Abstract
The National Center for Biotechnology Information (NCBI) recently announced ‘1000 prokaryotic genomes are now completed and available in the Genome database’. The increasing trend will provide us with thousands of sequenced microbial organisms over the next years. However, this is only the first step in understanding how cells survive, reproduce and adapt their behavior while being exposed to changing environmental conditions. One major control mechanism is transcriptional gene regulation. Here, striking is the direct juxtaposition of the handful of bacterial model organisms to the 1000 prokaryotic genomes. Next-generation sequencing technologies will further widen this gap drastically. However, several computational approaches have proven to be helpful. The main idea is to use the known transcriptional regulatory network of reference organisms as template in order to unravel evolutionarily conserved gene regulations in newly sequenced species. This transfer essentially depends on the reliable identification of several types of conserved DNA sequences. We decompose this problem into three computational processes, review the state of the art and illustrate future perspectives.
INTRODUCTION
The National Center for Biotechnology Information (NCBI) recently announced ‘1000 prokaryotic genomes are now completed and available in the Genome database’. In fact, we may download the genome annotations of 1024 fully sequenced microbial organisms from the NCBI database (1). Thanks to the next-generation sequencing techniques, the cost of DNA sequencing was reduced by over two orders of magnitude and has thus become a routine and widespread method to unravel the genetic repertoire of numerous species (2,3). The increasing trend will provide us with thousands of sequenced organisms over the next years. This genomic revolution in molecular biology leaves us with complete genomes of numerous microbes with varied ecological, economic and medical significance (4).
The availability of genome sequences, however, is only the first step in understanding how cells survive, reproduce and adapt their behavior while being exposed to varying environmental conditions. One major control mechanism is transcriptional gene regulation. The most important components of the cell’s transcriptional regulatory apparatus are the so-called transcription factors (TFs)—DNA-binding proteins that are able to detect intra- and extracellular signals. By binding to operator sequences, the transcription factor binding sites (TFBSs), they repress or stimulate the expression of other genes (target genes, TGs) and thereby decisively influence genetic programs like growth, survival and reproduction (5). Supplementary Figure S1 illustrates this transcriptional machinery. Depending on the surrounding and internal conditions of a cell, certain fractions of the total set of TFs are operating to control the expression of their TGs. Some regulators only control the expression of a single gene, whereas others organize the activation or repression of numerous TGs. Regulatory networks emerge. They are modeled as graphs, where nodes correspond to genes and directed edges represent transcriptional regulatory interactions. The reconstruction of these networks, i.e. the identification of the spatial and temporal regulatory interactions between TFs and their targets, is one of the most important goals in molecular systems biology (6).
Nowadays, high-throughput experimental techniques exist for the wet-lab reconstruction of gene regulatory networks. The two primary methods, genome-wide measurement of mRNA expression levels and the identification of TFBS locations, are widely available. The application of microarray and RNA-seq technology has opened the way to investigate organism-wide gene expression under different conditions in order to provide hypotheses about putative transcriptional gene regulatory interactions (7). Especially, studying genetic expression in response to the deletion of TF-encoding genes has been successfully utilized to identify potential TGs for numerous TFs in many microbial organisms. Subsequent identification of the TF binding-site location in the promoter regions of the putative TGs provides further evidence for the respective TF-TG interactions. Wet-lab determination of TFBS locations is done by electrophoretic mobility shift assays (EMSA) (8), DNAse footprinting (9), ChIP-chip (10) or ChIP-seq (11). By combining gene expression studies and TFBS location analysis, transcriptional gene regulatory interactions are reconstructed and the emerging networks are stored in publicly available databases (12,13). For prokaryotes, popular reference databases are RegTransBase (14), RegulonDB (15) and EcoCyc (16) for Escherichia coli, DBTBS (17) for Bacillus subtilis, MtbRegList (18) for Mycobacterium tuberculosis, PRODORIC (19) mainly for Pseudomonas aeruginosa but also E. coli and B. subtilis and CoryneRegNet (20) for corynebacteria (mainly Corynebacterium glutamicum).
Although inevitable for understanding the behavior and the complexity of microbial cells, the reconstruction of transcriptional gene regulatory networks is far from being complete. Even for the model organism E. coli, with the largest currently available experimentally validated knowledge of any free-living organism (21), we have some information about the transcriptional regulation of only around one-third of the genes (15). Network reconstruction and standardized data access is complicated by several problems: technical and procedural difficulties comprise, for example, the fabrication of TF-deletion mutants, the noise in gene expression data, the identification of concealed combinatorial effects caused by co-acting TFs and the determination of TFBS locations accurately to one base pair. Consequently, economic aspects arise: Replicated experiments are inevitable to provide statistical significance but drastically increase the amount of necessary temporal and monetary resources. Finally, successfully discovered gene regulatory interactions are published in scientific journals. This is an organizational issue since it requires curation teams to find and extract this knowledge from the literature manually instead of having it available in online repositories for direct, well-structured data access (22). In the light of these technical, monetary and structural difficulties, we conclude that a wet-lab reconstruction of gene regulatory networks is impossible to perform for any sequenced prokaryote separately. Striking is the direct juxtaposition of the six abovementioned reference repositories for E. coli, B. subtilis, M. tuberculosis, P. aeruginosa and C. glutamicum to the 1000 microbial genomes, which we may download from the NCBI. Recent advances in high-throughput genome sequencing will further widen this gap drastically.
