Abstract
It is known that transcription can induce DNA recombination, thus compromising genomic stability. RECQ5 DNA helicase promotes genomic stability by regulating homologous recombination. Recent studies have shown that RECQ5 forms a stable complex with RNA polymerase II (RNAPII) in human cells, but the cellular role of this association is not understood. Here, we provide evidence that RECQ5 specifically binds to the Ser2,5-phosphorylated C-terminal repeat domain (CTD) of the largest subunit of RNAPII, RPB1, by means of a Set2–Rpb1-interacting (SRI) motif located at the C-terminus of RECQ5. We also show that RECQ5 associates with RNAPII-transcribed genes in a manner dependent on the SRI motif. Notably, RECQ5 density on transcribed genes correlates with the density of Ser2-CTD phosphorylation, which is associated with the productive elongation phase of transcription. Furthermore, we show that RECQ5 negatively affects cell viability upon inhibition of spliceosome assembly, which can lead to the formation of mutagenic R-loop structures. These data indicate that RECQ5 binds to the elongating RNAPII complex and support the idea that RECQ5 plays a role in the maintenance of genomic stability during transcription.
INTRODUCTION
The numerous processes that occur in the nucleus during cell proliferation have to be tightly coordinated to ensure genome integrity and faithful genome propagation. Transcription is known to stimulate DNA recombination, thus affecting genome stability (1). This phenomenon, called transcription-associated recombination (TAR), has been linked to replication fork pausing that results from the convergence of transcription and replication. TAR has also been linked to the formation RNA : DNA hybrids (R-loops) between the nascent transcript and the template DNA strand, which increases the susceptibility of the non-transcribed strand to damage or to the formation of secondary structures that impair replication fork progression (1). R-loops are formed when the co-transcriptional assembly of mRNA-particle complexes is impaired (1). For example, it has been shown that inactivation of the human SR protein ASF/SF2, which is required for spliceosome assembly, results in DNA fragmentation, cell-cycle arrest and genomic instability as a consequence of R-loop formation (2). ASF/SF2 depletion also leads to accumulation of stalled replication forks, and chromosome breaks caused by ASF/SF2 deficiency occur specifically in S-phase, preferentially at gene-rich regions (3). These data suggest that TAR results as a consequence of replication fork collapse at R-loops (3).
RECQ5 belongs to the RecQ family of DNA helicases that play critical roles in the maintenance of genomic stability and cancer suppression (4). Recent studies in mammalian cells have established RECQ5 as an important anti-recombination factor that acts by controlling the assembly of the RAD51 filament on single-stranded DNA (ssDNA), which catalyses the homology search and strand invasion during homologous recombination (HR) (5,6). RECQ5 binds directly to the RAD51 recombinase and disrupts the RAD51-ssDNA filament in a reaction driven by ATP hydrolysis, thus preventing homologous duplex invasion during HR (6). In accordance with this finding, RECQ5-deficient cells versus RECQ5-proficient cells show an increased efficiency of HR-mediated DNA double-strand break (DSB) repair, an elevated frequency of sister chromatid exchange, a prolonged persistence of RAD51 foci in response to DNA damage and an increased rate of chromosomal rearrangements (5,6). Moreover, RECQ5 has been shown to accumulate at sites of DSBs and sites of replication arrest in a manner dependent on the MRE11–RAD50–NBS1 complex, a key player in DNA damage signaling and repair (7).
A number of recent proteomic studies have revealed that RECQ5 forms a stable complex with RNA polymerase II (RNAPII) in human cells (7–9). The RECQ5–RNAPII interaction is direct and is mediated by the largest subunit of RNAPII, RPB1 (8). Knockdown of the RECQ5 transcript in human cells has been found to increase the transcription of several genes (9). Likewise, RECQ5 has been shown to inhibit RNAPII transcription in an in vitro system reconstituted using purified proteins (10). Despite these findings, the function of RECQ5 during the RNAPII transcription cycle remains elusive.
Here, we provide evidence that RECQ5 associates with RNAPII during the productive elongation phase of transcription through direct binding to the C-terminal repeat domain (CTD) of RPB1. Moreover, we show that depletion of RECQ5 reduces the cellular sensitivity to diospyrin, a plant-derived bisnaphthoquinonoid, which interferes with spliceosome assembly, presumably by inhibiting DNA topoisomerase I (Top1)-mediated phosphorylation of ASF/SF2, and hence is likely to promote formation of R-loops during RNAPII transcription (3,11). These findings are discussed in light of a possible role for RECQ5 in promoting genomic stability at sites of RNAPII transcription.
MATERIALS AND METHODS
Plasmids, proteins and antibodies
The vector pTXB1 (New England Biolabs) was used for bacterial expression of wild-type and mutant forms of human RECQ5 as fusions with the self-cleaving chitin-binding domain (CBD) tag. Construction of these plasmids was previously described (12). The expression vector for RECQ5 Δ908–954 (ΔSRI) was constructed in the same way as the vector for RECQ5 Δ640–653 (12). Point mutations in RECQ5 were made using QuickChange Site-Directed Mutagenesis Kit (Stratagene). The vector pLexA-Km (Dualsystems) was used for construction of the yeast two-hybrid bait plasmids carrying different parts of the human RECQ5 cDNA. RECQ5 cDNA was amplified by PCR and ligated in pLexA-Km via EcoRI and SalI sites in frame with a LexA DNA-binding domain. Internal deletions and point mutations of RECQ5 were subcloned from the pTXB1 constructs. To express RECQ5 and its mutants in human cells as N-terminal fusions with green fluorescent protein (GFP), the respective RECQ5 cDNA cloned in the pTXB1 vector was PCR amplified using Phusion high-fidelity DNA polymerase (Finnzymes) and the following set of primers: 5′-AAACTCGAGCTATGAGCAGCCACCATACC-3′ and 5′ AAAGAATTCTCTCTGGGGGCCACACAGGCCTAA-3′. The resulting amplicon was digested and cloned into pEGFP-C1 (Clontech) between the XhoI and EcoRI sites. The RECQ5 protein and its mutants were produced in bacteria as fusions with CBD and purified as described previously (13). The antibodies used in this study are listed in Supplementary Table S1.
