Abstract
T-cell intracellular antigen (TIA)-proteins are known regulators of alternative pre-mRNA splicing. In this study, pull-down experiments and mass spectrometry indicate that TIAR/TIAL1 and hnRNP C1/C2 are associated in HeLa nuclear extracts. Co-immunoprecipitation and GST-pull-down assays confirmed this interaction. Interestingly, binding requires the glutamine-rich (Q-rich) C-terminal domain of TIAR and the leucine-rich plus acidic residues-rich C-terminal domains of hnRNP C1/C2. This interaction also occurs in an RNA-dependent manner. Recombinant GFP-TIAR and RFP-hnRNP C1 proteins display partial nuclear co-localization when overexpressed in HeLa cells, and this requires the Q-rich domain of TIAR. hnRNP C1 overexpression in the presence of rate-limiting amounts of TIAR in HeLa and HEK293 cells affects alternative splicing of Fas and FGFR2 minigenes, promoting Fas exon 6 and FGFR2 exon K-SAM skipping, respectively. The repressor activity of hnRNP C1 on Fas exon 6 splicing is mediated by Hu antigen R (HuR). Experiments involving tethering approaches showed that the repressor capacity of hnRNP C1 is associated with an exonic splicing silencer in Fas exon 6. This effect was reversed by splice-site strengthening and is linked to its basic leucine zipper-like motif. These results suggest that hnRNP C1/C2 acts as a bridge between HuR and TIAR to modulate alternative Fas splicing.
INTRODUCTION
Alternative pre-mRNA splicing is a major regulatory process for generating transcriptome and proteome diversity, with >95% of human multi-exon genes expressed from alternatively spliced mRNAs (1,2). Alternative splicing makes a significant contribution to the temporal and spatial diversity of protein isoforms in higher eukaryotes (3). The alternatively spliced regions of these isoforms include functional domains affecting features such as binding to DNA, RNA or other proteins, intracellular localization, enzymatic activity, regulation and stability (3). Thus, this phenomenon effectively expands the coding capacity of the human genome.
The vast majority of human introns do not contain bona fide consensus sequences at their splice sites, branch point or polypyrimidine tract, and instead have weak splicing signals with non-consensus features (4). Additional sequence elements located in exons or introns are involved in the differential use of splice sites. They are divided into four categories according to their position and function: exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs), intronic splicing enhancers (ISEs) and intronic splicing silencers (ISSs). These elements are often needed to identify authentic splicing signals within long-non-coding introns and, thus, to achieve correct splicing of exons. The fine balance between positive and negative regulation of splice site selection is likely to involve the combinatorial and coordinated action of cis-acting RNA sequence elements and trans-acting protein regulators. These intricate protein–RNA networks can be modulated in response to physiological and pathological conditions (5).
Heterogeneous nuclear ribonucleoproteins (hnRNPs) are a family of RNA-binding proteins that participate in both constitutive and alternative pre-mRNA splicing in mammalian cells (6). They are among the most abundant RNA-binding proteins, forming the core of the ribonucleoprotein complex that associates with nascent transcripts in eukaryotic cells, packaging them into hnRNP particles. The family has several members and some of them are subject to alternative transcript splicing and post-translational modifications. This structural variation is paralleled by wide functional diversity, largely involving interactions with DNA or, more commonly, RNA. They also recruit regulatory proteins associated with DNA and RNA metabolism, and appear to accompany transcripts throughout the life of the mRNA (5,6).
T-cell intracellular antigen (TIA) proteins are involved in controlling the metabolism and dynamics of both nuclear and cytoplasmic transcriptomes through mechanisms involving transcription, alternative pre-mRNA splicing and regulation of mRNA stability and translation (7). TIA-1 and TIAR/TIAL1 (‘TIA-1 related/like protein’) are structurally close to hnRNPs and composed of three RNA recognition motifs (RRMs) and a glutamine-rich (Q-rich) C-terminal domain (8,9). TIA proteins regulate the alternative pre-mRNA splicing of ∼15% of alternative cassette human exons as well as 5′-splice-site selection of Alu exons through binding to U-rich intronic stretches, facilitating atypical 5′-splice site recognition by U1 small nuclear ribonucleoprotein (10–15). In addition, these proteins regulate turnover and translation of cytoplasmic mRNAs (7,16–19).
This work addresses the molecular mechanisms underlying the regulation of Fas exon 6 splicing, an alternative splicing event that leads to the production of mRNA encoding either a membrane-bound form of the Fas receptor that promotes programmed cell death or a soluble form that inhibits apoptosis. The process is regulated in a number of physiological situations, including activation-mediated T-cell death, and failure to undergo the switch between isoforms is responsible for certain autoimmune lymphoproliferative disorders (13 and references therein). To improve our understanding of the mechanisms and networks governing splice-site selection, this study set out to identify nuclear proteins that interact with TIA-proteins and determine the contribution of these auxiliary proteins to the regulation of alternative splicing. The author reports that the hnRNP C1/C2 protein inhibits splicing through a mechanism involving HuR and propose a model in which hnRNPC1/C2 and TIAR play opposite roles in the regulation of alternative pre-mRNA splicing.
MATERIALS AND METHODS
Cell cultures and plasmid constructs
HeLa and HEK293 cells were cultured under standard conditions (20). Plasmids containing human hnRNP C1, TIAR, TIARΔQ (lacking the Q-rich C-terminal domain) cDNAs were generated by PCR using Pfu Turbo DNA polymerase (Stratagene, USA). The wt, mutant U-20C, and m0 Fas minigenes, and also the wt Fas 2xMS2 minigene and its derivatives were generated as described earlier (11,21,22). The FGFR2 minigenes RK97 and A7 were kindly provided by R. Breathnach (10,23).
HeLa nuclear extracts, preparation of recombinant proteins and in vitro transcription and translation assays
HeLa nuclear extracts (NEs) were purchased from 4C Biotech (Seneffe, Belgium). GST, MBP, GST-hnRNP C1, GST-TIAR, MBP-HuR, and their mutant derivatives were expressed in and purified as described earlier (22). In vitro transcription and translation assays were performed with a coupled transcription and translation system (Promega, USA) (24). HeLa NEs were incubated with calf intestinal alkaline phosphatase (calf intestinal phosphatase, CIP; Roche, USA) according to the manufacturer’s instructions.
In-gel digestion of proteins and sample preparation for mass spectrometric analysis
Protein spots were excised manually and then digested automatically using a Proteineer DP protein digestion station (Bruker-Daltonics, Bremen, Germany). The digestion protocol used was that of Schevchenko et al. (25), with minor variations (extended version in Supplementary Data).
MALDI peptide mass fingerprinting, LIFT TOF/TOF acquisition and database searching
Peptide mass fingerprint spectra were measured on a Bruker Ultraflex TOF/TOF MALDI mass spectrometer (Bruker-Daltonics, USA) (26) in positive ion reflector mode (extended version in supplementary material).
Immunoprecipitation
Anti-TIA1, anti-TIAR and anti-hnRNP C1/C2 (Santa Cruz Biotechnology, USA) and anti-U2AF65 (MC3 clone provided kindly by J. Valcárcel), antibodies were added to 10 µl of NE in the absence of RNase A complemented with buffer D to a total volume of 25 µl and incubated for 1 h on ice. After addition of 20 µl of protein G-Sepharose beads (1/1) and 40 µl of IPP 100 buffer (10 mM Tris pH 8.0, 100 mM NaCl, 0.1% NP-40), the reaction was incubated for 1 h on a rotating wheel at 4°C. A 10 µl aliquot of each sample was removed as loading control, and the beads were collected at 1000 rpm and washed four times with ice-cold IPP 100 buffer. The loading control and pellet were analysed by western blotting.