However, recently developed bioinformatics approaches have proven to be helpful here. The similarity of the gene regulatory networks between two organisms correlates with the grade of evolutionary and taxonomical conservation between them (23,24). Hence, the main idea is to use the gene regulatory network of one of the few reference organisms as template (source) in order to unravel evolutionarily conserved gene regulations in newly sequenced species (targets). This transfer of transcriptional regulatory interactions between source and target organisms essentially depends on the reliable discovery of conserved DNA sequence patterns. In the following, we decompose this process into three computational aspects: (i) orthology detection, (ii) TF binding-site prediction and (iii) a combination of both. We investigate recently published studies to illustrate the state of the art. Finally, we will identify open challenges and suggest future directions.
NETWORK TRANSFER
Conserved genes
In first studies, scientists concentrated on the conservation of the most apparent genetic elements: the genes. The assumption is that orthologous transcription factors regulate orthologous target genes. Babu et al. (25) used bi-directional best BLAST hits (BBHs) (26,27) as orthology detection to transfer the gene regulatory network of E. coli (112 TFs, 755 TGs, 1295 interactions) to 175 fully sequenced prokaryotes. They claim that ‘it is now generally accepted that in the majority of cases’ a transcriptional gene regulatory interaction is conserved if the participating genes, i.e. the TF and the TG, are conserved. For eukaryotes, Yu et al. (28) came to the same conclusion. They found this method to be ‘fairly robust’ in their studies.
However, this topic is discussed controversially in the scientific community. Price et al. studied whether putative orthologous TFs, identified by BBHs, have evolutionary conserved functions, i.e. whether they regulate conserved TGs (29). They showed that, especially for distantly related species, TFs identified as orthologs via BBHs often have different functions, respond to different signals and regulate different TGs. In conclusion, Price et al. finally suggest utilizing phylogenetic trees for the identification of putative orthologs, rather than BBHs.
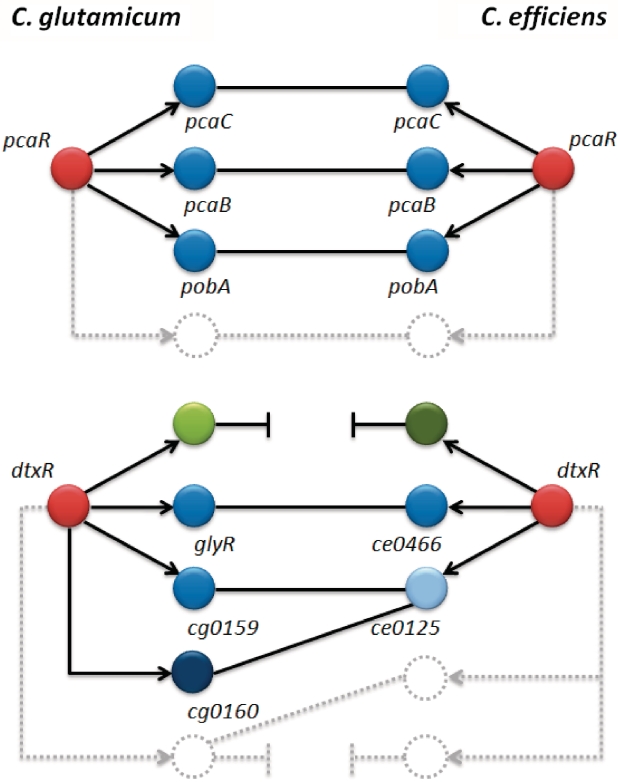
Figure 1 illustrates the general problem by means of the two regulators DtxR (30) and PcaR (31) of C. glutamicum and the taxonomically closely related organism C. efficiens. The regulons as well as putative orthologous genes of the two TFs were extracted from the CoryneRegNet database (32,33). All 12 TGs of PcaR are conserved in both organisms. In contrast, DtxR regulates 64 genes in C. glutamicum but only 27 in C. efficiens. From these TGs, only nine are clearly evolutionarily conserved, the others cannot be assigned unambiguously to exactly one putative homologous partner gene in the other organism (34).
Figure 1.
Illustration of the orthology detection problem. To demonstrate this problem, we compare the regulons of the transcription factors pcaR and dtxR of Corynebacterium glutamicum (CG) and Corynebacterium efficiens (CE). The red nodes represent the respective regulators, the others their target genes. Directed edges correspond to transcriptional regulatory interactions. Undirected edges symbolize putative orthologies due to sequence-based similarity. While for pcaR all 12 target genes are conserved in both organisms, for dtxR multiple problems occur: dtxR regulates 64 genes in CG but only 27 in CE. From these target genes, only nine are clearly evolutionarily conserved, i.e. one-to-one relationship, such as glyR and ce0466. The others are either inhomologous (green nodes) or show multiple, ambiguous sequence-based similarities, i.e. one-to-many or many-to-many relationship; cg0159, cg0160 (CG), and ce0125 (CE) may serve as an example here.
