Abstract
O6-Methylguanine (O6-meG), which is produced in DNA following exposure to methylating agents, instructs human RNA polymerase II to mis-insert bases opposite the lesion during transcription. In this study, we examined the effect of O6-meG on transcription in human cells and investigated the subsequent effects on protein function following translation of the resulting mRNA. In HEK293 cells, O6-meG induced incorporation of uridine or cytidine in nascent RNA opposite the adduct. In cells containing active O6-alkylguanine-DNA alkyltransferase (AGT), which repairs O6-meG, 3% misincorporation of uridine was observed opposite the lesion. In cells where AGT function was compromised by addition of the AGT inhibitor O6-benzylguanine, ∼58% of the transcripts contained a uridine misincorporation opposite the lesion. Furthermore, the altered mRNA induced changes to protein function as demonstrated through recovery of functional red fluorescent protein (RFP) from DNA coding for a non-fluorescent variant of RFP. These data show that O6-meG is highly mutagenic at the level of transcription in human cells, leading to an altered protein load, especially when AGT is inhibited.
INTRODUCTION
Radiation and chemicals can give rise to reactive species that damage DNA, producing strand breaks, abasic sites, and altered bases, sugars and phosphate groups. These structural modifications to the genetic material often exert detrimental effects on replication and gene expression, compromising DNA’s role as the repository of cellular information. DNA-dependent DNA polymerases can stall at damaged sites in the template, giving rise to collapsed replication forks, or synthesize incorrect products as they progress past the lesions, causing mutations in the daughter DNA (1–3). DNA-dependent RNA polymerases can also stall at damaged bases in DNA or bypass the lesions, resulting in altered transcripts in a process called transcriptional mutagenesis (4–8). To avert the deleterious effects of DNA damage, cells have evolved an elaborate array of DNA repair pathways that maintain and preserve DNA structure. But when genomic maintenance is compromised due to aberrant DNA repair, the accumulated damage and associated effects on replication and transcription result in pathology that can include cancer and developmental deficits (9,10).
Among the chemical agents that damage DNA are those that alkylate the purines, pyrimidines and phosphate groups. Methylating agents, such as the chemical N-methyl-N-nitrosourea (MNU) or the endogenous metabolite S-adenosylmethionine, produce two abundant lesions, 7-methylguanine, which is relatively innocuous, and 3-methyladenine, which is cytotoxic. Methylating agents that tend to be highly carcinogenic, which include MNU, also form significant amounts of O6-methylguanine (O6-meG) (11–14). Furthermore, the O6-position of guanine is a target site for alkylation by certain chemotherapeutic drugs, such as 1,3-bis (2-chloroethyl)-1-nitrosourea (BCNU) and temozolomide (15,16). BCNU adds a chloroethyl group to the O6-position of guanine, which subsequently rearranges and forms a cytotoxic interstrand DNA crosslink (17). Temozolomide spontaneously degrades, giving rise to a methyldiazonium ion that methylates DNA, including the O6-position of guanine (18). In both cases, the formation of the O6-alkylated guanine plays an important role in the cytotoxic mechanism of these drugs.
O6-MeG is highly mutagenic, instructing DNA polymerases to incorporate thymine instead of cytosine opposite the lesion, resulting in GC to AT transitions during replication (19–21). O6-MeG is also cytotoxic because it invokes DNA mismatch repair that results in a cycle of futile attempts at clearing the lesion, ultimately producing DNA strand breaks and inducing apoptosis (22). The mutagenic and cytotoxic consequences of O6-meG are ameliorated by the presence of the protein O6-alkylguanine–DNA alkyltransferase (AGT) (23–25). AGT is found in both prokaryotes and eukaryotes and works in a similar way in each. The protein binds to O6-meG, removes the methyl group and transfers it to a cysteine located within the AGT active site, generating S-methylcysteine in the protein and restoring guanine in DNA. AGT’s action is not enzymatic: Its activity cannot be regenerated after it has repaired a single lesion. AGT can also remove longer chain alkyl groups from the O6-position of guanine; hence, it should come as no surprise that AGT interferes with the chemotherapeutic efficacy of agents that alkylate the O6-position of guanine. In the case of BCNU, AGT repairs the O6-chloroethylguanine intermediate, thwarting the formation of the cytotoxic DNA interstrand crosslink (26). For temozolomide, AGT repairs O6-meG, thus clearing a cytotoxic DNA lesion that induces apoptosis (18,27).