GST/MBP-pull-down and western blot analyses
A 1–2 µg of recombinant GST, MBP, GST-TIAR, GST-hnRNP C1, MBP-HuR, or their mutant derivatives was incubated with either 5 µl of NE, 1 µg of purified recombinant protein, 3 µl of in vitro translated hnRNP C1, or partial fragments in rabbit reticulocyte lysates (RRL, Promega, USA) in a total volume of 25 µl, complemented with buffer D (20 mM HEPES pH 8.0, 20% glycerol, 0.2 mM EDTA, 100 mM KCl), for 30 min at 4°C. After addition of 60 µl of IPP 100 and 10 µl of glutathione-Sepharose 4B beads (GE Healthcare BioScience, UK) pre-equilibrated in the same buffer, the reaction was incubated for 1 h on a rotating wheel at 4°C. A 10 µl aliquot of the reaction was removed as a loading control and the pellet was washed four times with 400 µl of ice-cold IPP 100 buffer. After sedimentation by centrifugation at 1000 rpm for 1 min, the pellet was resuspended in SDS loading buffer, fractionated together with an aliquot of supernatant by electrophoresis on a 10% or 4–12% SDS-polyacrylamide gel (Invitrogen, USA). The following antibodies were used: anti- hnRNP C1/C2 and anti-TIAR (Santa Cruz Biotechnology, USA), anti-Stat3 (Phospho-Stat3 [Tyr705] [D3A7] XPTM) rabbit mAb (Cell Signalling, USA), Stat3 (C-20) rabbit polyclonal antibody (sc-482) (Santa Cruz Biotechnology, USA), anti-MS2 coat protein (kindly provided by Peter G. Stockley), anti-GFP (JL-8, Clontech, USA), and anti-α-tubulin (B512, Sigma-Aldrich, USA).
Fluorescence microscopy
After transfection (24 h), the cells were analysed by fluorescence confocal microscopy with GFP and RFP excitation filters. Fixation, permeabilization, To-pro-3 staining and visualization were performed as described earlier (22).
Transfections
Exponentially grown HeLa and HEK293 cells were transfected with 0.2–2.0 µg of plasmid DNA as described previously (10,11). Depletion of hnRNP C1/C2 gene expression was performed by pSUPER RNAi System (Oligoengine, USA) using two independent specific shRNAs against the 3′-untranslated region of the hnRNP C1/C2 mRNA as reported (27,28).
RNA purification and analysis
Cytoplasmic RNA was isolated by using an RNeasy kit (Qiagen, USA) and was analysed by RT–PCR and electrophoresis. Control experiments with different input amounts of RNA and different ratios between the alternatively spliced mRNAs indicated that the amplification of the species was quantitative under these conditions (13,20).
RNA–protein interaction assays
Band shift and UV-mediated cross-linking assays were performed as described earlier (13,22).
RESULTS
Polypeptides associated with TIAR in HeLa NEs
GST-pull down assays with recombinant GST alone and GST-TIAR proteins were used to identify major nuclear proteins interacting with TIAR from HeLa NEs. Precipitates were separated on SDS–polyacrylamide gels and stained with silver nitrate. Bands with increased intensity compared with GST and GST-TIAR controls in the absence of NEs (Figure 1A, compare lanes 2 and 3 with 4 and 5) were analysed by matrix-assisted laser desorption ionization time-of-flight mass spectroscopy (MALDI-TOF-MS). Four polypeptides of around 125 (weakly stained), 52, 45 and 37 kDa met the criteria (Figure 1A and Supplementary Table IS). These proteins were identified as matrin 3, an unnamed protein, beta-actin and hnRNP C1/C2 (Figure 1B). The protein score in MALDI-TOF-MS, accession number in NCBInr, and role of these proteins in the nucleus are summarised in Figure 1B. No other proteins were found to be co-precipitated with TIAR following GST-pull down. This may have been due to insufficient amounts of starting material or a failure to discriminate proteins with similar molecular sizes by SDS–PAGE.
Figure 1.
hnRNP C1/C2 is associated with TIAR in HeLa NEs. (A) Identification of TIAR precipitates from GST pull-downs. HeLa NEs were subjected to GST pull-down using either GST alone or GST-TIAR (GST-R). An aliquot (1 µl) of NE and pulled down proteins were separated on SDS-polyacrylamide gels and visualized by silver nitrate staining. The positions of recombinant GST-TIAR and GST proteins as well as the four putative protein products and their apparent molecular sizes are indicated. Protein molecular weight markers are shown to the left. (B) TIAR-interacting proteins identified by MALDI-TOF-MS. The protein products indicated in (A) were removed and identified by mass spectrometry. The identities of the corresponding protein products are indicated. Four proteins were identified: matrin 3 (125 kDa, weakly stained), an unnamed protein (52 kDa), beta-actin (45 kDa) and hnRNP C1/C2 (37 kDa). Their apparent molecular weights on SDS–PAGE are indicated. The protein score in MALDI-TOF-MS, accession number in NCBInr, and role in the nucleus are also summarised. (C–E) Validation of the association between hnRNP C1/C2 and TIAR. (C) Co-immunoprecipitation of hnRNP C1/C2 and TIA-proteins. HeLa NEs in the absence of RNase were processed for immunoprecipitation using either beads alone or antibodies indicated and analysed by immunoblotting using the same specific antibodies. An aliquot (10%) of HeLa NE prior to immunoprecipitation served as a control for the detection of protein immunoprecipitates. (D) Precipitation of hnRNP C1/C2 by GST-TIAR. GST pull-down assays were carried out after addition of GST alone or GST-TIAR to NEs. (E) Precipitation of TIAR by GST-hnRNP C1. GST pull-down assays were carried out after addition of GST alone or GST-hnRNP C1 to NEs. In both GST pull-down analyses (D and E), aliquots of the HeLa NEs (25%) prior to pull-down assays, the corresponding supernatants (SN) (10%) and pulled down proteins were analysed by western blotting in parallel using antibodies indicated. The positions of hnRNP C1/C2, TIAR and molecular weight markers are indicated.
Co-immunoprecipitation and GST pull-down of hnRNP C1/C2 and TIAR
To validate the association of hnRNP C1/C2 with TIAR, hnRNP C1/C2 was immunoprecipitated from HeLa NEs in the absence of RNase A treatment using specific antibodies against TIA1, TIAR and hnRNP C1/C2, and the presence of these proteins in the precipitates was analysed by western blot (Figure 1C). As a control, the presence of the splicing factor U2 snRNP auxiliary factor 65 kDa (U2AF65) was also tested. One-tenth of the HeLa NE prior to immunoprecipitation was also included as a control for detection of all immunoprecipitated proteins (Figure 1C, lane 1). Significant levels of hnRNP C1/C2 and TIA proteins were detected reproducibly in corresponding precipitates, but not in precipitates using beads or anti-U2AF65 antibody (Figure 1C, compare lane 2 and 6 with 3–5). To confirm the results obtained in co-immunoprecipitation experiments, GST-pull-down assays were also used. GST alone or a GST fusion containing full-length TIAR were added to NEs and their association with hnRNP C1/C2 analysed by western blotting. hnRNP C1/C2 was pulled down by GST-TIAR but not by GST alone (Figure 1D, compare lanes 2 and 3 with 4 and 5). Reciprocal experiments were also consistent with a specific association between hnRNP C1 and TIAR. Western blot analysis showed that TIAR was present in GST-hnRNP C1 precipitates, whereas TIAR was not precipitated by GST alone (Figure 1E, compare lanes 2 and 3 with 4 and 5). In all GST pull-down experiments, aliquots of the HeLa NEs (25%) and supernatants (10%) prior to precipitation were analysed in parallel. These results confirm the specificity of the association between TIAR and hnRNP C1/C2 in HeLa NEs and suggest that only a small portion of the total TIAR or hnRNP C1/C2 is involved in this interaction.
The Q-rich region of TIAR and the bZLP motif of hnRNP C1/C2 are necessary for TIAR-hnRNP association
Next, experiments were performed to identify the domain within TIAR responsible for the interaction with hnRNP C1/C2. TIAR contains three RRMs and a Q-rich C-terminal domain (Figure 2A) (8,9). The Q-rich domains of TIA proteins are reported to be essential for their protein–protein interactions (12,23,29). A TIAR deletion mutant lacking the Q-rich domain (TIARΔQ) was therefore generated. Recombinant GST fusions of wt TIAR and TIARΔQ were expressed in Echerichia coli and verified by immunoblot (Figure 2B). In GST pull-down assays, only GST-TIAR was able to precipitate significant amounts of hnRNP C1/C2 when added to NEs. To test whether TIAR contacts hnRNP C1 directly, 35S-labeled hnRNP C1 was generated by in vitro translation in RRL (Figure 2D, lanes 1 and 2), incubated with GST-TIAR and GST-TIARΔQ and, after precipitation using glutathione–agarose beads, the presence of hnRNP C1 in the precipitates was assessed by fractionation in denaturing SDS gels and autoradiography. hnRNP C1 co-precipitated strongly with GST-TIAR (Figure 2D, compare lanes 3 and 4 with 5 and 6) but only marginally with GST-TIARΔQ (Figure 2D, lanes 7 and 8). Similar results were obtained in the presence and absence of RNase A treatment (Figure 2E). RNase A activity was verified by analysing degradation of the RRL RNA present in the extract (Figure 2E, lower panel). However, when the TIAR-hnRNP C1/C2 interaction was analysed using HeLa NEs, the TIAR-hnRNP C1/C2 association appeared to be dependent upon RNA intermediates (Figure 2F). These results suggest that there is a protein–protein interaction mediated by the Q-rich domain of TIAR and also an RNA-mediated interaction between TIAR and hnRNP C1/C2. In addition, pull-down hnRNP C1 by GST-TIAR from RRL does not necessarily mean that the interaction is direct. It could be via other proteins present in the RRL.