In a another study about the human pathogen M. tuberculosis, Balazsi et al. (35) reconstructed the largest known gene regulatory network of this organism. All interactions have been integrated from the MtbRegList database, by extensive literature research, and by transferring data from E. coli to M. tuberculosis based on the evolutionary conservation of TFs and TGs in both organisms. For the last approach, they found that only 54 of the 410 orthology-based links match with the 581 interactions known from the literature. Additionally, Venancio and Aravind recently observed a lack of successfully identified potential transcription factor encoding genes (4), at least in M. tuberculosis. Different publications mention different numbers of TFs [150 and 194 in refs. (35,36)] while Vanancio and Aravind’s ‘careful profile-based searches’ suggest 235 TFs (4). In contrast, Wilson et al. predicted 172 TFs for M. tuberculosis by using their profiles to construct the DBD database (37). In any case, we still do not even have much knowledge about the 150 definite TFs. Besides, note that this method strongly depends on highly accurate genome annotations. These are often based on computer predictions and subsequently uploaded to the NCBI genome database, a risky procedure. For instance, Bakke et al. (38) compared three different genome annotation systems and found that only 47.7% of the predicted protein-coding genes were covered by all three systems. Furthermore, most approaches concentrate on the identification of conserved genes amongst different organisms but neglect genome shuffling and reorganization effects. Here, a major problem is gene duplication resulting in multiple putative orthologs in the target genome. One could avoid this by incorporating surrounding genes in the comparison, for instance with gene cluster detection; see e.g. (39,40).
We conclude that utilizing information about conserved genes between different organisms may be enough for studying general evolutionary dynamics of gene regulatory networks; but using this information alone may lack reliability for detailed reconstructions and subsequent analyses of the cell’s ability to organize dynamic behavior by means of finely controlling gene expression. Still, the identification of putative orthologous genes is one major step toward an automatic inter-species network transfer.
Conserved binding sites
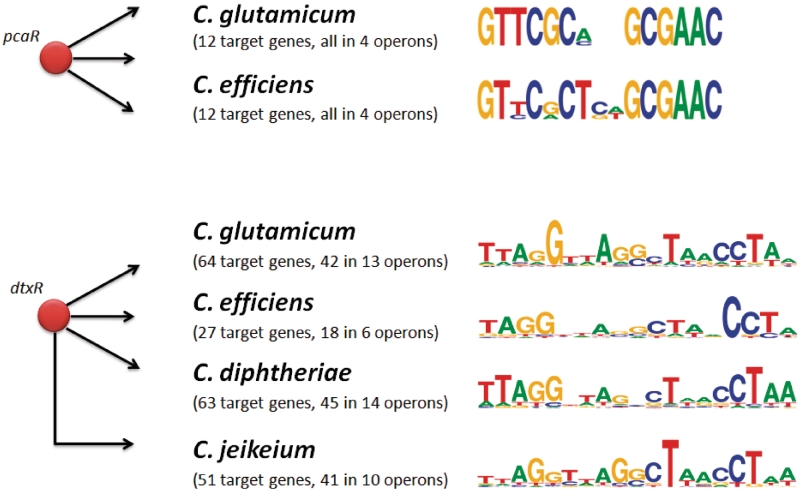
A different approach is to utilize knowledge about identified TF binding sites in the source organism. These TFBSs may be converted into computational models for subsequent profile-based predictions of gene regulatory interactions of orthologous TFs in the target organisms. This process is complicated by the comparably small length of the TFBSs (5–50 bp) resulting in computational difficulties regarding the statistical significance of detected putative TFBSs (41). One disadvantage is the necessity of knowledge about TFBSs for the respective TFs in the source organism. However, the main advantage is the potential to unravel regulatory interactions in target organisms that were not previously observed in the source organism, i.e. the TGs do not have to be conserved. However, it is known that orthologous TFs may regulate orthologous TGs through divergent TFBSs, especially in taxonomically distantly related organisms (4). In Figure 2, again the transcriptional regulators PcaR (for C. glutamicum and C. efficiens) and DtxR (for C. glutamicum, C. efficiens, C. diphtheriae and C. jeikeium) are used to illustrate the problem of TFBS conservation. To provide some numbers: Baumbach et al. (42) employed known TFBSs to move with the regulatory network of DtxR from C. glutamicum to the human pathogen C. diphtheriae. For the later bacterium, they pretended not to know the DtxR binding sites and target genes in order to evaluate the bioinformatics prediction performance. For a restrictive significance threshold they found three out of 32 TFBSs (9%) in C. diphtheriae with no false positive and, for an intermediate threshold, seven TFBSs (22%) with one false positive. With a comparably weak significance cutoff, they found 24 of the 32 TFBSs (75%) but paid a high price: 59 false positives. The statistics suggest that generally one should be able to find more true regulated TGs in C. diphtheriae (coverage), but we need to keep in mind that we are using the C. glutamicum TFBSs for predictions of binding sites in C. diphtheriae, where the DtxR binding motif is slightly different (see Figure 2). This is the price to be paid for moving from one organism to a different one with having the source organism’s TFBSs as only information source. On top of that, real TFBSs do not behave according to probabilistic sequence models, and therefore the expected coverage prediction can only be true up to an order of magnitude (42). Another well-studied example for the evolutionary divergence of binding sites is the DNA damage-response regulator LexA. Its TFBSs, termed SOS box, are similar among taxonomically closely related species but different in others (43). Hence, for instance, the LexA regulons of C. glutamicum [48 TGs (44)] and E. coli [25 TGs (15)] only share six orthologous genes (6).
Figure 2.