Cell culture
HeLa, HEK293 and HEK293T cells were maintained in DMEM (Gibco) containing 5% fetal calf serum (Gibco) and 100 U/ml penicillin/streptomycin. Transfections of HEK293 cells were carried out using the TransIT®-LT1 reagent according to the manufacturer’s instructions (Invitrogen). Transfection efficiency was >90% as determined by visualization of GFP expression using a fluorescence microscope 48 h post-transfection. Transfection of siRNA oligonucleotides (Microsynth) was carried out using Lipofectamine RNAiMAX (Invitrogen) according to manufacturer’s instructions. The following oligonucleotides were used: siRECQ5#1 (sense strand): 5′-CAGGAGGCUGAUAAAGGGUUA-3′; siRECQ5#2 (sense strand): 5′-GGAGAGUGCGACCAUGGCU-3′; and siCtrl (sense strand): 5′-CGUACGCGGAAUACUUCGA-3′.
Yeast two-hybrid assay
The yeast two-hybrid screen for proteins that interact with RECQ5 was carried out using Saccharomyces cerevisiae strain L40 (MATa trp1 leu2 his3 LYS2::lexA-HIS3 URA3::lexA-lacZ), which was sequentially transformed with the bait plasmid encoding the LexA-RECQ5 411-991 and a randomly primed human peripheral blood cDNA library cloned into the BglII sites of pACT (Clontech). Among the 1.4 × 107 transformants tested for histidine prototrophy and β-galactosidase staining, 198 positive clones were found. Clones carrying the bait and prey plasmids were tested for β-galactosidase activity using a pellet X-gal (PXG) assay as previously described (14).
Far western assay
Far western assays were performed as described previously (15). Briefly, duplicate samples of ∼1.5 µg of the purified proteins were separated on a 4–20% Criterion gel (Bio-Rad). One half of the gel was stained with Coomassie brilliant blue and the second half was transferred to nitrocellulose membrane. The membrane was incubated overnight at 4°C in blocking/renaturation buffer, containing 1× PBS, 3% non-fat dry milk, 0.2% Tween 20, 0.1% PMSF, 5 mM NaF and 2 mM dithiothreitol. The nitrocellulose was then probed with ≥300 000 cpm [32P]-labeled GSTyCTD fusion protein that had been hyperphosphorylated with CTDK-I for 4 h at 4°C. After extensive washing, the nitrocellulose was air-dried and subjected to autoradiography.
Immobilized CTD peptide binding assay
Synthetic biotinylated peptides were dissolved in PBS and incubated with 300 µl of TetraLink Tetrameric Avidin Resin (Invitrogen) for 45 min at room temperature (RT). The peptide concentrations in the column onput, flow-through and wash fractions were monitored by absorbance at 280 nm and these values were used to approximate the amount of peptide on the column. Generally, 30–50 µg of biotinylated peptide was conjugated to the 300 µl column. The peptide columns were stored in PBS at 4°C and were stable for ∼2 months. Approximately 10–15 µg of purified protein and 50 mg insulin were dissolved in PBS for a final volume of 500 µl as the onput; 450 µl of the onput was applied to the peptide resin and incubated for 20 min at RT with mixing every for 5 min. The flow-through and two washes with half-column volumes of PBS were collected. The resin was then extensively washed with 5 ml of buffer containing 25 mM HEPES (pH 7.6), 8% glycerol, 150 mM NaCl and 0.1 mM EDTA (HGNE150). The bound protein was then eluted with four half-column volumes of HGNE300 and four half-column volumes of HGNE1000. The resin was regenerated with 5 ml HGNE1000 and 5 ml PBS. Samples were analyzed by SDS–PAGE followed by Coomassie blue staining.
CBD pull-down assay
CBD-tagged RECQ5 and its variants were produced in Escherichia coli BL21-CodonPlus(DE3)-RIL cells (Stratagene) and immobilized on chitin beads (20 µl; New England Biolabs) as previously described (7). Beads were incubated for 2 h at 4°C with a total extract from HEK293 T cells (600 µg of protein) in a volume of 500 µl of buffer TN2 [50 mM Tris–HCl (pH 8), 120 mM NaCl, 20 mM NaF, 15 mM sodium pyrophosphate and 0.5% (v/v) NP-40] supplemented with 1 mM benzamidine, 0.2 mM PMSF, 0.5 mM sodium orthovanadate and protease inhibitor cocktail (Roche) and then were washed three times with buffer TN2. Bound proteins were released from the beads by addition of 25 µl of 3× SDS-loading buffer followed by incubation at 95°C for 7 min. Eluted proteins were separated by 10% SDS–PAGE and analyzed by western blotting using the indicated antibodies. RECQ5 was detected by Ponceau S staining.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed using the ChIP-IT Express kit from Active Motif (Carlsbad, CA, USA) according to manufacturer’s instructions. Briefly, HeLa or 293 (mock or GFP-RECQ5 transfected) cells grown to 70-80% confluency were crosslinked with 1% formaldehyde (Sigma) at RT for 10 min, and glycine (0.125 M) was added to stop the reaction. Isolated nuclei were then sheared using a Bioruptor sonicator (Diagenode, Liege, Belgium) to obtain chromatin fragments between 200 and 500 bp. Ten percent of the sonicated chromatin was reserved for use as an input DNA control. For each ChIP reaction, ∼6.3 µg of crosslinked chromatin were immunoprecipitated overnight at 4°C with 4 µg of a specific antibody or control IgG. After elution of immune complexes and reversion of crosslinking, DNA was recovered by using the QIAquick PCR Purification Kit (Qiagen).