Figure 2.
The Q-rich C-terminal domain of TIAR interacts with hnRNP C1/C2. (A) Schematic structure of human TIAR protein. The protein sequence of TIAR contains three RRM 1–3 and a Q-rich C-terminal domain. RRMs are represented by boxes and the Q-rich domain by an oval. The positions of specific residues limiting each domain are indicated. (B) Western-blot analysis of recombinant GST-TIAR wt and mutant versions. Equal amounts (50 ng) of GST-TIAR and GST-TIARΔQ (deletion mutant of TIAR without the Q-rich domain) were analysed by immunoblotting. (C) The Q-rich domain of TIAR is required for precipitation of hnRNP C1/C2 by TIAR. GST pull-down assays were carried out after addition of GST alone, GST-TIAR or GST-TIARΔQ to NEs, and the pellets were analysed by western blotting for the presence of hnRNP C1/C2. Aliquots of the HeLa NEs (25%) prior to pull-down assays and the corresponding supernatants (SN) (10%) were also analysed in parallel. (D) Interaction between GST-TIAR, GST-TIARΔQ, and in vitro translated hnRNP C1. Pull-down assays were carried out as in (C), using 35S-labeled hnRNP C1 translated in vitro. Pelleted proteins (P) and the pulled-down supernatant (SN) were analysed by SDS–PAGE and autoradiography. (E) Interaction between TIAR and in vitro translated hnRNP C1 is RNA-independent in RRL. Assays were carried out as in (D), using 35S-labeled hnRNP C1 translated in vitro in the presence (+) or absence (−) of RNase A (10 µg of RNase A for 15 min at 37°C). The lower panel shows verification of the RNase A treatment by agarose gel electrophoresis and ethidium bromide staining of the remaining RNA in the binding reaction. The positions of 35S-labeled hnRNP C1, endogenous RNA in RRL, and molecular weight markers are indicated. (F) RNA-dependent interaction between hnRNP C1/C2 and TIAR in HeLa NEs. GST pull-down assays were carried out after addition of GST alone or GST-TIAR to NEs without (−) or with (+) RNase A treatment prior to the pull-down assays, as indicated in (E). The presence of hnRNP C1/C2 in the corresponding supernatants (SN) and pellets (P) was analysed as in (C). In each case, the positions of hnRNP C1/C2 and molecular weight markers are indicated. (G) Phosphorylation-dependent association between hnRNP C1/C2 and TIAR in HeLa NEs. GST pull-down assays were performed with HeLa NEs incubated with (+) or without (−) calf intestinal phosphatase (CIP) for 1 h at 37°C. The presence of hnRNP C1/C2 in the corresponding supernatants (SN) and pellets (P) was analysed as before. The lower panel shows verification of the CIP treatment by western-blot analysis of relative protein expression levels of phosphorylated (Tyr705) and total transcription factor Stat3. The positions of hnRNP C1/C2, phosphorylated, and total Stat3 and molecular weight markers are indicated.
HnRNP C1/C2 is present in HeLa NEs as a phosphoprotein (30). To test whether the phosphorylation status of hnRNP C1/C2 is implicated in the binding activity mediated by TIAR, HeLa NEs were in vitro dephosphorylated with CIP. In GST pull-down assays, GST-TIAR was unable to precipitate significant amounts of hnRNP C1/C2 when added to CIP-treated NEs (Figure 2G). Protein dephosphorylation was verified by western blotting to determine the relative protein expression levels of the phosphorylated (Tyr705) and total transcription factor Stat3 in untreated (−) HeLa NEs and extracts treated with CIP (+) (Figure 2G, lower panel). This result suggests that phosphorylation-dephosphorylation cycles can regulate the degree of association between hnRNP C1/C2 and TIAR.
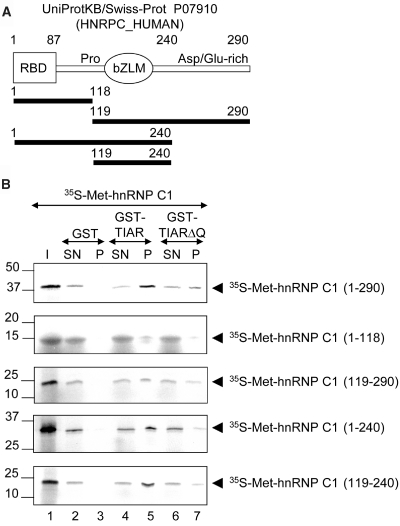
The protein sequence of hnRNP C1/C2, which is highly conserved among vertebrates, contains an RNA-binding domain (RBD), a basic leucine zipper (bZIP)-like motif (bZLP), both involved in RNA binding, and an acidic (Asp-Glu-rich) C-terminal domain (Figure 3A) (31–33). To define the fragment of hnRNP C1 that interacts with TIAR, in vitro translated hnRNP C1 and fragments containing functional domains of hnRNP C1 (Figure 3A) were tested in pull-down assays using GST-TIAR or GST-TIARΔQ. The results suggest that the hnRNP C1 fragments containing the bZLM and/or Asp/Glu-rich C-terminal domains are more important than the N-terminal residues in the interaction with TIAR (Figure 3B). Interestingly, these results were confirmed when GST-TIARΔQ was used to test the binding to the same fragments of hnRNP C1. As shown in Figure 3B, GST-TIARΔQ was marginally co-precipitated with separated fragments of 1–240 and 119–240 (Figure 3B, compare lanes 1–5 with 6 and 7). These results suggest that the contact surface between TIAR and hnRNP C1 mainly comprises the Q-rich domain of TIAR and the bZLM motif of hnRNP C1.
Figure 3.
The leucine zipper (bZIP)-like motif (bZLM) of hnRNP C1/C2 interacts with TIAR. (A) Schematic representation of the domains of human hnRNP C1 protein. The protein sequence of hnRNP C1 contains an RNA RBD, a bZIP-like motif (bZLM), and an acidic domain. The RBD and bZLM domains are represented by a rectangle and an oval, respectively. The white bar represents the proline (Pro) and Asp/Glu-rich C-terminal domains of hnRNP C1. The black bars show schematic representation of deletion mutants of human hnRNP C1. Amino acid numbers are indicated. (B) Interaction of hnRNP C1 with TIAR involves the bZLM and C-terminal regions of hnRNP C1 and the Q-rich domain of TIAR. Pull-down assays with GST, GST-TIAR and GST-TIARΔQ and in vitro translated full-length and deleted versions of hnRNP C1 were performed as in Figure 2D. The positions of full-length and deleted versions of 35S-labeled hnRNP C1 and molecular weight markers are indicated.
TIAR and hnRNP C1 co-localize partially in the nucleus of HeLa cells
HnRNP C1/C2 mostly accumulates in the nuclear compartment of HeLa cells (34), whereas TIAR shows a dual nuclear-cytoplasmic localization pattern (22). To test the TIAR-hnRNP C1 association and the participation of the Q-rich domain of TIAR, their subcellular distribution was compared by confocal microscopy. Expression constructs containing TIAR and hnRNP C1 fused with GFP and RFP, respectively, were transfected into HeLa cells and the subcellular localization of the fusion proteins analysed (Figure 4). HnRNP C1 was only observed in the cell nucleus, whereas TIAR was detected in both the nucleus and cytoplasm (Figure 4A, details in lower panels). Interestingly, TIAR and hnRNP C1 had a broad distribution in the nucleus and were present in a diffuse pattern in the surrounding nucleoplasm. The proteins appeared to be excluded from the nucleoli. Merging the fluorescence signals from GFP-TIAR and RFP-hnRNP C1 indicated that TIAR and hnRNP C1 can co-localize in the nucleus (Figure 4A, detail in Merge). To verify the role played by the Q-rich domain of TIAR, GFP-TIARΔQ and RFP-hnRNP C1 were co-expressed (Figure 4B). GFP-TIARΔQ displayed a similar nuclear-cytoplasmic distribution to GFP-TIAR, but there was only a partial overlap with RFP-hnRNP C1 (Figure 4B, detail in Merge, and Figure 4C). These results are consistent with our findings in co-immunoprecipitation and GST pull-down analyses, suggesting the capacity of TIAR to interact with hnRNP C1 through its Q-rich domain.