In biochemical studies, O6-meG induces transcriptional mutagenesis when RNA polymerases bypass the lesion, resulting in significant incorporation of uridine into nascent RNA (28,29). However, the impact of this observation has not been demonstrated in human cells, either at the level of altered transcripts or on the ensuing effects on protein function. To examine this, a strategy was used in which a site-specific O6-meG was positioned on the transcribed strand of a gene encoding a fluorescence-defective red fluorescent protein (RFP) variant in a plasmid that does not contain any known mammalian origin of replication. Transcription past the lesion that resulted in cytidine incorporation produced mRNA that encoded proteins with no fluorescent activity. In contrast, incorporation of uridine resulted in transcripts that encoded the sequence of a functional, fluorescent protein providing a clear demonstration that transcriptional mutagenesis can produce altered RNA that affects protein function following translation.
MATERIALS AND METHODS
Chemicals and biochemicals
PspOMI, DpnI, BfuAI, NotI, SalI, T4 PNK, T4 DNA ligase and helper phage M13K07 were purchased from New England Biolabs (Waltham, MA, USA). Esp3I was purchased from Fermentas (Glen Burnie, MD, USA). Plasmids and HEK293 Tet-Off cells were procured from Clontech (Mountain View, CA, USA). KOD Hot Start Master Mix was purchased from EMD Biosciences (Gibbstown, NJ, USA). DNA oligomers were made by Sigma-Aldrich (St Louis, MO, USA). All media and supplements for cell culture were purchased from Mediatech (Manassas, VA, USA) and Clontech (Mountain View, CA, USA). Chemical reagents were purchased from Fisher Scientific (Pittsburgh, PA, USA).
Vectors
Details of construction of pTT2-RΔ-G-F1(+), which provided the core vector for the synthesis of pRFPwt, pRFPm, pRinsert and pRFPm-O6meG, can be found under Supplementary data. Each vector contained two tetracycline response element-tight (TRE-tight) promoters in tandem, which control the expression of the fluorescent reporters AcGFP [green fluorescent protein (GFP)] and DsRed-Express (RFP), and an F1 origin of replication, which directs production of single-strand DNA (ssDNA) that corresponds to the coding strands of the RFP and GFP genes. Each vector was named according to the RFP sequence it contained. pRFPwt contained the entire DsRed-Express coding sequence. pRFPm contained the DsRed-Express coding sequence with the mutation S21P (TCC → CCC). pR-insert contained the DsRed-Express coding sequence with a 73 bp insert, which when digested with the enzyme Esp3I is excised, linearizing the plasmid with ends flanking an 11-bp gap centered on the first C in the proline codon CCC of pRFPm. pRFPm-O6meG contains the damaged base O6-meG opposite the first C in the proline codon CCC.
ssDNA production
The vector pRFPm was propagated in One Shot OmniMax2 T1 R bacteria (Invitrogen) containing the F′ episome. These cells were super-infected with helper phage M13K07 at a final concentration of >1 × 109 pfu/ml culture. Bacterial cells were pelleted, the culture supernatant was filtered through a 0.22-μm filter, and the filtered media was treated with 1 U/ml Dnase (Worthington Biochemical) to remove contaminating DNA from lysed bacteria. ssDNA was recovered by polyethylene glycol precipitation of phage particles followed by phage lysis with proteinase K and DNA purification by phenol extraction and isopropanol precipitation [Invitrogen online protocol, modified from (30)]. ssDNA was used directly after precipitation without further purification.
Production of pRFPm-O6meG
pRFPm-O6meG was produced in vitro by combining pRFPm ssDNA with Esp3I-digested pRFP-insert DNA, which formed a gapped duplex. A DNA oligomer containing O6-meG was ligated into the gap (Supplementary Figure S4). To form a gapped duplex, 300 μg of pRFP-insert was digested with Esp3I at 37°C for 2 h, removing the 73-bp fragment. pRFP-insert was mixed with 1 mg pRm ssDNA and denatured with 90% formamide, 1 mM EDTA at 45°C for 30 min. The sample was subsequently dialyzed against 50% formamide, 200 mM Tris–HCl, pH 7.8 at 37°C for at least 3 h to induce renaturation and formation of gapped duplex DNA. This was followed by consecutive dialyses against 100 mM Tris–HCl, 100 mM NaCl, 1 mM EDTA, pH 7.8 at 4°C overnight and 10 mM Tris–HCl, 1 mM EDTA pH 7.8 (TE buffer) at room temperature for 5 h, respectively (31). The annealed DNA was precipitated, resuspended in 10 mM Tris–HCl pH 7.8 and analyzed by agarose gel electrophoresis to confirm the appearance of double-stranded, open circular (gapped) plasmid DNA.