Quantitative real-time PCR
The amount of immunoprecipitated DNA in each ChIP reaction was measured by quantitative real-time PCR (qPCR) using a Roche LightCycler 480 Real-Time PCR System and a Roche LightCycler 480 DNA SYBR Green I Master. Primer sequences used for the qPCR reactions are listed in Supplementary Table S2. Specificity of target amplification was confirmed by agarose gel electrophoresis and melting curve analysis. For each primer pair, a series of 5-fold dilutions of input DNA were used to generate a linear standard curve in which the crossing point was plotted versus log10 of template concentration. Primer pair efficiency was calculated from these data as E = 10(−1/slope), and primer pairs with E > 1.8 were used for qPCR. For data analysis, Pfaffl’s method was used (16). Fold enrichment of immunoprecipitated target regions was expressed as ratio of the amount of DNA estimated for a specific antibody versus the amount of DNA estimated for the control IgG antibody. All qPCR reactions were performed at least six times using DNA template obtained from two independent ChIP experiments. The data were plotted using GraphPad Prism software as mean ± SEM.
Helicase assay
PAGE-purified DNA and RNA oligonucleotides used for helicase assays were purchased from Microsynth. The sequences are: deoxy-18 (d18): 5′-tcc cag tca cga cgt tgt-3′; ribo-18 (r18): 5′-ucc cag uca cga cgu ugu-3′. The oligonucleotides were 5′-end labeled with T4 polynucleotide kinase and γ[32P]ATP (GE HealthCare) and annealed to M13mp2 ssDNA at a 1 : 1 (mol/mol) ratio in the presence of 20 mM Tris–HCl (pH 7.4) and 150 mM NaCl. For helicase assays, RECQ5 at concentrations ranging from 0 to 360 nM was incubated either with 1 nM M13mp2/d18 or with 1 nM M13mp2/r18 substrate for 30 min at 37°C in 10 µl of 33 mM Tris–acetate buffer (pH 7.9) containing 10 mM Mg-acetate, 66 mM K-acetate, 0.1 mg/ml BSA and 2 mM ATP. Helicase reactions were stopped by addition of 2.5 µl of 5× stop dye (100 mM EDTA, 60% glycerol, 1.5% SDS, 0.1% bromophenol blue and 0.1% xylene cyanol). Products were separated by electrophoresis on 15% native polyacrylamide gels in TBE buffer and visualized by autoradiography.
Cell viability assay
Cell viability was measured using Resazurin Fluorimetric Cell Viability Assay Kit (Biotium, Inc.) according to the manufacturer’s instructions. Briefly, HeLa cells were seeded at the confluency of 20% in 10-cm plates. After 24 h, cells were transfected with appropriate siRNA as described above. One day after transfection, siRNA-treated cells were harvested and seeded in a 96-well plate at a density of 10 000 cells/well in a volume of 100 µl of DMEM containing fetal calf serum and penicillin/streptomycin. Next day, cells were treated with different concentrations of diospyrin (compound D1; obtained from Dr Banasri Hazra of Jadavpur University, Kolkata) ranging from 0 to 100 µM. Experiments were carried out in hexaplicates for each drug concentration. After 24 h, cells were washed gently once with pre-warmed PBS, and then a mixture of resazurin and DMEM (100 µl) in a ratio 1 : 10 was added in to each well. After 4 h of incubation at 37°C, cell viability was monitored by measuring fluorescence with excitation wavelength at 540 nm and emission wavelength at 590 nm in a SpectraMax reader M5 (Molecular Devices). The fluorescent signal generated from the assay is directly proportional to the number of living cells in the sample. The percentage of survival of diospyrin-treated cells was calculated relative to mock (DMSO)-treated cells and plotted using GraphPad Prism as mean ± SD
RESULTS
RECQ5 binds to the CTD of RNAPII through a conserved motif
In a yeast two-hybrid screen for proteins that interact with the non-conserved C-terminal region of human RECQ5 (amino acids 411–991 fused to the LexA DNA-binding domain), we isolated a clone from a human peripheral blood cell cDNA library that encoded the CTD of the largest subunit of RNAPII, RPB1 (amino acids 1266–1970). This domain consists of evolutionary conserved repeats of the heptapetide sequence YSPTSPS that serve as a binding platform for proteins involved in transcription and mRNA processing (17–19). Thus, our finding suggests that RECQ5 is a novel RNAPII CTD-binding protein.
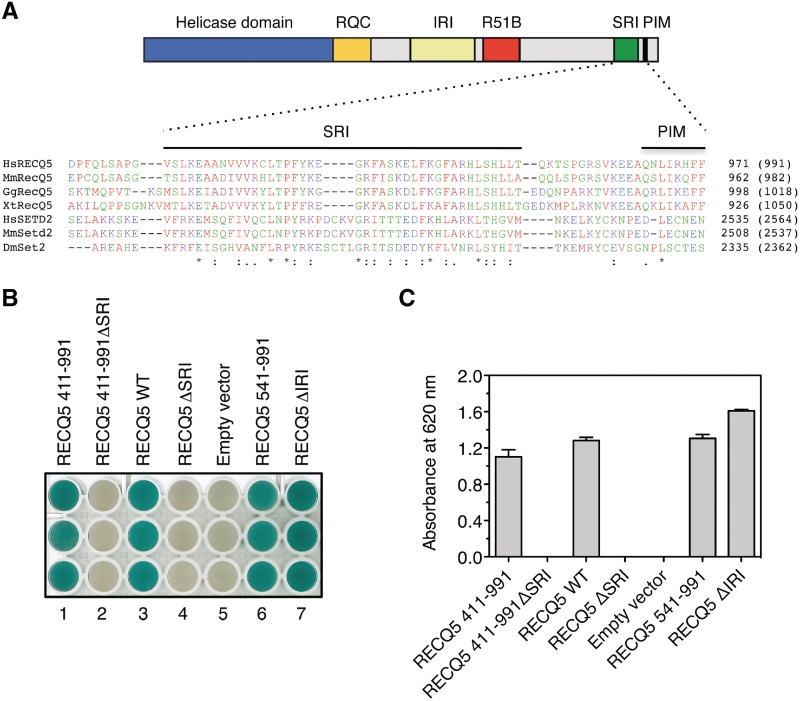
During transcription, the CTD undergoes dynamic phosphorylation on serine residues, producing different phosphorylation patterns that predominate in individual stages of the transcription cycle and determine the recruitment of a specific set of proteins (17–19). Using the MotifScan software, we found that the extreme C-terminal region of RECQ5 (amino acids 910–950) contains a putative phospho-CTD (P-CTD)-binding motif, called a Set2–Rpb1-interacting (SRI) domain (Figure 1A). This motif was initially identified as a Ser2,5 P-CTD-binding module in the yeast methyltransferase Set2, which catalyses histone H3 lysine 36 methylation during transcription (20). NMR studies on the SRI domains of the yeast Set2 and its human homologue SETD2 have revealed a novel CTD-binding fold consisting of a left-turned three-helix bundle (21,22). RECQ5 appears to contain only helices 1 and 2 that form the CTD-binding surface.