Figure 4.
Sub-cellular distribution and partial co-localization of hnRNP C1 and TIAR in the nucleus of HeLa cells. (A) Subcellular localization of hnRNP C1 and TIAR. HeLa cells were transiently transfected with RFP-hnRNP C1 (red) and GFP-TIAR (green) constructs. After 24 h post-transfection, the cells were fixed and the protein distribution was analysed by fluorescence confocal microscopy. (B) Partial co-localization of hnRNP C1 and TIAR is mediated by the Q-rich domain. HeLa cells were transiently transfected with RFP-hnRNP C1 (red) and GFP-TIARΔQ (green) constructs and processed as in (A). From left to right in (A) and (B), images show cells examined using phase contrast (Phase), To-pro-3 staining (To-pro-3), fluorescence from RFP (Cherry-hnRNP C1), fluorescence from GFP (GFP-TIAR and GFP-TIARΔQ), and merged pictures. In (A) and (B), lower panels show detailed images. The micrographs were taken with a laser-scanning confocal microscopy using a 63× objective. (C) Quantification of hnRNP C1 and TIAR/TIARΔQ colocalization in HeLa cells transfected with either RFP-hnRNP C1-GFP-TIAR or RFP-hnRNP C1-GFP-TIARΔQ constructs. Represented values are means ± SE for two independent experiments with at least seven counted fields in each of the experimental conditions analysed.
Functional relevance of the TIAR-hnRNP C1 interaction in the control of alternative splicing
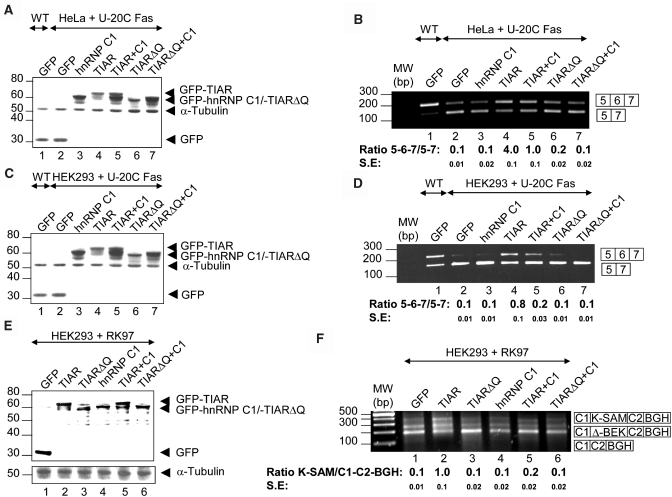
To assess the functional significance of the TIAR-hnRNP C1 interaction in the control of alternative pre-mRNA splicing, two TIAR-dependent splicing events were investigated using human Fas apoptosis-promoting receptor (Fas) and fibroblast growth factor receptor-2 (FGFR2) minigenes. The Fas minigene contains human Fas exons 5, 6 and 7 and introns 5 and 6 (11 and Supplementary Figure 1SA). The 3′-splice sites of intron 5 is weak and highly U2AF dependent, whereas the intron 6 3′-splice site is strong and less U2AF dependent. Experiments were performed to assess the response of these introns to the presence of both proteins. The wt/mutant Fas minigenes [U-20C, Fas minigene harboring a mutation at the polypyrimidine tract of intron 5 that increases exon 6 skipping (13)] and derivative plasmids expressing GFP, GFP-hnRNP C1, GFP-TIAR and /or GFP-TIARΔQ proteins were co-transfected in HeLa (Figure 5A and B) and HEK293 (Figure 5C and D) cells. The patterns of alternatively spliced Fas products were monitored by RT–PCR and the expression levels of recombinant GFP fusion proteins by western blotting (Figure 5A–D). The results showed that overexpression of hnRNP C1 alone did not affect splicing of Fas exon 6 (Figure 5A–D, compare lanes 1 and 2 with 3). However, consistent with previous results (13), overexpression of TIAR promoted a significant inclusion of Fas exon 6 (Figure 5A–D, compare lanes 2 and 3 with 4). Co-overexpression of both hnRNP C1 and TIAR proteins reduced the effect of TIAR on Fas splicing, resulting in a significant skipping of Fas exon 6 (Figure 5A–D, compare lanes 2 and 3 with 4 and 5). This effect was lost when TIARΔQ was overexpressed alone or in combination with hnRNP C1 (Figure 5A–D, compare lanes 3–5 with 6 and 7). Taken together, these results are consistent with a repressor activity of hnRNP C1 on Fas exon 6 splicing mediated by TIAR.
Figure 5.
Functional impact of the TIAR-hnRNP C1 association on alternative pre-mRNA splicing. (A–D) Effect of hnRNP C1 and/or TIAR overexpression on alternative splicing of mutant U-20C Fas reporter minigene in HeLa (A and B) and HEK293 (C and D) cells. (A and C) Western-blot analyses (20 µg of protein extracts in lanes 1–7) of the expression levels of GFP (lanes 1 and 2), GFP-hnRNP C1 (lane 3), GFP-TIAR (lane 4), GFP-TIAR plus GFP-hnRNP C1 (lane 5), GFP-TIARΔQ (lane 6) and GFP-TIARΔQ plus GFP-hnRNP C1 (lane 7) and endogenous α-Tubulin (lanes 1-7) in co-transfected HeLa (A) and HEK293 (C) cells. (B and D) Analyses of alternatively spliced products derived from wt and mutant U-20C Fas minigenes (200 ng) co-transfected with pEGFP-C1 (lanes 1 and 2; 500 ng), pEGFP-C1-hnRNP C1 (lane 3; 500 ng), pEGFP-C1-TIAR (lane 4; 500 ng), pEGFP-C1-TIAR plus hnRNP C1 (lane 5; 500 ng of each plasmid), pEGFP-C1-TIARΔQ (lane 6; 500 ng), or pEGFP-C1-TIARΔQ plus hnRNP C1 (lane 7; 500 ng of each plasmid) plasmids in HeLa (B) and HEK293 (D) cells. In all samples, the total DNA amount was adjusted with empty pcDNA3.1 plasmid. The values of the ratios between 5-6-7 and 5-7 amplification products are means ± SE for two to three independent experiments. (E and F) Effects of overexpression of hnRNP C1 and/or TIAR on alternative splicing of the FGFR2 RK97 reporter minigene in HEK293 cells. (E) Western-blot analysis (20 µg of protein extracts in lanes 1–6) of the expression levels of GFP (lane 1), GFP-TIAR (lane 2), GFP-TIARΔQ (lane 3), GFP-hnRNP C1 (lane 4), GFP-TIAR plus GFP-hnRNP C1 (lane 5), GFP-TIARΔQ plus GFP-hnRNP C1 (lane 6) and endogenous α-tubulin in co-transfected HEK293 cells. (F) Analysis of alternatively spliced products derived from the RK97 minigene (200 ng) co-tranfected with pEGFP-C1 (lanes 1; 500 ng), pEGFP-C1-TIAR (lane 2; 500 ng), pEGFP-C1-TIARΔQ (lane 3; 500 ng), pEGFP-C1-hnRNP C1 (lane 4; 1000 ng), pEGFP-C1-TIAR plus hnRNP C1(lane 5; 500 and 1000 ng of each plasmid, respectively), or pEGFP-C1-TIARΔQ plus hnRNP C1 (lane 6; 500 and 1000 ng of each plasmid, respectively) plasmids in HEK293 cells. In all samples, the total DNA amount was adjusted with empty pcDNA3.1 plasmid. The values of the ratios between C1-K-SAM-C2-BGH and C1-C2-BGH amplification products are means ± SE for two to three independent experiments. Positions of predicted alternatively spliced products are indicated with boxes on the right.
