P. G. Devi for help with plasmid construction and B. T. Chait for access to mass spectrometry facilities.
REFERENCES
- 1.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 2.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 3.Selby CP, Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J. Biol. Chem. 1990;265:21330–21336. [PubMed] [Google Scholar]
- 4.Witkin EM. Time, temperature, and protein synthesis: a study of ultraviolet-induced mutation in bacteria. Cold Spring Harbor Symp. Quant. Biol. 1956;21:123–140. doi: 10.1101/sqb.1956.021.01.011. [DOI] [PubMed] [Google Scholar]
- 5.Witkin EM. Radiation-induced mutations and their repair. Science. 1966;152:1345–1353. doi: 10.1126/science.152.3727.1345. [DOI] [PubMed] [Google Scholar]
- 6.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 7.Selby CP, Witkin EM, Sancar A. Escherichia coli mfd mutant deficient in “mutation frequency decline” lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc. Natl Acad. Sci. USA. 1991;88:11574–11578. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, Darst SA. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124:507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 9.Pakotiprapha D, Liu Y, Verdine GL, Jeruzalmi D. A structural model for the damage-sensing complex in bacterial nucleotide excision repair. J. Biol. Chem. 2009;284:12837–12844. doi: 10.1074/jbc.M900571200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. I. Structural domains and binding properties. J. Biol. Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 11.Gorbalenya AE, Koonin EV, Domchenko AP, Blinov VM. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4729. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy MN, Gong P, Ralto K, Manelyte L, Savery NJ, Theis K. An N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor Mfd. Nucleic Acids Res. 2009;37:6042–6053. doi: 10.1093/nar/gkp680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith AG, Szczelkun MD, Savery NJ. Controlling the motor activity of a transcription-repair coupling factor: autoinhibition and the role of RNA polymerase. Nucleic Acids Res. 2007;35:1802–1811. doi: 10.1093/nar/gkm019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith AJ, Savery NJ. RNA polymerase mutants defective in the initiation of transcription-coupled DNA repair. Nucleic Acids Res. 2005;33:755–764. doi: 10.1093/nar/gki225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc. Natl. Acad. Sci. USA. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park J-S, Marr MT, Roberts JW. E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 17.Cayuela ML, Elias-Arnanz M, Penalver-Mellado M, Padmanabhan S, Murillo FJ. The Stigmatella aurantiaca homolog of Myxococcus xanthus high-mobility-group A-type transcription factor CarD: insights into the functional modules of CarD and their distribution in bacteria. J. Bacteriol. 2003;185:3527–3537. doi: 10.1128/JB.185.12.3527-3537.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Moreno D, Abellon-Ruiz J, Garcia-Heras F, Murillo FJ, Padmanabhan S, Elias-Arnanz M. CdnL, a member of the large CarD-like family of bacterial proteins, is vital for Myxococcus xanthus and differs functionally from the global transcriptional regulator CarD. Nucleic Acids Res. 2010;38:4586–4598. doi: 10.1093/nar/gkq214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stallings CL, Stphanou NC, Chu L, Hochschild A, Nickels BE, Glickman MS. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell. 2009;138:149–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams PD, Pannu NS, Read RJ, Brunger AT. Cross-validated maximum likelihood enhances crystallographic simulated annealing refinement. Proc. Natl Acad. Sci. USA. 1997;94:5018–5023. doi: 10.1073/pnas.94.10.5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strokopytov BV, Fedorov A, Mahoney NM, Kessels M, Drubin DG, Almo SC. Phase translation function revisited: structure solution of the cofilin-homology domain from yeast actin-binding protein 1 using six-dimensional searches. Acta Crystallogr. D Biol. Crystallogr. 2005;61:285–293. doi: 10.1107/S0907444904033037. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.Dove SL, Joung K, Hochschild A. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature. 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 24.Thibodeau SA, Fang R, Joung JK. High-throughput beta-galactosidase assay for bacterial cell-based reporter systems. Biotechniques. 2004;36:410–415. doi: 10.2144/04363BM07. [DOI] [PubMed] [Google Scholar]
- 25.Lane WJ, Darst SA. Molecular evolution of multi-subunit RNA polymerases: structural analysis. J. Mol. Biol. 2009;395:686–704. doi: 10.1016/j.jmb.2009.10.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 27.Campbell EA, Darst SA. The anti-σ factor SpoIIAB forms a 2:1 complex with σF, contacting multiple conserved regions of the σ factor. J. Mol. Biol. 2000;300:17–28. doi: 10.1006/jmbi.2000.3838. [DOI] [PubMed] [Google Scholar]
- 28.Sali A, Potterton L, Yuan F, van-Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 29.Holmes SF, Foster JE, Erie DA. Kinetics of multisubunit RNA polymerases: experimental methods and data analysis. Methods Enzymol. 2003;371:71–81. doi: 10.1016/S0076-6879(03)71005-2. [DOI] [PubMed] [Google Scholar]
- 30.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 31.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl Acad. Sci. USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- 33.Kuznedelov K, Minakhin L, Niedziela-Majka A, Dove SL, Rogulja D, Nickels BE, Hochschild A, Heyduk T, Severinov K. A role for interaction of the RNA polymerase flap domain with the sigma subunit in promoter recognition. Science. 2002;295:855–857. doi: 10.1126/science.1066303. [DOI] [PubMed] [Google Scholar]
- 34.Murakami K, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 35.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 36.Gregory BD, Nickels BE, Garrity SJ, Severinova E, Minakhin L, Urbauer RJ, Urbauer JL, Heyduk T, Severinov K, Hochschild A. A regulator that inhibits transcription by targeting an intersubunit interaction of the RNA polymerase holoenzyme. Proc. Natl Acad. Sci. USA. 2004;101:4554–4559. doi: 10.1073/pnas.0400923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nechaev S, Kamali-Moghaddam M, Andre E, Leonetti J-P, Geiduschek EP. The bacteriophage T4 late-transcription coactivator gp33 binds the flap domain of Escherichia coli RNA polymerase. Proc. Natl Acad. Sci. USA. 2004;101:17365–17370. doi: 10.1073/pnas.0408028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deighan P, Diez CM, Leibman M, Hochschild A, Nickels BE. The bacteriophage lambda Q antiterminator protein contacts the eta-flap domain of RNA polymerase. Proc. Natl Acad. Sci. USA. 2008;105:15305–15310. doi: 10.1073/pnas.0805757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laptenko O, Lee J, Lomakin I, Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 2003;23:6322–6334. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- 41.Sosunova E, Sosunov V, Kozlov M, Nikiforov V, Goldfarb A, Mustaev A. Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc. Natl. Acad. Sci. USA. 2003;100:15469–15474. doi: 10.1073/pnas.2536698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nickels BE, Hochschild A. Regulation of RNA polymerase through the secondary channel. Cell. 2004;118:281–284. doi: 10.1016/j.cell.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 43.Perederina A, Svetlov V, Vassylyeva M, Tahirov T, Yokoyama S, Artsimovitch I, Vassylyev D. Regulation through the secondary channel – structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 44.Sevostyanova A, Svetlov V, Vassylyev DG, Artsimovitch I. The elongation factor RfaH and the initiation factor s bind to the same site on the transcription elongation complex. Proc. Natl Acad. Sci. USA. 2008;105:865–870. doi: 10.1073/pnas.0708432105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nickels BE. Genetic assays to define and characterize protein-protein interactions involved in gene regulation. Methods. 2009;47:53–62. doi: 10.1016/j.ymeth.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 46.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK – a program to check the stereochemical quality of protein structures. J. appl. Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.