Figure 1.
RECQ5 binds to the CTD of RNAPII through a SRI motif. (A) RECQ5 contains a CTD-binding motif. Top panel: domain organization of RECQ5. R51B, RAD51-binding domain; PIM, PCNA-interacting motif. Bottom panel: alignment of RECQ5 homologues with selected Set2 homologues carried out using ClustalX. The locations of the SRI and PIM motifs are indicated by black lines. (B) Interaction of RECQ5 and its mutants with the RNAPII CTD in the yeast two-hybrid system. Clones carrying the bait (RECQ5) and prey (CTD) plasmids were tested for β-galactosidase activity using the pellet X-gal assay. Blue color indicates positive interaction. (C) Plot of absorbance at 620 nm measured after 35 min in individual wells of the ELISA plate shown in (B) relative to wells with empty bait vector. The values represent the mean of three experiments.
To show that the putative SRI motif of RECQ5 is important for RECQ5 binding to the CTD of RNAPII, we deleted it (amino acids 908–954) from the bait plasmids carrying the full-length RECQ5 and RECQ5 411–991, respectively. The resulting constructs were tested for yeast two-hybrid interaction with the CTD prey plasmid isolated in our yeast two-hybrid screen. We found that neither of these mutants interacted with RNAPII CTD (Figure 1B and C). In contrast, the full-length RECQ5 as well as RECQ5 411-991 showed positive yeast two-hybrid interaction with CTD (Figure 1B and C). A previous study demonstrated that human CTD was phosphorylated when expressed in yeast (23). Thus, our data suggest that RECQ5 binds to the P-CTD of RNAPII by means of the SRI domain.
It has been shown that RECQ5 binds to RNAPII through a region including amino acids 396–617 (9). Using a series of internal deletion variants of RECQ5, we mapped more precisely the boundaries of this domain to amino acids 515 and 640, respectively (Supplementary Figure S1). Hereafter, this domain is referred to as the Internal RNAPII-interacting (IRI) domain. A deletion of the proximal part of the IRI domain of RECQ5 that spans amino acids 515–568 (ΔIRI) completely abolished binding of RECQ5 to the hypophosphorylated form of RNAPII (RNAPIIA) (Supplementary Figure S1C, Lane 4). In contrast, yeast two-hybrid analysis indicated that the RECQ5 ΔIRI mutant bound to the RNAPII CTD to a similar extent as wild-type RECQ5 (Figure 1B and C). Likewise, an N-terminally truncated variant of RECQ5 of amino acids 541–991 was found to interact with the RNAPII CTD in the yeast two-hybrid system, although it was defective in interacting with RNAPIIA (Figure 1B and C, and data not shown).
RECQ5 specifically binds to Ser2,5-phosphorylated CTD heptapeptide repeats
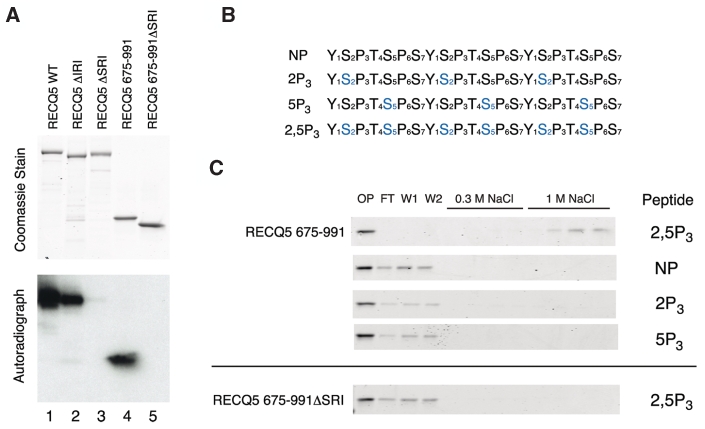
As an initial test of P-CTD binding, purified recombinant versions of RECQ5 were subjected to SDS–PAGE and far western analysis using a 32P-labeled GSTyCTD fusion protein phosphorylated on Ser2 and Ser5 by yeast CTDK-I (binding probe is expected to carry ∼25 doubly phosphorylated repeats). The full-length RECQ5 bound very well to the P-CTD fusion protein (Figure 2A, Lane 1), whereas RECQ5 deleted for the SRI domain did not bind appreciably (Figure 2A, Lane 3). Likewise, a C-terminal fragment containing the SRI domain (residues 675–991) bound to P-CTD-like full-length RECQ5, but a similar fragment lacking the SRI domain did not show any binding (Figure 2A, Lanes 4 and 5). Notably, RECQ5 missing only the IRI domain bound P-CTD almost as well as the wild-type protein (Figure 2A, Lane 2 versus 1). These data indicate that RECQ5 binding to the P-CTD requires the SRI domain.
Figure 2.