To address whether hnRNP C1 also affects splicing of other pre-mRNA substrates, FGFR2 splicing was tested using the RK97 minigene (10; Supplementary Figure 1SC). GFP-tagged proteins were expressed at similar levels in HEK293 and HeLa cells (Figure 5A and C). Overexpression of TIAR caused a significant and reproducible increase in FGFR-2 K-SAM exon inclusion, whereas overexpression of hnRNP C1 alone had no effect (Figure 5F, compare lanes 1 with 2 and 4). However, co-overexpression of TIAR and hnRNP C1 inhibited K-SAM exon inclusion (Figure 5F, compare lanes 1, 2 and 4 with 5). This effect was lost when TIARΔQ was overexpressed alone or in combination with hnRNP C1 (Figure 5F, compare lane 2 with 3 and 6). These results suggest that hnRNP C1 functions as a trans-acting repressor of TIAR-regulated alternative splicing.
Regulation of Fas splicing by hnRNP C1 is mediated by HuR
FGFR2 exon K-SAM and Fas exon 6 splicing is completely dependent on the presence of IAS1 and URI6 sequences close to the 5′-splice sites and on the recognition of these sequences by U1 snRNP and TIA-proteins, which facilitate intron–exon definition and splicing of the downstream introns (10,11,13; Supplementary Figure 1SA and C). Given that hnRNP C1 binds RNA through U-rich stretches and both sequences contain cis-acting elements with these features (Supplementary Figure 2SA), experiments were performed to test whether FGFR2 and Fas RNAs can be efficiently bound by hnRNP C1 (Supplementary Figure 2SA). In vitro assays were carried out using recombinant purified GST, GST-hnRNPC1 and GST-TIAR fusion proteins. RNA binding was tested by electrophoretic mobility shift assay (EMSA; Supplementary Figure 2SC and D) and UV-crosslinking (Supplementary Figure 2SE and F). The results showed that addition of the same amount of purified recombinant TIAR or hnRNP C1 proteins (Supplementary Figure 2SB) results in a significant increase in direct binding of TIAR to both FGFR2 and wt/mutant (the m0 minigene, with replacement of a uridine-rich sequence on exon 6 [URE6] (13; Supplementary Figure 1SA)) Fas RNAs (Supplementary Figures 2SC and D and 3S). This preferential binding of TIAR to both RNAs was also observed in UV-crosslinking assays (Supplementary Figure 2SE and F). hnRNP C1 exists as a stable heterotetramer [(C1)3 (C2)1] that binds RNA cooperatively in the nucleus (31–33). However, the results of EMSA and UV-crosslinking assays suggest that hnRNP C1 has a limited but reproducible affinity for URE6. Therefore, these results suggest that FGFR2 and Fas U-rich sequences close to both 5′-splice sites function as high affinity sites, mainly facilitating TIAR rather than hnRNP C1 recruitment. However, the experimental approach is limited by the use of in vitro RNA-binding assays, meaning that hnRNP C1 could bind to both FGFR2 IAS1 and Fas URE6 RNA sequences.
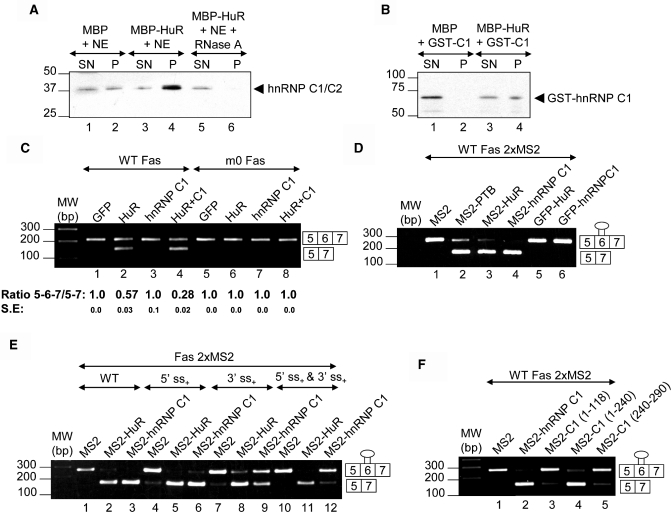
TIA proteins stimulate Fas exon 6 inclusion by facilitating recognition of its associated 5′-splice site by U1 snRNP, which in turn enhances recognition of the upstream 3′-splice site by U2AF and therefore exon definition. hnRNP I [also known as Polypyrimidine Tract-Binding (PTB) protein] and Hu antigen R (HuR) promote exon skipping through an ESS by inhibiting molecular events that lead to exon definition via stabilization of U2AF65 (13,22). A recent report showed extensive association between HuR and hnRNP family components, for example, hnRNP C1/C2 in nuclear hnRNP/mRNP complexes, consistent with a broader participation in mRNA processing events (35). To explore this possibility, MBP pull-downs were used to confirm the interaction between HuR and hnRNP C1/C2. MBP alone or an MBP fusion containing full-length HuR were added to HeLa NEs and their association with hnRNP C1/C2 was analysed by immunoblot for endogenous hnRNP C1/C2 in the precipitates. hnRNP C1/C2 was pulled down by MBP-HuR but not by MBP alone (Figure 6A, compare lanes 1 and 2 with 3 and 4). Co-precipitation of HuR with hnRNP C1/C2 in NEs was RNA-dependent, since it was completely lost when extracts were pre-treated with RNAse A (Figure 6A, compare lanes 3 and 4 with 5 and 6). However, pull-downs using recombinant purified MBP-HuR and GST-hnRNP C1 suggested that a protein–protein interaction is also possible (Figure 6B). To analyse a potential in vivo interaction between HuR and hnRNP C1/C2, nuclear co-localization studies were performed using recombinant GFP-HuR and RFP-hnRNP C1. The proteins were highly colocalized in the nucleoplasm but excluded from the nucleolus (Supplementary Figure 4S). These results support the existence of a genuine association between a nuclear pool of HuR and hnRNP C1/C2 proteins, in agreement with earlier results (35).
Figure 6.
hnRNP C1 represses Fas exon 6 inclusion through the URE6 exonic silencer. (A and B) Precipitation of endogenous and recombinant purified hnRNP C1 by MBP-HuR. (A) MBP pull-down assays were carried out after addition of MBP alone (lanes 1 and 2) or MBP-HuR to NEs without (lanes 3 and 4) or with (lanes 5 and 6) RNase A treatment, and the resulting pellets (P) and the corresponding supernatants (SN) (10%) were analysed in parallel by immunobloting with anti-hnRNP C1/C2 antibody. (B) MBP pull-down assays were performed after addition of MBP alone (lanes 1 and 2) or MBP-HuR (lanes 3 and 4) to recombinant purified GST-hnRNP-C1 protein and analysed as described in (A). (C) The effects of HuR (500 ng) and hnRNP C1 (1000 ng) overexpression on alternative splicing of wt/mutant (m0) Fas minigenes (200 ng) in HeLa cells. Analysis of alternatively spliced products derived from Fas reporter minigenes co-transfected with GFP-tagged plasmids. In all samples, the total DNA amount was adjusted with empty pcDNA3.1 plasmid. The values of the ratios between 5-6-7 and 5-7 amplification products are means ± SE for two to three independent experiments. (D) Tethering of PTB, HuR or hnRNP C1 to Fas exon 6 through heterologous RBDs promotes exon skipping. The Fas 2xMS2 minigene (300 ng) containing two MS2 stem-loop-binding sites in tandem adjacent to the URE6 sequence was co-transfected with plasmids (1000 ng) expressing the MS2 fusion proteins indicated. (E) The Fas 2xMS2 minigene was modified by mutagenesis to strength the intron 6 5′-splice site, intron 5 3′-splice site, or both, generating three Fas 2xMS2-derived versions identified as 5′-ss+, 3′-ss+ and 5′-ss+ plus 3′-ss+, respectively. The Fas 2xMS2 minigenes (300 ng) were co-transfected with plasmids (1000 ng) expressing MS2, MS2-HuR, or MS2-hnRNP C1. (F) The Fas 2xMS2 minigene (300 ng) was co-transfected with plasmids (1000 ng) expressing empty vector MS2, MS2-hnRNP C1 (1–290), MS2-hnRNP C1 (1–118), MS2-hnRNP C1 (1–240), or MS2-hnRNP C1 (240–290).