Illustration of the binding-sites detection problem. Here, we demonstrate the problem when moving from one organism to another by investigating the evolutionary conservation of transcription factor binding sites. As in Figure 1, we study the transcriptional regulators pcaR in Corynebacterium glutamicum (CG) and Corynebacterium efficiens (CE) as well as the regulator dtxR in CG, CE, Corynebacterium diphtheriae (CD) and Corynebacterium jeikeium (CJ). For pcaR, all 12 target genes are conserved as are the transcription factor binding sites (TFBSs), depicted by the sequence logos (74) at the right side. It is more complicated with dtxR. The regulons are not conserved, ranging from 27 target genes in CE to 64 targets in CG. The sequence logos for DtxR are also slightly different for CG, CE, CD, and CJ.
To sum up, TFBS prediction has the potential to provide us with knowledge about new gene regulatory interactions. However, we still need to know some TFBSs of a particular TF from the source organism. The major drawback is the poor trade-off between sensitivity and specificity.
Combining both, orthologous genes and conserved binding sites
With the insights gained through the above-introduced studies, we now concentrate on recent approaches that combine both, the identification of orthologous genes as well as the detection of conserved TF binding sites. Under the assumption that a TF-DNA binding within the promoter region of a TG generally affects the co-transcription and co-expression of all genes within the TG’s operon, we may further extend our predictions. For a given conserved TF-TG regulation, we extend the set of TGs in the respective regulon of the target organism by all genes within the TG’s operon (35). Apparently, we need careful operon predictions for this step; refer to (45) for a summary of the state of the art. An overview of the combined inter-species network transfer procedure that utilizes orthology detection and TFBS prediction together with operon extension is depicted in Supplementary Figure S2. We start with the genome annotation data for source and target organisms. Together with the template regulatory network and the known TFBSs from the source organism, we may compute potentially conserved TFs, TGs and TFBSs. In the next step, we assume a TF-TG regulatory interaction to be conserved if the TF, the TG and the TFBS are evolutionarily conserved. In addition, if a TG encodes for the first gene within an operon in the target organism, we extend the regulon of the TF-ortholog by all the genes within the operon. The main advantage of combining TFBS prediction with orthology detection is the drastically decreased false positive rate.
The bioinformatics tool Regulogger serves as our first example here, where specificity was increased up to 25-fold over approaches that solely rely on the identification of conserved TF binding sites (46). Alkema et al. predicted 125 conserved regulogs in Staphylococcus aureus, i.e. sets of co-regulated genes with conserved regulatory sequence across multiple species. They utilized the COG (47) database as orthology detection and a combination of Gibbs Sampling (48) and the TFBS (49) software for binding site predictions. The promoter region is defined as the sequence 250 bp upstream of a putative target gene. Operons are defined as genes with the same orientation and with an intergenic distance of <50 bp, following a suggestion from ref. (50). Note that using the COG database may be impracticable for future studies since COG annotations for newly sequenced species are not available.
In ref. (51) and a subsequent follow-up study (52), the TRACTOR_DB (53) database was used to study conserved regulatory networks in 30 gamma-proteobacteria by using the network of E. coli as template. The number of predicted interactions (regulons) ranges from 6 (3) for Xanthomonas axonopodis to 1901 (69) for Salmonella typhimurium. Here, BBHs were utilized for the detection of orthologous genes. The promoter region was defined as the sequence ranging from −400 to +50 bp relative to the putative target gene start site. For the prediction of operons and TFBSs, the TRACTOR_DB (53) database and PATSER (54) were used.
In a feasibility study for taxonomically closely related species, four corynebacteria, the attempt to transfer data from C. glutamicum to C. efficiens, C. jeikeium and C. diphtheriae yielded 530 new gene regulations (55). The database content of the underlying CoryneRegNet database was increased by factor 4.2 for the three target organisms. Reliable knowledge for ∼40% of the common transcription factors was made available, compared with ∼5% for which knowledge was available before. Here, a promoter region was defined −560 to +20 bp relative to the putative target gene start site. The software packages PoSSuMsearch (56) and Transitivity Clustering (57,58) were used for TFBS predictions and orthology detections, respectively. A disadvantage is the usage of the operon database VIMSS (59). Since the update frequency is limited by technical restrictions, there is a delay for operon annotations of newly sequenced species. Table 1 summarizes the results of the transfer exemplarily for the transcriptional regulators GlxR (60), LexA (44), RamB (61), McbR (62) and DtxR (30). For the latter case, we know the regulons of all four organisms, the source as well as the three targets. Here, the transfer pipeline reconstructed almost half of the DtxR regulons with no false positives.
Table 1.
Examples for regulons transferred between corynebacteriaa
| GlxR | LexA | RamB | McbR | DtxR | ||
|---|---|---|---|---|---|---|
| CG | 99 | 20 | 47 | 46 | 64 | |
| TP | FP | |||||
| CD | 35 | 9 | 27 | 11 | 25 of 63 (40%) | 0 |
| CE | 104 | 14 | 22 | 26 | 18 of 27 (67%) | 0 |
| CJ | 33 | 4 | 13 | 12 | 21 of 51 (41%) | 0 |
aThe table shows the number of known and predicted target genes for five transcription factors that are conserved among the species C. glutamicum (CG), C. diphtheriae (CD), C. efficiens (CE), and C. jeikeium (CJ). CG served as source organism, while CD, CE, and CJ are the target organisms. A combination of orthology detection, binding-site conservation and operon extension has been used for the inter-species transfer procedure(55). The DtxR regulons of CD, CE and CJ have been known in advance allowing us to judge the prediction performance, i.e. we may give numbers for true positives (TP) and false positives (FP).