Effect of CTD phosphorylation status on RECQ5–CTD interaction. (A) Binding of RECQ5 and its mutants to in vitro phosphorylated CTD. Top panel: Coomassie stained gel. Bottom panel: far western blot probed with 32P-labeled GSTyCTD fusion protein hyperphosphorylated with CTDK-I. (B) Depiction of synthetic three-repeat CTD peptides used for the binding assay. The serines in blue are phospho-serines. (C) Immobilized peptide-binding assay of RECQ5 675-991 on the four synthetic peptides. Fractions were analyzed by SDS–PAGE with Coomassie blue staining. OP, onput; FT, flow-through; W1 and W2, washes.
To check whether the SRI domain of RECQ5 has the same binding specificity as the SRI domain of Set2 (22), synthetic biotinylated peptides with three consensus CTD heptapeptide repeats were immobilized on avidin-coated beads to create affinity resins. Because the full-length RECQ5 showed significant background interaction with the TetraLink resin, we used the C-terminal fragment of RECQ5 with and without the SRI domain for the peptide-binding experiments. Binding was evaluated with four different phosphorylated peptides: (i) a non-phosphorylated peptide (NP); (ii) a peptide with phospho-serines at position 2 of each repeat (2P3); (iii) a peptide with phospho-serines at position 5 of each repeat (5P3); and (iv) a peptide with phospho-serines at positions 2 and 5 of each repeat (2,5P3) (Figure 2B). The RECQ5 675-991 protein bound well to the resin with the 2,5P3 peptide: the protein was depleted from the column flow-through fraction and was only eluted from the resin by 1 M NaCl (Figure 2C, top panel). In contrast, RECQ5 675–991 did not bind detectably to resins with the NP, 2P3 or 5P3 peptides (Figure 2C, top panel). Moreover, RECQ5 675-991ΔSRI was unable to bind to the 2,5P3 column, confirming that the SRI domain of RECQ5 is required for interaction with the phosphorylated CTD (Figure 2C, bottom panel). These results substantiate that the SRI domain of RECQ5, like the SRI domain of Set2, binds directly and specifically to CTD repeats with phospho-serines at positions 2 and 5.
The SRI, but not IRI, domain of RECQ5 mediates its binding to the hyperphoshorylated form of RNAPII
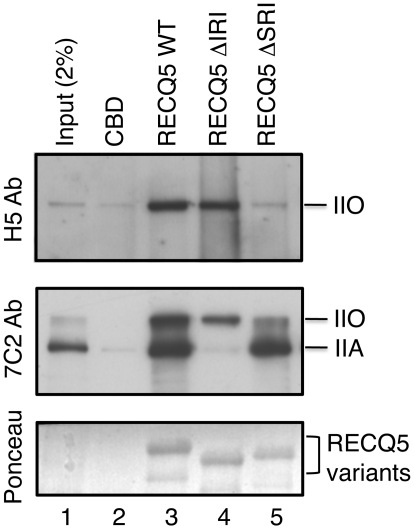
Next we compared the binding capabilities of wild-type and mutant forms of RECQ5 to RNAPII holoenzyme from human cells. To do so, RECQ5 proteins were produced in bacteria as fusions with a CBD, bound to chitin beads and incubated with a total extract of HEK293T cells. RNAPII binding was analyzed by western blotting using two different mouse monoclonal antibodies against the RNAPII CTD: (i) H5 that recognizes CTD repeats containing phospho-Ser2 and hence selectively detects the hyperphosphorylated form of RNAPII (IIO); and (ii) 7C2 that recognizes CTD repeats irrespective of their phosphorylation status and, therefore, can detect both RNAPIIO and RNAPIIA. As expected, wild-type RECQ5 was found to bind both forms of RNAPII (Figure 3, Lane 3). RECQ5 ΔIRI was impaired in binding to RNAPIIA but exhibited binding to RNAPIIO with an extent similar to that of wild-type RECQ5 (Figure 3, Lane 4). On the contrary, RECQ5 ΔSRI showed binding to RNAPIIA but was impaired in interacting with RNAPIIO (Figure 3, Lane 5). These data indicate that the binding of RECQ5 to the hyperphosphorylated form of RNAPII is mediated by the SRI domain of RECQ5.
Figure 3.
Binding of RECQ5 and its mutants to RNAPIIA and RNAPIIO. Chitin beads coated with either wild-type (WT) or mutant forms (ΔIRI or ΔSRI) of RECQ5 produced in bacteria as fusions with the CBD were incubated with HEK293 T cell extract. RNAPII binding was analyzed by western blotting using the H5 antibody that recognizes RNAPIIO (top panel) and 7C2 antibody that recognizes both RNAPIIA and RNAPIIO (middle panel). RECQ5 proteins were visualized by Ponceau S staining (bottom panel). Lane 1, 2% of input material.