A DNA 11-mer containing O6-meG (5′ CACGG [O6-meG]GCCCT) was phosphorylated with T4 PNK and annealed to the gapped duplex at a ratio of at least 5 : 1 by heating to 45°C and allowing to cool to 4°C in a refrigerator. The mixture was incubated with T4 DNA ligase (8 U/μg dsDNA) for 24 h at 14°C producing covalently closed circular, double-strand DNA containing a single O6-meG at a defined position (vector pRFPm-O6meG, Supplementary Figure S4). The closed-circular DNA was purified by two consecutive CsCl (1.01 g/ml) gradient centrifugations (331 000g for 24 h in a Beckman 70.1Ti rotor at 21°C) in the presence of ethidium bromide (0.4 mg/ml). The recovered closed-circular DNA was extracted with water-saturated butanol, dialyzed against TE buffer at 4°C, ethanol precipitated and resuspended in TE buffer.
Restriction digests with the enzyme PspOMI were used to confirm the presence of O6-meG. pRFPm and pRFPm-O6meG were digested with PspOMI for 1 h at 37°C. The products were resolved on a 0.8% agarose gel containing ethidium bromide and visualized using a KODAK Gel Logic100 gel imaging system.
Cell culture and transfection
Stably transfected Tet-Off HEK293 cells were grown in Eagle’s minimal essential medium (EMEM) supplemented with 10% (v/v) Tet system approved fetal bovine serum, 1 mM sodium pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin and 100 μg/ml G418 and maintained in a humidified incubator at 37°C under 5% CO2. Cells were transiently transfected using PolyFect Transfection Reagent from Qiagen (Valencia, CA, USA) according to protocol. In brief, 48 h before transfection, cells were plated in a six-well cell culture cluster (106 cells/well). The plasmids (2 µg) were pre-incubated with PolyFect Transfection Reagent, mixed with EMEM and added to the cells. In order to block repair of O6-meG by cellular AGT, O6-benzylguanine dissolved in dimethyl sulfoxide (DMSO) was added to cells where appropriate 1 h before transfection. The cells were subsequently incubated at 37°C and 5% CO2 and assayed after 24 h.
Plasmid replication in human cells
Methylated or unmethylated control DNA (pRFPm) from bacteria, or DNA (pRFPm) isolated from human cell nuclei was digested with DpnI for 1 h in the absence or presence of 200 mM NaCl along with non-specific carrier DNA (32). Following incubation with DpnI, linear DNA was digested using exonuclease III as described (33). An aliquot of the digested DNA containing 0.05 ng (after DpnI digestion) or 0.008 ng (after DpnI and exonuclease III digestion) was used in a PCR reaction (GoTaq green master mix, Promega) containing primers (RdF: 5′-CAGCGGCCCTTCTCTCTTA-3′; RR: 5′-CGCTACAGGAACAGGTGGTG-3′) whose product spans 7 DpnI sites.
Fluorescence microscopy
For fluorescence microscopy, 2 × 105 cells were plated on collagen type I coated four-well culture slides (BD BioCoat, BD Biosciences) and transfected 24 h later with 1 µg plasmid as described above. After 24 h, cells were washed with PBS, fixed with 4% formaldehyde and covered with anti-fade reagent (Invitrogen) and a cover slip. Fluorescent cells were visualized using a Nikon Microphot-SA equipped with a super high-pressure 100W mercury lamp. Digital images were produced using SPOT software. ImageJ and GIMP software were used to further process images for publication.
Flow cytometry
Flow cytometry and cell sorting were performed on at least three experimentally independent cell populations on a BD-FACSAria cell sorter (BD Biosciences). GFP and RFP were excited with a 488 nm laser, and detected through spectral filters at 530/30 and 610/20, respectively. Every fluorescence-activated cell sorting (FACS) analysis recorded 1 000 000 events. For cell sorting, gates for GFP- and RFP-positive cells were drawn by hand around untransfected cells. GFP-expressing cells were collected in RLT Lysis buffer (Qiagen).