Note that the presented list of case studies and examples is explicitly not claimed to be exhaustive. We concentrated on specific examples that highlight genome-wide approaches and provide clear and easily interpretable results allowing us to receive an impression of the state of the art. For instance the usage of corynebacterial data throughout this work is not biased but based on practical considerations: We have some proven knowledge about four taxonomically closely related organisms (32), which allowed us to judge and investigate typical problems.
In summary, we conclude that solely using orthologous TFs and TGs is too unreliable. It overestimates the inter-species conservation of TF-TG interactions and underestimates the amount of new regulations that have not been observed in the reference organism before. The TFBS-based approach is capable of identifying new regulations in the target organisms but suffers from high false positive rates if used in isolation. The combination of both is reliable. Essentially, we filter the comparably unspecific TFBS-based results by adding further evidence predicated on conserved TGs. Still, we neglect the underestimation of new regulations of a specific TF in the target organism, i.e. conserved TFBSs but no conserved TGs; something to be discussed in the next section.
OPEN CHALLENGES AND FUTURE DIRECTIONS
The major problem with all approaches is the dependency on a successful discovery of evolutionarily conserved sequences. In contrast to the promoter sequences, TFBSs are comparably short. Furthermore, their variation is comparably high, even within the same organism. This may result in low information content, i.e. an unfavorable signal-to-noise ratio, and is one of the main reasons for the high false positive rates of computational TFBS-identification methods. In addition, we observe a reduced sensitivity when moving from one organism to another one, even if closely related (42). Orthology detection methods are integrated to reduce these error rates, which generally hinders unraveling regulatory interactions in target organisms with many inorthologous TGs. New gene regulations for unconserved TGs may not be identified anymore. We propose an additional step, depicted at the bottom right of Supplementary Figure S2, to counter this problem. After the identification of conserved interactions between source and target, we should not use the TFBSs of the source organism but use the conserved TFBSs in the target organism. These are expected to be more precise since they are putative true binding sites from the target organism itself. Revised computational profiles, constructed from these TFBSs, could subsequently be utilized to scan for further TFBSs in the target organism. Note that we still risk a number of false positive predictions. This can be reduced by applying restrictive significance thresholds and by re-adjusting (fine-tuning) the TFBSs by using motif discovery tools (see below). One possible tool to integrate with a network transfer pipeline may be PhyloGibbs. It identifies conserved sequences motifs and additionally accounts for phylogenetic distances (63,64). We may further decrease the number of false positives by not scanning upstream sequences with fixed positions relative to a TGs start sites. Instead, we might want to use more reasonable promotor sequences by integrating software dedicated to the discovery of transcription start-sites (65).
Another problem with TFBSs is the annotation procedure, where data is transferred manually from the literature to the reference databases. In a recent study about the TFBSs of seven TFs from E. coli, Keilwagen et al. found that 34.5% of the 536 TFBS annotations are questionable; 51 are suggested to be removed, 134 to be shifted by some base pairs (66). The incorporation of so-called sequence motif discovery tools helps with identifying such annotation problems, subsequent TFBS readjusting and finally with the fine-tuning and discovery of new binding motifs in the target organism. A summary and review of corresponding tools is available in a paper from Tompa et al. (67), newer tools may be found e.g. in (66,68,69).
Although the identification of orthologous genes and proteins is a long-standing challenge in computational biology, classical sequence-based approaches neglect to incorporate methods to distinguish between groups of sequences that share common ancestry from groups that share inserted domains but are otherwise unrelated. This protein domain shuffling problem was recently introduced and attacked with a method called Neighborhood Correlation (70,71). However, we suggest performing more research about the discovery of protein domain architecture and its impact on TF-DNA binding and TG conservation; after all, we are still interested in predicting reliable gene regulatory networks here, but not necessarily in unraveling the path of evolution itself.
Besides technical difficulties we also face organizational problems. While nowadays sequenced genomes and their annotations are stored in a well-structured manner, e.g. with the NCBI repositories, gene regulatory interactions, binding sites, operon annotations, homology information, etc., are not. Instead, this data is scattered over numerous publications, not utilizing standardized vocabularies to describe the content for subsequent processing. Text Mining tools are necessary to retrieve relevant literature suggestions (72,73). In addition, even if the data are stored in public databases, it is often not available through standard interfaces. Furthermore, software packages usually need to be compiled, installed and configured locally, often a difficult and time-consuming task. We recommend that the community should follow the advices of Philippi and Köhler (22); primarily, we propose enforcing standardization by making it a requirement for publication in scientific journals.