RECQ5 associates with RNAPII-transcribed genes within the region of productive elongation
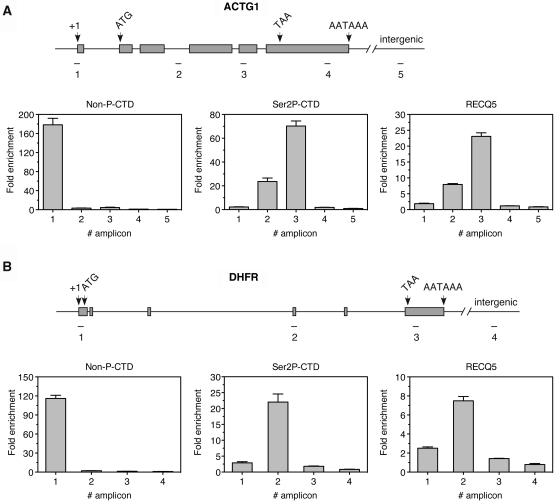
RNAPII binds to the promoter with a CTD in the non-phosphorylated state. CTD phosphorylation on Ser5 is one of the first steps in transcription initiation and leads to the movement of the transcription complex to a promoter-proximal pausing site. The escape of RNAPII from the pausing site and subsequent productive elongation of the transcript is associated with Ser2 phosphorylation of the CTD repeats (19). To investigate the interaction between RECQ5 and RNAPII during the transcription cycle, we used ChIP to characterize the distribution of RECQ5 along constitutively expressed genes. We chose two genes, the ACTG1 gene encoding γ-actin (3.1 kb) and the DHFR gene encoding dihydrofolate reductase (30 kb) (Figure 4A and B, top panels), which were used previously to dissect the distribution of RNAPII and its phosphorylated isoforms along the transcriptional unit (24). Exponentially growing HeLa cells were crosslinked with formaldehyde. Isolated chromatin fraction was sonicated to shear the genomic DNA into small fragments ranging from 200 to 500 bp and subjected to immunoprecipitation using the following antibodies: (i) 8WG16 that specifically recognizes non-phosphorylated CTD repeats; (ii) ab5095 that specifically binds to phospho-Ser2 in CTD; and (iii) anti-RECQ5 antibody raised against the C-terminal fragment of RECQ5 spanning amino acids 675–991. After reversing the crosslinks, ChIP-enriched DNA fragments were subjected to qPCR analysis using primers amplifying the core promoter, different regions within the transcriptional unit and an intergenic region located downstream of each gene (Figure 4A and B, top panels). Amplicon sizes ranged between 109 and 174 bp. In agreement with the published data (24), we found that the non-phosphorylated RNAPII was bound exclusively to the core promoter of the tested genes (Figure 4). Ser2-phosphorylation of RNAPII was nearly absent at the core promoter and accumulated in the body of each gene (Figure 4). On both genes, the phospho-Ser2 signal dramatically dropped in the 3′-untranslated region (3′-UTR) and reached intergenic background levels (Figure 4). Although RECQ5 could bind to the hypophosphorylated form of RNAPII, it showed a relatively low occupancy at the promoter region (Figure 4). Notably, the RECQ5 distribution pattern was almost identical to that of phospho-Ser2-CTD (Figure 4). Thus, our data suggest that, during the RNAPII transcription cycle, RECQ5 associates with the productive elongation complex most likely through binding to Ser2,5 P-CTD.
Figure 4.
Distribution of RECQ5 and RNAPII along the ACTG1 and DHFR genes. (A) and (B) Top panels: schematic depiction of the ACTG1 and DHFR genes showing the locations of the transcription start site (+1), exons (grey boxes) and introns (lines connecting the grey boxes), the start codon (ATG), the stop codon (TAA) and the polyadenylation signal (AATAAA). The location of the amplicons used in qPCR analysis is also shown. The positions of the central base pair of each amplicon relative to the transcription start site are given in Supplementary Table S2. (A) and (B) Bottom panels: plots of data from qPCR analysis. ChIPs of HeLa cells were performed with the 8WG16 antibody that specifically recognizes the non-phosphorylated CTD (non-P-CTD) of RNAPII, the ab5095 antibody that specifically recognizes the Ser2 phosphorylated CTD (Ser2 P-CTD) of RNAPII and anti-RECQ5 antibody. Fold enrichment was calculated as a ratio of the amount of DNA estimated for a specific antibody versus the amount of DNA estimated for the control IgG. The amplification of an intergenic region served as an internal background control.
The SRI, but not IRI, domain of RECQ5 mediates its association with RNAPII-transcribed genes
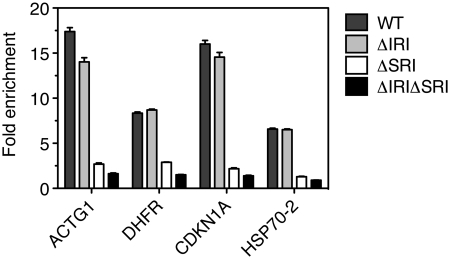
To investigate which of the two RNAPII-binding domains of RECQ5 is responsible for the observed accumulation of RECQ5 at the coding regions of the ACTG1 and DHFR genes, the full-length RECQ5, RECQ5 ΔIRI, RECQ5 ΔSRI and RECQ5 ΔIRIΔSRI, respectively, were ectopically expressed in HEK293 cells as fusions with GFP (Supplementary Figure S2) and subjected to ChIP analysis using an anti-GFP antibody. In control ChIP experiments, cells were transfected with empty vector expressing GFP only. The results obtained from the qPCR analysis clearly indicated that the SRI domain of RECQ5 was essential for the binding of RECQ5 to the coding regions of the ACTG1 and DHFR genes (Figure 5). In contrast, the IRI domain of RECQ5 was found to be dispensable for its association with these genes (Figure 5). To confirm our finding, we also examined the binding of RECQ5 and its mutants to the coding regions of two other genes, namely CDKN1A (encoding p21) and HSP70-2. As expected, GFP-tagged wild-type RECQ5 was largely enriched at the coding region of these two genes (Figure 5). The mutants lacking the SRI domain, RECQ5 ΔSRI and RECQ5 ΔIRIΔSRI, failed to accumulate at these regions, while the RECQ5 ΔIRI mutant showed similar fold of enrichment as wild-type RECQ5 (Figure 5).
Figure 5.
The SRI domain of RECQ5 mediates its association with RNAPII-transcribed genes. HEK293 cells were transfected either with empty vector (mock) or with vectors expressing wild-type (WT) or mutant (ΔIRI, ΔSRI or ΔIRIΔSRI) forms of RECQ5 as fusions with GFP. Forty-eight hours post-transfection, chromatin was immunoprecipitated by anti-GFP antibody and subjected to qPCR analysis using primer pairs complementary to the coding regions of the ACTG1, DHFR, CDKN1A and HSP70-2 genes (Supplementary Table S2). Fold enrichment was calculated as a ratio of the qPCR values obtained with RECQ5-transfected cells versus mock-transfected cells.
Point mutagenesis studies on the SRI domain of human SETD2 identified several residues as critical for P-CTD binding (22). We generated mutations in equivalent residues of RECQ5 (F938L, K939A and R943A). Out of these, the R943A mutation completely abolished the interaction of RECQ5 411–991 with the CTD in the yeast two-hybrid system (Supplementary Figure S3A). Likewise, the R943A mutation abolished the association of RECQ5 with the coding region of the ACTG1 gene (Supplementary Figure S3B).