To assess whether this association has functional consequences for Fas exon 6 splicing, HeLa cells were co-transfected with wt Fas minigene and plasmids expressing GFP, GFP-hnRNP C1 and /or GFP-HuR. HuR ovexpression with the wt Fas minigene promoted exon 6 skipping, whereas GFP and hnRNP C1 overexpression had no effect on Fas exon 6 splicing (Figure 6C, compare lane 1 with 2 and 3; Supplementary Figure 5SA). However, HuR and hnRNP C1 co-overexpression promoted a significant and reproducible increase in Fas exon 6 exclusion (Figure 6C, lane 4; Supplementary Figure 5SA). Nevertheless, the mutant m0 Fas minigene, where URE6 was replaced by a random sequence (13; Supplementary Figure 1SA), was insensitive to equivalent levels of GFP, HuR, or hnRNP C overexpression (Figure 6C, compare lanes 1–4 with 5–8; Supplementary Figure 5SA). These results suggest that hnRNP C1 functions as an HuR-dependent corepressor of Fas exon 6 splicing.
To further confirm that hnRNP C1 displays a repressor activity mediated by the URE6 RNA silencer, the URE6 sequence was substituted with two MS2 stem-loop-binding sites in tandem from the bacteriophage MS2 protein (Fas 2xMS2 minigene) (13; Supplementary Figure 1SB and D). Co-transfection with a plasmid expressing an MS2-hnRNP C1 fusion protein led to significant exon 6 skipping (Figure 6D, lane 4), whereas expression of MS2 protein alone did not (Figure 6D, lane 1; Supplementary Figure 5SB). As described previously (22), expression of MS2-PTB or MS2-HuR fusion proteins also led to efficient exon 6 skipping (Figure 6D, lanes 2 and 3; Supplementary Figure 5SB). However, the URE6 substitution rendered the minigene insensitive to GFP-HuR or GFP-hnRNP C1 overexpression (Figure 6D, compare lanes 3 and 4 with 5 and 6). The straightforward interpretation of these results is that hnRNP C1 represses exon 6 inclusion in a URE6-dependent manner. Nevertheless, using an MS2 tethering approach with FGFR2 minigene A7 (Supplementary Figures 1SD and 6S) in HEK293 cells, the data obtained in these experiments suggest, perhaps not surprisingly, a limited contribution of hnRNP C1 alone, because the K-SAM exon is already fully skipped in HEK293 cells. However, the results also show that MS2-hnRNP C1 can compete with MS2-TIAR to modulate K-SAM splicing, illustrating that hnRNP C1 displays a repressor activity mediated by TIAR (Supplementary Figure 6S).
To analyse repressor effect of hnRNP C1 per se, the tethering analysis was extended using modified Fas 2xMS2 minigenes, which contain a strengthened intron 6 5′-splice site, intron 5 3′-splice site, or both (22). In this scenario, hnRNP C1 functioned as a potent repressor of RNA substrates with weak 5′-and 3′-splice sites and with a stronger 5′-splice site, but not of those with a stronger 3′-splice site (Figure 6E, compare lanes 1–3 and 4–6 with 7–9 and 10–12; Supplementary Figure 5SC). These results suggest that the repressor capacity of hnRNP C1 on Fas splicing acts by targeting molecular events that lead to exon definition, in agreement with the previous observations for PTB (13) and HuR (22). The results obtained using a tethering approach also show that the leucine-rich domain had sufficient repressor activity to promote Fas exon 6 skipping (Figure 6F, compare lanes 1, 3 and 5 with 2 and 4; Supplementary Figure 5SD). Thus, the inhibitory capacity of hnRNP C1 appears to reside in its bZLM motif.
DISCUSSION
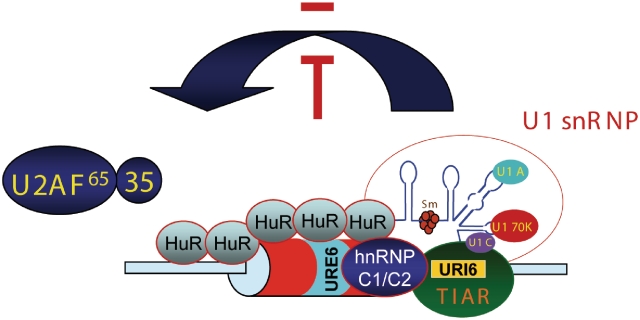
The results of this study show that TIAR/TIAL1, hnRNP C1/C2 and HuR proteins can play opposite and cooperative roles in the control of Fas alternative splicing. The splice-enhancing activity of TIAR that facilitates recruitment of U1 snRNP to the weak intron 6 5′-splice site is repressed by the cooperative interaction of HuR and hnRNP C1/C2 through an ESS by inhibiting molecular events that lead to exon definition (Figure 7). These observations indicate a functional association of TIA proteins and HuR with hnRNPs to modulate the molecular bridging that leads to splicing control.
Figure 7.
Model for the regulation of Fas exon 6 splicing by hnRNP C1/C2, HuR and TIAR. Molecular interactions lead to hnRNP C1/C2-HuR-dependent exclusion of Fas exon 6. TIAR promotes U1 snRNP recruitment to the 5′-splice site of Fas intron 6. This facilitates exon definition by enhancing U2AF binding to the 3′-splice site of Fas intron 5, therefore leading to Fas exon 6 inclusion. hnRNP C1/C2 cooperates with HuR to promote Fas exon 6 skipping through the ESS URE6.
HnRNP C1/C2 forms the core of the 40S hnRNP particle in combination with hnRNP A and hnRNP B proteins. hnRNP C1 and hnRNP C2 form a stable heterotetramer, (C1)3(C2)1. hnRNP C2 (303 amino acids) is an alternatively spliced variant of hnRNP C1 (290 amino acids), resulting of the inclusion of 13 amino acids between glycine 106 and serine 107 (6,31,32). The results described here indicate that, in HeLa NEs, hnRNP C1/C2 and TIAR are associated with other nuclear proteins, such as Matrin 3 and a beta-actin isoform. Proteomic analyses of immunopurified human RNA polymerase II (Pol II) complexes have also demonstrated that hnRNPs, Matrin 3 and nuclear actin are associated with the phosphorylated C-terminal domain of the largest subunit of Pol II (36).
The protein sequence of hnRNP C1/C2 is highly conserved among vertebrates and contains an RBD and a bZLM motif, both involved in RNA binding (33) and an acidic (Asp-Glu-rich) C-terminal domain (Figure 3A) (31,32). The results of this study suggest that the protein–protein and RNA–protein associations between TIAR and hnRNP C1 mainly involve the Q-rich domain of TIAR and the bZLM and acidic C-terminal domains of hnRNP C1, whereas the RBD may be irrelevant for these interactions. Interactions between TIAR and hnRNP C1/C2 proteins are further documented by the colocalization of recombinant GFP-TIAR and RFP-hnRNP C1 proteins in the nucleus of HeLa cells (Figure 4A and C). This co-localization is lost when the Q-rich domain of TIAR is deleted (Figure 4B and C). The ability of hnRNP C1 to repress Fas exon 6 skipping is linked to the bZLM domain, suggesting that this motif can have a dual role, promoting both protein–protein and RNA–protein interactions. However, it is still unclear how this domain functions as a repressor of Fas exon 6 splicing. The capacity of hnRNP C1/C2, TIAR and HuR binding sites to cooperate in site splice selection may be suggestive of interactions between C1/C2, HuR and TIA proteins. However, although this and other studies indicate that at least a fraction of these proteins exist in similar complexes (33,36), their ability to establish functional interactions has never been documented. It is tempting to suggest that this hnRNP C1/C2 fragment may also counteract the bridging interaction leading to exon definition by reinforcing the repressor activity mediated by HuR (Figure 7; 22). Here, HuR binding to a high-affinity site located in the URE6 sequence might enhance the association and increase the affinity for nearby sequences. This would facilitate the association between hnRNP C1/C2 and adjacent silencers, allowing Fas alternative splicing to be regulated by creating a zone of silencing. HuR and hnRNP C1/C2 might function like PTB (37) or hnRNP A1 and hnRNP H (38). However, the role of hnRNP C1/C2 in alternative splicing control is versatile. For example, it has been reported that hnRNP C1/C2 down-regulation by small interfering RNAs promotes exon skipping or the use of more internal 3′-splice sites, suggesting that hnRNP C1/C2 acts as a splicing enhancer and enforces exon inclusion in a cell-type-dependent manner (28). In our hands, silencing of hnRNP C1/C2 gene expression mediated by specific shRNAs against the 3′-untranslated region of the hnRNP C1/C2 mRNA powerfully inhibits HeLa cell proliferation but has no effect on alternative splicing patterns of Fas endogenous and ectopic (minigene) pre-mRNAs (data not shown), a finding that is consistent with previous results (27). This observation is also consistent with Fas exon 6 inclusion being prevalent in the HeLa cell line (13,22). Interestingly, these observations agree with the recently described hnRNP C1/C2 splicing ‘code/map’ (39), showing that hnRNP C1/C2 recruitment to alternative exons results in silencing of exon inclusion, whereas binding to the preceding intron enhances the inclusion of alternative exons.