CONCLUSIONS
Despite all the technical and organizational problems, we conclude that the inter-species transfer of knowledge about gene regulatory networks from model organisms to reference organisms is generally feasible. Reference networks for some prokaryotes are publicly available and can be used for automatic annotations, at least for somewhat related species. In principle, we have all the necessary computational tools available but we are not using them as integral part of standard data-processing pipelines. The performance is limited in terms of sensitivity, which can be improved, for instance, by incorporating phylogenetic sequence motif discovery tools. However, predicted regulations are reliable if the integrated tools are combined appropriately. Hence, we suggest to define standard pipelines similar to the one depicted in Supplementary Figure S2. Furthermore, we motivate their compulsory application to any new genome sequence. Database providers for the reference organism networks could (i) allow uploading whole-genome sequence annotations or (ii) automatically integrate all new genomes from NCBI. After inter-species transferring, potential gene regulations for the target organism may be downloaded, visualized or post-processed. Researchers are automatically provided with new promising wet-lab targets for further studies. This would significantly reduce the gap between existing bacterial genome sequences and the knowledge about gene regulatory networks, a big step in systems biology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for this work was provided by the German Academic Exchange Service (DAAD), the Cluster of Excellence for Multimodel Computing and Interaction (MMCI) of the German Research Foundation (DFG) and the Boehringer Ingelheim Fonds (BIF). Funding for open acces charge: MMCI.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Metzker ML. Sequencing technologies – the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 3.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 4.Venancio TM, Aravind L. Reconstructing prokaryotic transcriptional regulatory networks: lessons from actinobacteria. J. Biol. 2009;8:29. doi: 10.1186/jbiol132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pabo CO, Sauer RT. Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 6.Baumbach J, Tauch A, Rahmann S. Towards the integrated analysis, visualization and reconstruction of microbial gene regulatory networks. Brief Bioinform. 2009;10:75–83. doi: 10.1093/bib/bbn055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Vliet AH. Next generation sequencing of microbial transcriptomes: challenges and opportunities. FEMS Microbiol. Lett. 2009;302:1–7. doi: 10.1111/j.1574-6968.2009.01767.x. [DOI] [PubMed] [Google Scholar]
- 8.Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007;2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galas DJ, Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun LV, Chen L, Greil F, Negre N, Li TR, Cavalli G, Zhao H, Van Steensel B, White KP. Protein-DNA interaction mapping using genomic tiling path microarrays in Drosophila. Proc. Natl. Acad. Sci. USA. 2003;100:9428–9433. doi: 10.1073/pnas.1533393100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonneau R. Learning biological networks: from modules to dynamics. Nat. Chem. Biol. 2008;4:658–664. doi: 10.1038/nchembio.122. [DOI] [PubMed] [Google Scholar]
- 13.Herrgard MJ, Covert MW, Palsson BO. Reconstruction of microbial transcriptional regulatory networks. Curr. Opin. Biotechnol. 2004;15:70–77. doi: 10.1016/j.copbio.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Kazakov AE, Cipriano MJ, Novichkov PS, Minovitsky S, Vinogradov DV, Arkin A, Mironov AA, Gelfand MS, Dubchak I. RegTransBase–a database of regulatory sequences and interactions in a wide range of prokaryotic genomes. Nucleic Acids Res. 2007;35:D407–D412. doi: 10.1093/nar/gkl865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gama-Castro S, Jimenez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Penaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muniz-Rascado L, Martinez-Flores I, Salgado H, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keseler IM, Bonavides-Martinez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37:D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sierro N, Makita Y, de Hoon M, Nakai K. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 2008;36:D93–D96. doi: 10.1093/nar/gkm910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacques PE, Gervais AL, Cantin M, Lucier JF, Dallaire G, Drouin G, Gaudreau L, Goulet J, Brzezinski R. MtbRegList, a database dedicated to the analysis of transcriptional regulation in Mycobacterium tuberculosis. Bioinformatics. 2005;21:2563–2565. doi: 10.1093/bioinformatics/bti321. [DOI] [PubMed] [Google Scholar]
- 19.Grote A, Klein J, Retter I, Haddad I, Behling S, Bunk B, Biegler I, Yarmolinetz S, Jahn D, Munch R. PRODORIC (release 2009): a database and tool platform for the analysis of gene regulation in prokaryotes. Nucleic Acids Res. 2009;37:D61–D65. doi: 10.1093/nar/gkn837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumbach J, Wittkop T, Kleindt CK, Tauch A. Integrated analysis and reconstruction of microbial transcriptional gene regulatory networks using CoryneRegNet. Nat. Protoc. 2009;4:992–1005. doi: 10.1038/nprot.2009.81. [DOI] [PubMed] [Google Scholar]
- 21.Salgado H, Gama-Castro S, Peralta-Gil M, Diaz-Peredo E, Sanchez-Solano F, Santos-Zavaleta A, Martinez-Flores I, Jimenez-Jacinto V, Bonavides-Martinez C, Segura-Salazar J, et al. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 2006;34:D394–D397. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Philippi S, Kohler J. Addressing the problems with life-science databases for traditional uses and systems biology. Nat. Rev. Genet. 2006;7:482–488. doi: 10.1038/nrg1872. [DOI] [PubMed] [Google Scholar]