Taken together, our results indicate that out of the two RNAPII-interacting domains of RECQ5, the SRI domain is necessary and sufficient to mediate the association of RECQ5 with the coding regions of RNAPII-transcribed genes. Moreover, these data imply that the recruitment of RECQ5 to the sites of transcription is dependent on Ser2,5 phosphorylation of the RNAPII CTD.
RECQ5 negatively affects cell viability upon inhibition of spliceosome assembly
To understand the functional role of RECQ5 during RNAPII transcription, we explored the possibility that RECQ5 is involved in the cellular response to co-transcriptionally formed R-loops. To this end, we first examined whether RECQ5 could unwind an RNA : DNA hybrid duplex prepared by annealing of a synthetic 18-mer RNA oligonucleotide to M13mp2 ssDNA. We found that RECQ5 failed to unwind this structure although it could unwind the corresponding DNA : DNA duplex (Supplementary Figure S4). This finding suggests that RECQ5 does not disrupt R-loops in vivo.
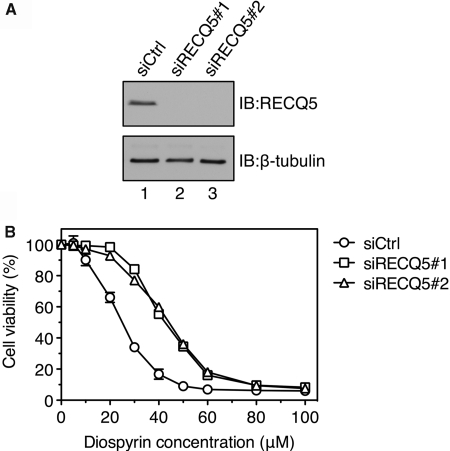
Next, we evaluated the effect of RECQ5 depletion on cell survival upon inhibition of spliceosome assembly, which leads to the formation of R-loops. To do so, RECQ5 was down-regulated in HeLa cells by RNA interference using two different siRNAs (Figure 6A). RECQ5-proficient and RECQ5-deficient cells were treated with diospyrin that blocks spliceosome assembly. It was shown that this Top1 kinase inhibitor caused the same phenotypic defects as ASF/SF2 depletion (3). Surprisingly, we found that RECQ5-proficient cells displayed a much higher sensitivity to diospyrin than RECQ5-deficient cells (Figure 6B). These data suggest that RECQ5 negatively affects cell viability in conditions leading to the formation of R-loops.
Figure 6.
RECQ5 depletion alleviates sensitivity of human cells to diospyrin. (A) Western blot analysis of extracts of HeLa cells transfected with RECQ5 siRNA (siRECQ5 #1 and 2) and control siRNA (siCtrl), respectively. Cells were harvested 72 h post-transfection. Blots were probed with antibodies against RECQ5 and β-tubulin (loading control). (B) Graph showing the percentage survival of RECQ5-proficient and RECQ5-deficient cells after treatment with different concentrations of diospyrin. Cell viability assays were performed as described in ‘Materials and Methods’ section. Each data point represents mean ± SD (n = 6). IB, immunoblotting.
DISCUSSION
Genetic ablation of RECQ5 helicase in mice results in genomic instability and cancer susceptibility (6). Studies in mouse and human cells have suggested that RECQ5 regulates HR by promoting disassembly of the RAD51 presynaptic filament (6). However, RECQ5 was also found to interact with RNAPII in human cells, suggesting a role in transcription (7–9). In this work, we provide evidence that RECQ5 associates with RNAPII during the productive elongation phase of transcription through direct binding to the Ser2,5 P-CTD of RPB1 by means of a SRI motif located at the C-terminus of RECQ5. Although RECQ5 could also bind to hypophosphorylated form of RNAPII through an additional domain, herein referred as IRI domain, it showed relatively low occupancy at the promoter regions of RNAPII-transcribed genes, excluding a role for RECQ5 during transcription initiation. In addition, we show that RECQ5 negatively affects cell viability upon inhibition of spliceosome assembly, which can induce the formation of mutagenic R-loop structures. Moreover, we demonstrate that RECQ5 cannot unwind RNA : DNA duplexes in vitro, which excludes the possibility the RECQ5 directly disrupts R-loops. Together, these findings suggest that RECQ5 might play a role in the maintenance of genomic stability during RNAPII transcription.
Recently, Islam et al. (25) have also reported that the SRI domain of RECQ5 mediates its binding to the hyperphosphorylated form of RNAPII in human cells. In addition, these authors have found that the IRI domain of RECQ5 shares extensive homology with the so-called KIX domain identified as a protein interaction module in several RNAPII transcription regulators. In agreement with our results, they demonstrated that mutations at conserved residues in the IRI/KIX domain of RECQ5 abolished the interaction of RECQ5 with RNAPIIA, but not with RNAPIIO (25). However, in contrast to our observations, Islam et al. (25) found that the IRI/KIX domain of RECQ5 could bind to both RNAPIIA and RNAPIIO. This discrepancy may arise from the use of different experimental approaches. Whereas we analyzed binding of RNAPII from a cell extract to beads coated with recombinant RECQ5 protein produced in bacteria, Islam et al. (25) monitored RECQ5–RNAPII complex formation in vivo by immunoprecipitation. Thus, it is possible that efficient binding of the IRI/KIX domain of RECQ5 to RNAPIIO is dependent on a post-translational modification of RECQ5, which is absent if the protein is produced in bacteria. Nevertheless, our ChIP experiments clearly show that the IRI/KIX domain of RECQ5 does not play a role in the association of RECQ5 with the coding regions of RNAPII-transcribed genes, where the RNAPII CTD exists in the hyperphosphorylated state. Further studies are needed to elucidate the complex mode of the RECQ5–RNAPII interaction. In particular, the site of interaction for the IRI/KIX domain of RECQ5 on RNAPII needs to be determined.