Variations in the concentration of general splicing factors and regulators, including members of the hnRNP, TIA and Hu protein families, have been proposed as mechanisms to achieve cell-type or developmental stage-specific splicing. For example, three hnRNPs, including hnRNP I (also known as PTB), hnRNP A1 and hnRNP A2, are up-regulated by c-Myc and can bind to splicing signals flanking pyruvate kinase exon 9 and repress its inclusion (40,41). This results in exon 10 inclusion and generates the embryonic pyruvate kinase 2 isoform, which promotes aerobic glycolysis in embryonic and cancer cells. This mechanism is essential for cell growth, defining a pathway that regulates an alternative splicing event required for tumour cell proliferation. In another study, we reported that the relative abundance of TIA1, TIAR, and their isoforms differs in human tissues and cell lines, with TIAR showing a more restricted pattern of expression (20). The final splicing outcome will of course also depend on additional factors that synergize or antagonize TIA1/TIAR activities. For example, Hu proteins were recently reported to antagonize TIA1/TIAR to achieve neuron-specific (42) and Fas (22) alternative splicing. In addition, post-translational modifications can modulate the activity of regulatory factors. TIA1 phosphorylation by the Fas-activated serine/threonine kinase, for instance, increases TIA1 activity and enhances inclusion of Fas exon 6, thus creating a positive feedback loop by which Fas signalling induces higher levels of the proapoptotic form of the receptor (21). Despite hnRNP C1/C2 being ubiquitously expressed in mammalian cells, it is also subject to cell-cycle-dependent regulation (43) and can be phosphorylated under physiological conditions at Ser-225-228, Ser-240, Ser-247 and Ser-286 (44,45). Indeed, hnRNP C1/C2 was identified as the main substrate phosphorylated by casein kinases CK1α and CK2 (44,45). Furthermore, hnRNP C1/C2 undergoes a cycle of phosphorylation-dephosphorylation in HeLa NEs that modulates its binding to pre-mRNAs, and this hyperphosphorylation of hnRNP C1 is mediated by an RNA-dependent kinase activity in HeLa NEs (30). These observations and the results shown in Figure 2G might begin to explain the discrepancies between the TIAR-hnRNP C1 interaction in RRL versus HeLa NEs. Thus, the requirement for RNA-dependent phosphorylation of hnRNP C1/C2 may be modified in HeLa NEs upon RNase A treatment (Figure 2F), whereas the coupled transcription and translation system in RRL could generate sufficient phosphorylated hnRNP C1 protein. Likewise, endogenous phosphorylation of hnRNP C1/C2 in HeLa NEs could explain the greater efficiency of GST-pull down experiments with GST-TIAR than GST-hnRNP C1 (Figure 1D and E).
In general, transcriptome diversity is regulated by interactions between cis-acting DNA/RNA elements and specific trans-acting proteins and numerous studies suggest that transcription and post-transcriptional events (e.g. alternative splicing) are often co-regulated in the nucleus (46,47). Three very recent reports demonstrated RNA polymerase II-mediated cross-talk between chromatin structure organization and exon–intron architecture, implying that exon selection (i.e. ‘splicing code’) might be modulated by chromatin structure or nucleosome positioning as well as by histone methylation (48–50). Given the association of hnRNP C1/C2 with the SWI/SNF chromatin remodeling complex (51) and the role of other SWI/SNF components in alternative splicing (52), hnRNP C1/C2 may mediate some effects in a co-transcriptionally dependent manner. We now know that a key link between transcription and RNA splicing is provided by different families of highly conserved RNA-binding proteins known as hnRNPs or SR proteins (36). Given abundant evidence for co-transcriptional pre-mRNA splicing of most eukaryotic genes, it is widely accepted that hnRNPs participate in spliceosome assembly on nascent RNAs to catalyse intron removal before the completion of transcription, thus explaining the association of hnRNPs with active genes. An overall hypothesis has emerged that pre-mRNA-containing ribonucleoprotein complexes (pre-mRNPs) are the functional equivalent of ‘post-transcriptional operons’. Pre-mRNA splicing begins co-transcriptionally on chromatin and this feature influences the alternative splicing outcome; for example, changes in transcription elongation rates and pausing have an effect on exon inclusion (46,47). This suggests that the kinetics of splice-site synthesis and the time for interaction with splicing factors before synthesis of the next splice site have a role in the combinatorial and coordinated control of alternative splicing (53,54). These findings support a model in which hnRNP C1/C2, TIAR, HuR and U1 snRNP are co-transcriptionally recruited to nascent RNA Pol II transcripts to promote spliceosome assembly. The combination of these factors and the co-transcriptional recruitment would be expected to increase the fidelity of the earliest recognition of weak 5′splice sites in exons of nascent pre-mRNAs. Modifications in the amounts or activities of TIA1/TIAR, PTB, SPF45, RBM5, HuR and /or hnRNP C1/C2 (11,13,22,55,56) and their capacity to interact and modulate the splicing decision can regulate Fas exon 6 splicing to produce isoforms that activate or inhibit the extrinsic apoptosis pathway. Control of Fas splicing may be strongly dependent on the coordinated and combinatorial arrangement of several binding sites that coexist in very close proximity. This capacity to collaborate is not surprising, since these proteins display redundant binding specificity and have similar structures. The results of this study also raise the possibility that these proteins improve interactions with other hnRNPs, suggesting that associations via other RNA-binding proteins represent an efficient system to build specificity with non-specific RNA-binding proteins (57) and expand the range of ‘splicing code’ options (39,58). Many factors can both activate and repress splicing in different contexts, and they may have synergistic or antagonistic effects on the co-transcriptional decision involved in alternative splicing. More work will be required to ascertain the mechanism(s) through which hnRNP C1/C2 and other hnRNPs proteins influence splice site selection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fondo de Investigaciones Sanitarias-FEDER (PI051605); Ministerio de Ciencia e Innovación-FEDER (BFU2008-00354). An institutional grant from Fundación Ramón Areces, Spain (to CBMSO). Funding for open access charge: Ministerio de Ciencia e Innovación-European funds of regional development (FEDER) (BFU2008-00354).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The author thanks José Manuel Sierra and José Alcalde for encouragement. The author also grateful to Juan Valcárcel, Myriam Gorospe, Rafael Cuesta, Richard Breathnach, Federico Mayor Jr and Peter G. Stockley, for providing reagents.