- 23.Babu MM, Lang B, Aravind L. Methods to reconstruct and compare transcriptional regulatory networks. Methods Mol. Biol. 2009;541:163–180. doi: 10.1007/978-1-59745-243-4_8. [DOI] [PubMed] [Google Scholar]
- 24.Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA. Structure and evolution of transcriptional regulatory networks. Curr. Opin. Struct. Biol. 2004;14:283–291. doi: 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Madan Babu M, Teichmann SA, Aravind L. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006;358:614–633. doi: 10.1016/j.jmb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huynen MA, Bork P. Measuring genome evolution. Proc. Natl. Acad. Sci. USA. 1998;95:5849–5856. doi: 10.1073/pnas.95.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu H, Luscombe NM, Lu HX, Zhu X, Xia Y, Han JD, Bertin N, Chung S, Vidal M, Gerstein M. Annotation transfer between genomes: protein-protein interologs and protein-DNA regulogs. Genome Res. 2004;14:1107–1118. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price MN, Dehal PS, Arkin AP. Orthologous transcription factors in bacteria have different functions and regulate different genes. PLoS Comput. Biol. 2007;3:1739–1750. doi: 10.1371/journal.pcbi.0030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brune I, Werner H, Huser AT, Kalinowski J, Puhler A, Tauch A. The DtxR protein acting as dual transcriptional regulator directs a global regulatory network involved in iron metabolism of Corynebacterium glutamicum. BMC Genomics. 2006;7:21. doi: 10.1186/1471-2164-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brinkrolf K, Brune I, Tauch A. Transcriptional regulation of catabolic pathways for aromatic compounds in Corynebacterium glutamicum. Genet. Mol. Res. 2006;5:773–789. [PubMed] [Google Scholar]
- 32.Baumbach J. CoryneRegNet 4.0 - A reference database for corynebacterial gene regulatory networks. BMC Bioinformatics. 2007;8:429. doi: 10.1186/1471-2105-8-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baumbach J, Apeltsin L. Linking Cytoscape and the corynebacterial reference database CoryneRegNet. BMC Genomics. 2008;9:184. doi: 10.1186/1471-2164-9-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baumbach J, Wittkop T, Rademacher K, Rahmann S, Brinkrolf K, Tauch A. CoryneRegNet 3.0–an interactive systems biology platform for the analysis of gene regulatory networks in corynebacteria and Escherichia coli. J. Biotechnol. 2007;129:279–289. doi: 10.1016/j.jbiotec.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 35.Balazsi G, Heath AP, Shi L, Gennaro ML. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008;4:225. doi: 10.1038/msb.2008.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo M, Feng H, Zhang J, Wang W, Wang Y, Li Y, Gao C, Chen H, Feng Y, He ZG. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009;19:1301–1308. doi: 10.1101/gr.086595.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson D, Charoensawan V, Kummerfeld SK, Teichmann SA. DBD–taxonomically broad transcription factor predictions: new content and functionality. Nucleic Acids Res. 2008;36:D88–D92. doi: 10.1093/nar/gkm964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bakke P, Carney N, Deloache W, Gearing M, Ingvorsen K, Lotz M, McNair J, Penumetcha P, Simpson S, Voss L, et al. Evaluation of three automated genome annotations for Halorhabdus utahensis. PLoS ONE. 2009;4:e6291. doi: 10.1371/journal.pone.0006291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bocker S, Jahn K, Mixtacki J, Stoye J. Computation of median gene clusters. J. Comput. Biol. 2009;16:1085–1099. doi: 10.1089/cmb.2009.0098. [DOI] [PubMed] [Google Scholar]
- 40.Raghupathy N, Durand D. Gene cluster statistics with gene families. Mol. Biol. Evol. 2009;26:957–968. doi: 10.1093/molbev/msp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahmann S, Müller T, Vingron M. On the power of profiles for transcription factor binding site detection. Statistical Applications in Genetics and Molecular Biology. 2003;2 doi: 10.2202/1544-6115.1032. Article 7. [DOI] [PubMed] [Google Scholar]
- 42.Baumbach J, Brinkrolf K, Wittkop T, Tauch A, Rahmann S. CoryneRegNet 2: An Integrative Bioinformatics Approach for Reconstruction and Comparison of Transcriptional Regulatory Networks in Prokaryotes. Journal of Integrative Bioinformatics. 2006;3:24. [Google Scholar]
- 43.Mazon G, Erill I, Campoy S, Cortes P, Forano E, Barbe J. Reconstruction of the evolutionary history of the LexA-binding sequence. Microbiology. 2004;150:3783–3795. doi: 10.1099/mic.0.27315-0. [DOI] [PubMed] [Google Scholar]
- 44.Jochmann N, Kurze AK, Czaja LF, Brinkrolf K, Brune I, Huser AT, Hansmeier N, Puhler A, Borovok I, Tauch A. Genetic makeup of the Corynebacterium glutamicum LexA regulon deduced from comparative transcriptomics and in vitro DNA band shift assays. Microbiology. 2009;155:1459–1477. doi: 10.1099/mic.0.025841-0. [DOI] [PubMed] [Google Scholar]
- 45.Brouwer RW, Kuipers OP, van Hijum SA. The relative value of operon predictions. Brief Bioinform. 2008;9:367–375. doi: 10.1093/bib/bbn019. [DOI] [PubMed] [Google Scholar]
- 46.Alkema WB, Lenhard B, Wasserman WW. Regulog analysis: detection of conserved regulatory networks across bacteria: application to Staphylococcus aureus. Genome Res. 2004;14:1362–1373. doi: 10.1101/gr.2242604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson W, Rouchka EC, Lawrence CE. Gibbs Recursive Sampler: finding transcription factor binding sites. Nucleic Acids Res. 2003;31:3580–3585. doi: 10.1093/nar/gkg608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lenhard B, Wasserman WW. TFBS: Computational framework for transcription factor binding site analysis. Bioinformatics. 2002;18:1135–1136. doi: 10.1093/bioinformatics/18.8.1135. [DOI] [PubMed] [Google Scholar]