What is the exact role of RECQ5 during the elongation phase of RNAPII transcription? Our experiments with diospyrin suggest that RECQ5 might be involved in the suppression of the DNA-damaging effects of RNAPII transcription rather than in the repair of transcription-induced DNA damage. Interestingly, a recent study by Aygun et al. (10) showed that RECQ5 could directly inhibit RNAPII transcription in vitro in a manner dependent on its IRI/KIX domain, which led the authors to propose a model in which RECQ5 promotes genome stability by regulating RNAPII transcription itself. In line with this hypothesis, Islam et al. (25) demonstrated that the IRI/KIX, but not the SRI, domain of RECQ5 was required for suppression of sister chromatid exchange and resistance to camptothecin-induced DNA damage. However, it should be noted that the in vitro transcription system used by Aygun et al. (10) lacked CTD kinase activity responsible for CTD phosphorylation on Ser2 during transcription. Our experiments with the RECQ5 ΔSRI mutant have indicated that hyperphosphorylation of RNAPII prevents binding of RECQ5 to RNAPII via the IRI/KIX domain, suggesting that Ser2 CTD phosphorylation may alleviate the inhibitory effect of RECQ5 on RNAPII transcription so that RECQ5 could not affect RNAPII transcription during productive elongation in vivo. However, it is possible that a formation of an R-loop during transcription could trigger a domain interaction switch in the RECQ5–RNAPII complex to adopt the inhibitory arrangement, which would prevent further extension of the R-loop structure and hence diminish its DNA-damaging effect. Our finding of negative effect of RECQ5 on cell viability under conditions that favor R-loop formation is consistent with this hypothesis. Future work will clarify the molecular mechanism of this phenomenon to shed light on the biological processes that enforce genomic stability during transcription.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation (3100A0-116008); UBS AG; Indo-Swiss Research Programme; Sassella-Stiftung; Czech Science Foundation (GA204/09/0565); National Institutes of Health (USA) (GM040505). Funding for open access charge: UBS AG.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Jean Marc Egly for the RNAPII 7C2 antibody, Dr Banasri Hazra (Jadavpur University, Kolkata, India) for diospyrin, Stefano di Marco for help with cell culture and Christiane Koenig for excellent technical assistance.
REFERENCES
- 1.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 3.Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 5.Hu Y, Lu X, Barnes E, Yan M, Lou H, Luo G. Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol. Cell. Biol. 2005;25:3431–3442. doi: 10.1128/MCB.25.9.3431-3442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng L, Kanagaraj R, Mihaljevic B, Schwendener S, Sartori AA, Gerrits B, Shevelev I, Janscak P. MRE11 complex links RECQ5 helicase to sites of DNA damage. Nucleic Acids Res. 2009;37:2645–2657. doi: 10.1093/nar/gkp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aygun O, Svejstrup J, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc. Natl Acad. Sci. USA. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izumikawa K, Yanagida M, Hayano T, Tachikawa H, Komatsu W, Shimamoto A, Futami K, Furuichi Y, Shinkawa T, Yamauchi Y, et al. Association of human DNA helicase RecQ5beta with RNA polymerase II and its possible role in transcription. Biochem. J. 2008;413:505–516. doi: 10.1042/BJ20071392. [DOI] [PubMed] [Google Scholar]
- 10.Aygun O, Xu X, Liu Y, Takahashi H, Kong SE, Conaway RC, Conaway JW, Svejstrup JQ. Direct inhibition of RNA polymerase II transcription by RECQL5. J. Biol. Chem. 2009;284:23197–23203. doi: 10.1074/jbc.M109.015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tazi J, Bakkour N, Soret J, Zekri L, Hazra B, Laine W, Baldeyrou B, Lansiaux A, Bailly C. Selective inhibition of topoisomerase I and various steps of spliceosome assembly by diospyrin derivatives. Mol. Pharmacol. 2005;67:1186–1194. doi: 10.1124/mol.104.007633. [DOI] [PubMed] [Google Scholar]
- 12.Schwendener S, Raynard S, Paliwal S, Cheng A, Kanagaraj R, Shevelev I, Stark JM, Sung P, Janscak P. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J. Biol. Chem. 2010;285:15739–15745. doi: 10.1074/jbc.M110.110478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mockli N, Auerbach D. Quantitative beta-galactosidase assay suitable for high-throughput applications in the yeast two-hybrid system. Biotechniques. 2004;36:872–876. doi: 10.2144/04365PT03. [DOI] [PubMed] [Google Scholar]
- 15.Phatnani HP, Greenleaf AL. Identifying phosphoCTD-associating proteins. Meth. Mol. Biol. 2004;257:17–28. doi: 10.1385/1-59259-750-5:017. [DOI] [PubMed] [Google Scholar]
- 16.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 18.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Crit. Rev. Biochem. Mol. Biol. 2009;44:117–141. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vojnic E, Simon B, Strahl BD, Sattler M, Cramer P. Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. J. Biol. Chem. 2006;281:13–15. doi: 10.1074/jbc.C500423200. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Phatnani HP, Guan Z, Sage H, Greenleaf AL, Zhou P. Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc.Natl Acad. Sci. USA. 2005;102:17636–17641. doi: 10.1073/pnas.0506350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourquin JP, Stagljar I, Meier P, Moosmann P, Silke J, Baechi T, Georgiev O, Schaffner W. A serine/arginine-rich nuclear matrix cyclophilin interacts with the C-terminal domain of RNA polymerase II. Nucleic Acids Res. 1997;25:2055–2061. doi: 10.1093/nar/25.11.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng C, Sharp PA. RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and gamma-actin genes. Mol. Cell. Biol. 2003;23:1961–1967. doi: 10.1128/MCB.23.6.1961-1967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Islam MN, Fox D, 3rd, Guo R, Enomoto T, Wang W. RecQL5 promotes genome stabilization through two parallel mechanisms—interacting with RNA polymerase II and Acting as a helicase. Mol. Cell Biol. 2010;30:2460–2472. doi: 10.1128/MCB.01583-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.