qRT2-PCR
Quantitative real-time reverse transcriptase PCR (qRT2-PCR) analysis was performed with transfected cells acquired by sorting GFP-positive cells using FACS. RNA was purified using RNeasy micro kit (Qiagen) and further DNase treated using TURBO DNA-freeTM (Ambion) according to protocol. Subsequently, first strand cDNA synthesis was performed using an RT Script kit followed by quantitative real-time PCR performed in triplicate using HotStart-IT® SYBR® Green qPCR Master Mix (both from USB) on an iCycler MyiQ (Bio-Rad). Relative gene expression quantification was based on the comparative threshold cycle method (2−ΔΔCt) (34). The cycle parameters were 95°C for 2 min, and then 40 cycles at 95°C for 15 s and 60°C for 1 min followed by melt curve analysis. The sequences of the primers used are as follows: RFP (forward primer: 5′-CGGCTCCTTCATCTACAAGG-3′; reverse primer: 5′-CGCTACAGGAACAGGTGGTG-3′), GFP (forward primer: 5′-CACATGAAGCAGCACGACTT-3′; reverse primer: 5′-ATGTTGTGGCGGATCTTGA-3′) and GAPDH (forward primer: 5′-CGAGATCCCTCCAAAATCAA-3′; reverse primer: 5′-TTCACACCCATGACGAACAT-3′).
mRNA sequencing
RNA was purified using RNeasy micro kit (Qiagen) and further DNase treated using TURBO DNA-freeTM (Ambion) according to protocol. Subsequently, first strand cDNA synthesis was performed using an RT Script kit (USB). cDNA was amplified by PCR with KOD proofreading polymerase using primers: forward 5′-ATAGCATGTCGACGCCCTTCTCTCTTAAGGTAGCTACA-3′; and reverse 5′-ATAGCATGCGGCCGCTGCTTCACGTACACCTTGGAGCCGT-3′. These primers introduced SalI and NotI restriction sites into the 5′- and 3′-ends of the PCR product, respectively. The PCR products were digested with SalI and NotI and cloned into a SalI–NotI digested vector, pCI-neo, from Promega. Individual colonies were mini-prepped and sequenced using the M13 reverse primer. For each experimental replicate, 20 clones were sequenced.
Restriction enzyme assay
PspOMI digestion of reverse transcriptase PCR (RT–PCR) amplified cellular RFP transcripts was used to analyze the extent of transcriptional mutagenesis. Briefly, cDNAs obtained from cells transfected with pRFPm or pRFPm-O6meG were amplified by PCR using KOD Hot Start Master Mix and RdF (5′-CAGCGGCCCTTCTCTCTTA-3′) and RdR (5′-TGCTTCACGTACACCTTGGA-3′) primers and subsequently digested with PspOMI. A standard curve was created by a series of mixtures of pRFPwt and pRFPm plasmids using the same PCR and digestion protocol. The digested DNA was resolved on a 1% agarose gel and band intensities analyzed by plotting lanes and measuring the area under the curve for each band using ImageJ Software (NIH). The ratio of the area of the upper band, the mutated sequence, to the total area of both bands, combined wild-type (wt) and mutated sequences, was used to create a standard curve (Supplementary Figure S1). The calculated ratio from each cDNA was compared with the standard curve to determine the percentage of mutant transcripts under each condition from at least three independent experiments.
RESULTS
Synthesis of plasmids containing site-specific O6-MeG
The overall strategy for examining the effect of O6-meG on transcription in a cellular system relied on biochemical data showing that the lesion directs insertion of either cytidine or uridine into nascent RNA, producing normal or altered transcripts, respectively (28). When such changes occur within an mRNA’s translated region, the cellular protein synthesis machinery could be directed to incorporate amino acids into the growing peptide chains that result in aberrant proteins (5).
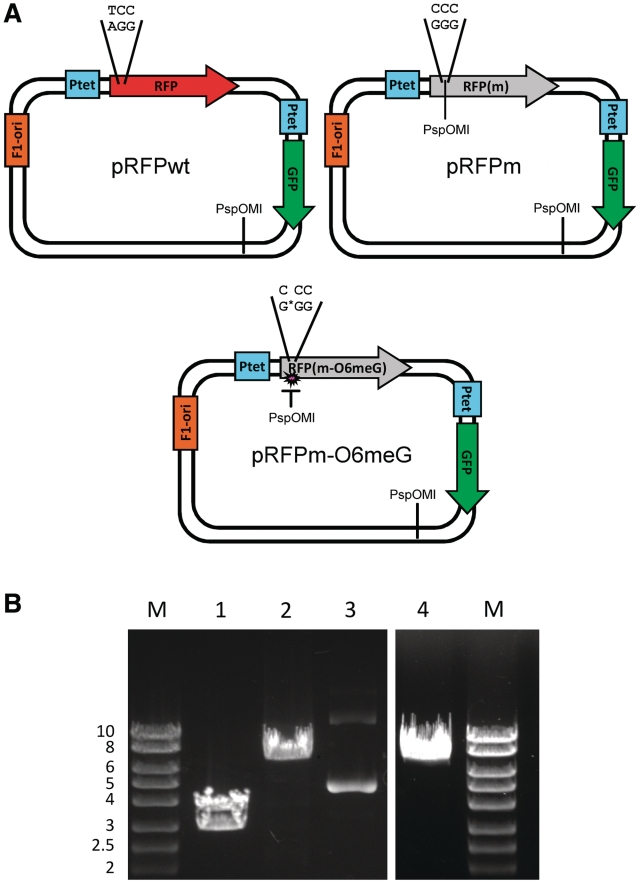
The approach required the assembly of three vectors: pRFPwt, pRFPm and pRFPm-O6meG (Figure 1A). Vector pRFPwt contained two reporter genes, one encoding a GFP and the other encoding wt RFP. The use of two fluorescent reporters added an important control for the experiments described here. Vector pRFPwt acted as a positive control with both GFP and RFP fluorescence. In pRFPm, the RFP gene contained a T to C point mutation, generating a S21P (TCC→CCC) substitution that resulted in a non-fluorescent RFP variant. This vector acted as a second control, exhibiting only GFP fluorescence.