- 50.Moreno-Hagelsieb G, Collado-Vides J. A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics. 2002;18(Suppl. 1):S329–336. doi: 10.1093/bioinformatics/18.suppl_1.s329. [DOI] [PubMed] [Google Scholar]
- 51.Espinosa V, Gonzalez AD, Vasconcelos AT, Huerta AM, Collado-Vides J. Comparative studies of transcriptional regulation mechanisms in a group of eight gamma-proteobacterial genomes. J. Mol. Biol. 2005;354:184–199. doi: 10.1016/j.jmb.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 52.Gonzalez Perez AD, Gonzalez Gonzalez E, Espinosa Angarica V, Vasconcelos AT, Collado-Vides J. Impact of Transcription Units rearrangement on the evolution of the regulatory network of gamma-proteobacteria. BMC Genomics. 2008;9:128. doi: 10.1186/1471-2164-9-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perez AG, Angarica VE, Vasconcelos AT, Collado-Vides J. Tractor_DB (version 2.0): a database of regulatory interactions in gamma-proteobacterial genomes. Nucleic Acids Res. 2007;35:D132–136. doi: 10.1093/nar/gkl800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 55.Baumbach J, Rahmann S, Tauch A. Reliable transfer of transcriptional gene regulatory networks between taxonomically related organisms. BMC Syst. Biol. 2009;3:8. doi: 10.1186/1752-0509-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beckstette M, Homann R, Giegerich R, Kurtz S. Fast index based algorithms and software for matching position specific scoring matrices. BMC Bioinformatics. 2006;7:389. doi: 10.1186/1471-2105-7-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wittkop T, Baumbach J, Lobo FP, Rahmann S. Large scale clustering of protein sequences with FORCE – A layout based heuristic for weighted cluster editing. BMC Bioinformatics. 2007;8:396. doi: 10.1186/1471-2105-8-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wittkop T, Emig D, Lange S, Rahmann S, Albrecht M, Morris JH, Bocker S, Stoye J, Baumbach J. Partitioning biological data with transitivity clustering. Nat. Methods. 2010;7:419–420. doi: 10.1038/nmeth0610-419. [DOI] [PubMed] [Google Scholar]
- 59.Price MN, Huang KH, Alm EJ, Arkin AP. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 2005;33:880–892. doi: 10.1093/nar/gki232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kohl TA, Baumbach J, Jungwirth B, Puhler A, Tauch A. The GlxR regulon of the amino acid producer Corynebacterium glutamicum: in silico and in vitro detection of DNA binding sites of a global transcription regulator. J. Biotechnol. 2008;135:340–350. doi: 10.1016/j.jbiotec.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 61.Gerstmeir R, Cramer A, Dangel P, Schaffer S, Eikmanns BJ. RamB, a novel transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J. Bacteriol. 2004;186:2798–2809. doi: 10.1128/JB.186.9.2798-2809.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rey DA, Nentwich SS, Koch DJ, Ruckert C, Puhler A, Tauch A, Kalinowski J. The McbR repressor modulated by the effector substance S-adenosylhomocysteine controls directly the transcription of a regulon involved in sulphur metabolism of Corynebacterium glutamicum ATCC 13032. Mol. Microbiol. 2005;56:871–887. doi: 10.1111/j.1365-2958.2005.04586.x. [DOI] [PubMed] [Google Scholar]
- 63.van Nimwegen E. Finding regulatory elements and regulatory motifs: a general probabilistic framework. BMC Bioinformatics. 2007;8(Suppl. 6):S4. doi: 10.1186/1471-2105-8-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siddharthan R, Siggia ED, van Nimwegen E. PhyloGibbs: a Gibbs sampling motif finder that incorporates phylogeny. PLoS Comput. Biol. 2005;1:e67. doi: 10.1371/journal.pcbi.0010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mendoza-Vargas A, Olvera L, Olvera M, Grande R, Vega-Alvarado L, Taboada B, Jimenez-Jacinto V, Salgado H, Juarez K, Contreras-Moreira B, et al. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS ONE. 2009;4:e7526. doi: 10.1371/journal.pone.0007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Keilwagen J, Baumbach J, Kohl TA, Grosse I. MotifAdjuster: a tool for computational reassessment of transcription factor binding site annotations. Genome Biol. 2009;10:R46. doi: 10.1186/gb-2009-10-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, Kent WJ, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 68.Baumbach J, Wittkop T, Weile J, Kohl T, Rahmann S. MoRAine – A web server for fast computational transcription factor binding motif re-annotation. Journal of Integrative Bioinformatics. 2008;5 doi: 10.2390/biecoll-jib-2008-91. [DOI] [PubMed] [Google Scholar]
- 69.Linhart C, Halperin Y, Shamir R. Transcription factor and microRNA motif discovery: the Amadeus platform and a compendium of metazoan target sets. Genome Res. 2008;18:1180–1189. doi: 10.1101/gr.076117.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Joseph JM, Durand D. Family classification without domain chaining. Bioinformatics. 2009;25:i45–53. doi: 10.1093/bioinformatics/btp207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song N, Joseph JM, Davis GB, Durand D. Sequence similarity network reveals common ancestry of multidomain proteins. PLoS Comput. Biol. 2008;4:e1000063. doi: 10.1371/journal.pcbi.1000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jensen LJ, Saric J, Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nat. Rev. Genet. 2006;7:119–129. doi: 10.1038/nrg1768. [DOI] [PubMed] [Google Scholar]
- 73.Winnenburg R, Wachter T, Plake C, Doms A, Schroeder M. Facts from text: can text mining help to scale-up high-quality manual curation of gene products with ontologies? Brief Bioinform. 2008;9:466–478. doi: 10.1093/bib/bbn043. [DOI] [PubMed] [Google Scholar]
- 74.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.