REFERENCES
- 1.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 3.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 7.Reyes R, Alcalde J, Izquierdo JM. Depletion of T-cell intracellular antigen proteins promotes cell proliferation. Genome Biol. 2009;10:R87. doi: 10.1186/gb-2009-10-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian Q, Streuli M, Saito H, Schlossman SF, Anderson P. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell. 1991;67:629–639. doi: 10.1016/0092-8674(91)90536-8. [DOI] [PubMed] [Google Scholar]
- 9.Dember LM, Kim ND, Liu KQ, Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- 10.Del Gatto-Konczak F, Bourgeois CF, Le Guiner C, Kister L, Gesnel MC, Stévenin J, Breathnach R. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 2000;20:6287–6299. doi: 10.1128/mcb.20.17.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Förch P, Puig O, Kedersha N, Martínez C, Granneman S, Séraphin B, Anderson P, Valcárcel J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 12.Förch P, Puig O, Martínez C, Seraphin B, Valcárcel J. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 2002;21:6882–6892. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izquierdo JM, Majós N, Bonnal S, Martínez C, Castelo R, Guigó R, Bilbao D, Valcárcel J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 14.Aznarez I, Barash Y, Shai O, He D, Zielenski J, Tsui LC, Parkinson J, Frey BJ, Rommens JM, Blencowe BJ. A systematic analysis of intronic sequences downstream of 5′ splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation. Genome Res. 2008;18:1247–1258. doi: 10.1101/gr.073155.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gal-Mark N, Schwartz S, Ram O, Eyras E, Ast G. The pivotal roles of TIA proteins in 5′ splice-site selection of alu exons and across evolution. PLoS Genet. 2009;5:1–13. doi: 10.1371/journal.pgen.1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.López de Silanes I, Galbán S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell. Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazan-Mamczarz K, Lal A, Martindale JL, Kawai T, Gorospe M. Translational repression by RNA-binding protein TIAR. Mol. Cell. Biol. 2006;26:2716–2727. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim HS, Kuwano Y, Zhan M, Pullmann R, Jr, Mazan-Mamczarz K, Li H, Kedersha N, Anderson P, Wilce MC, Gorospe M. Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol. Cell. Biol. 2007;27:6806–6817. doi: 10.1128/MCB.01036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamasaki S, Stoecklin G, Kedersha N, Simarro M, Anderson P. T-cell intracellular antigen-1 (TIA-1)-induced translational silencing promotes the decay of selected mRNAs. J. Biol. Chem. 2007;282:30070–30077. doi: 10.1074/jbc.M706273200. [DOI] [PubMed] [Google Scholar]
- 20.Izquierdo JM, Valcárcel J. Two isoforms of the T-cell intracellular antigen 1 (TIA-1) splicing factor display distinct splicing regulation activities. Control of TIA-1 isoform ratio by TIA-1-related protein. J. Biol. Chem. 2007;282:19410–19417. doi: 10.1074/jbc.M700688200. [DOI] [PubMed] [Google Scholar]
- 21.Izquierdo JM, Valcárcel J. Fas-activated serine/threonine kinase (FAST K) synergizes with TIA-1/TIAR proteins to regulate Fas alternative splicing. J. Biol. Chem. 2007;282:1539–1543. doi: 10.1074/jbc.C600198200. [DOI] [PubMed] [Google Scholar]
- 22.Izquierdo JM. Hu antigen R (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition. J. Biol. Chem. 2008;283:19077–19084. doi: 10.1074/jbc.M800017200. [DOI] [PubMed] [Google Scholar]
- 23.Gesnel MC, Theoleyre S, Del Gatto-Konczak F, Breathnach R. Cooperative binding of TIA1 and U1 snRNP in K-SAM exon splicing activation. Biochem. Biophys. Res. Commun. 2007;358:1065–1070. doi: 10.1016/j.bbrc.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 24.Izquierdo JM. Fas splicing regulation during early apoptosis is linked to caspase-mediated cleavage of U2AF65. Mol. Biol. Cell. 2008;19:3299–3307. doi: 10.1091/mbc.E07-11-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 26.Suckau D, Resemann A, Schuerenberg M, Hufnagel P, Franzen J, Holle A. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal. Bioanal. Chem. 2003;376:952–965. doi: 10.1007/s00216-003-2057-0. [DOI] [PubMed] [Google Scholar]
- 27.Hossain MN, Fuji M, Miki K, Endoh M, Ayusawa D. Downregulation of hnRNP C1/C2 by siRNA sensitizes HeLa cells to various stresses. Mol. Cell. Biochem. 2007;296:151–157. doi: 10.1007/s11010-006-9308-2. [DOI] [PubMed] [Google Scholar]
- 28.Venables JP, Koh CS, Froehlich U, Lapointe E, Couture S, Inkel L, Bramard A, Paquet ER, Watier V, Durand M, et al. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol. Cell. Biol. 2008;28:6033–6043. doi: 10.1128/MCB.00726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu C, York B, Wang S, Feng Q, Xu J, O'Malley BW. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol. Cell. 2007;25:765–778. doi: 10.1016/j.molcel.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fung PA, Labrecque R, Pederson T. RNA-dependent phosphorylation of a nuclear RNA binding protein. Proc. Natl. Acad. Sci. 1997;94:1064–1068. doi: 10.1073/pnas.94.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorlach M, Burd C, Dreyfuss G. The determinants of RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein C proteins. J. Biol. Chem. 1994;269:23074–23078. [PubMed] [Google Scholar]
- 32.McAfee J, Shahied-Milam L, Soltaninassab S, LeStourgeon W. A major determinant of hnRNP C protein binding to RNA is a novel bZIP-like RNA binding domain. RNA. 1996;2:1139–1152. [PMC free article] [PubMed] [Google Scholar]
- 33.Koloteva-Levine N, Amichay M, Elroy-Stein O. Interaction of hnRNP-C1/C2 proteins with RNA: analysis using the yeast three-hybrid system. FEBS Lett. 2002;523:73–78. doi: 10.1016/s0014-5793(02)02938-1. [DOI] [PubMed] [Google Scholar]
- 34.Nakielny S, Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J. Cell Biol. 1996;134:1365–1373. doi: 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papadopoulou C, Patrinou-Georgoula M, Guialis A. Extensive association of HuR with hnRNP proteins within immunoselected hnRNP and mRNP complexes. Biochim. Biophys. Acta. 2010;1804:692–703. doi: 10.1016/j.bbapap.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 36.Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 37.Spellman R, Smith CW. Novel modes of splicing repression by PTB. Trends Biochem. Sci. 2006;31:73–76. doi: 10.1016/j.tibs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Fisette JF, Toutant J, Dugré-Brisson S, Desgroseillers L, Chabot B. hnRNP A1 and hnRNP H can collaborate to modulate 5′ splice site selection. RNA. 2009;16:228–238. doi: 10.1261/rna.1890310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc. Natl. Acad. Sci. 2010;107:1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu H, Hinman MN, Hasman RA, Mehta P, Lou H. Regulation of neuron-specific alternative splicing of neurofibromatosis type 1 pre-mRNA. Mol. Cell. Biol. 2008;28:1240–1251. doi: 10.1128/MCB.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piñol-Roma S, Dreyfuss G. Cell cycle-regulated phosphorylation of the pre-mRNA-binding (heterogeneous nuclear ribonucleoprotein) C proteins. Mol. Cell. Biol. 1993;13:5762–5770. doi: 10.1128/mcb.13.9.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stone JR, Maki JL, Collins T. Basal and hydrogen peroxide stimulated sites of phosphorylation in heterogeneous nuclear ribonucleoprotein C1/C2. Biochemistry. 2003;42:1301–1308. doi: 10.1021/bi0268091. [DOI] [PubMed] [Google Scholar]
- 45.Kattapuram T, Yang S, Maki JL, Stone JR. Protein kinase CK1alpha regulates mRNA binding by heterogeneous nuclear ribonucleoprotein C in response to physiologic levels of hydrogen peroxide. J. Biol. Chem. 2005;280:15340–15347. doi: 10.1074/jbc.M500214200. [DOI] [PubMed] [Google Scholar]
- 46.Roberts GC, Gooding C, Mak HY, Proudfoot NJ, Smith CW. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 1998;26:5568–5572. doi: 10.1093/nar/26.24.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 48.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 2009;16:990–996. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 49.Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcárcel J, Guigó R. Nucleosome positioning as a determinant of exon recognition. Nat. Struct. Mol. Biol. 2009;16:996–1002. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 50.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahajan MC, Narlikar GJ, Boyapaty G, Kingston RE, Weissman SM. Heterogeneous nuclear ribonucleoprotein C1/C2, MeCP1, and SWI/SNF form a chromatin remodeling complex at the beta-globin locus control region. Proc. Natl Acad. Sci. 2005;102:15012–15017. doi: 10.1073/pnas.0507596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 53.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 54.Sapra AK, Ankö ML, Grishina I, Lorenz M, Pabis M, Poser I, Rollins J, Weiland EM, Neugebauer KM. SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol Cell. 2009;34:179–190. doi: 10.1016/j.molcel.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 55.Corsini L, Bonnal S, Basquin J, Hothorn M, Scheffzek K, Valcárcel J, Sattler MJ. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat. Struct. Mol. Biol. 2007;14:620–629. doi: 10.1038/nsmb1260. [DOI] [PubMed] [Google Scholar]
- 56.Bonnal S, Martínez C, Förch P, Bachi A, Wilm M, Valcárcel J. RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol. Cell. 2008;32:81–95. doi: 10.1016/j.molcel.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 57.Singh R, Valcárcel J. Building specificity with nonspecific RNA-binding proteins. Nat. Struct. Mol. Biol. 2005;12:645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- 58.Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.