Figure 1.
Plasmids. (A) The vectors used in these studies contained two reporter genes, RFP and GFP, under the control of independent, identical tetracycline-responsive promoters Ptet. The vectors also contained an F1 origin of replication, which directs production of ssDNA. pRFPm contained a single base change from pRFPwt, which resulted in an S21P mutation that coded for a non-fluorescent RFP. In pRFPm-O6meG, O6-meG replaced G opposite the 5′ C in the proline codon. The PspOMI restriction sites used for vector analysis are also indicated. (B) The presence of O6-meG blocks PspOMI digestion. The vectors were incubated with PspOMI and the products were resolved by agarose gel electrophoresis. 1: pRFPm + PspOMI. 2: pRFPm-O6meG + PspOMI. 3: pRFPm-O6meG uncut plasmid. 4: pRFPwt + PspOMI. Marker in kilobases. See text for details.
The third vector, pRFPm-O6meG, contained a single O6-meG opposite the 5′ C in proline codon 21. pRFPm-O6meG was the experimental vector used to determine the effect of O6-meG on transcription and protein function. In accordance with biochemical data, transcription past the site-specific O6-meG lesion should produce RNA containing either 5′-CCC-3′, which would encode proline and a functionless RFP, or 5′-UCC-3′, which would encode serine and restore RFP activity. Hence, in this scenario, base misincorporation reverts the RFP RNA to the wt sequence that restores fluorescence activity in response to transcriptional mutagenesis.
A restriction digest with PspOMI was performed to verify the presence of a site-specific O6-meG in vector pRFPm-O6meG. PspOMI cuts DNA at the sequence 5′-GGGCCC-3′, but does not cut when one of the guanines is replaced with an O6-meG, as is the case for one of the two PspOMI sites in pRFPm-O6meG. For the unmodified pRFPm, digestion with PspOMI should produce DNA fragments 3654 and 2865 bp in length. When pRFPm was incubated with PspOMI, two bands were observed with the expected sizes (Figure 1B, Lane 1). Furthermore, no closed circular plasmid was seen, indicating that the digest went to completion. In contrast, when pRFPm-O6meG was incubated with PspOMI, only a single DNA band was produced (Figure 1B, Lane 2), indicating that only one PspOMI site was cut, producing a pattern identical to that of PspOMI-digested pRFPwt (Figure 1B, Lane 4). These results showed that a site-specific O6-meG was present in the purified pRFPm-O6meG vector.
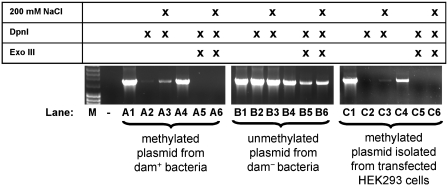
Plasmid replication was not observed in HEK293 cells
Detection of transcriptional mutagenesis using site-specific DNA damage in plasmids is predicated on the notion that replication does not occur, thus preventing formation of mutated daughter DNA that would confound the results observed during transcription. To eliminate replication in the experiments reported here, plasmids lacking any known mammalian origin of replication were used. However, plasmid replication in human cells has been reported even when the vectors did not contain a specific known origin of replication (35). In fact, transcription itself has been shown to induce replication in some cases but not others (36,37).
To test for plasmid replication, methylated and unmethylated control DNA samples and plasmid DNA isolated from HEK293 cells were left untreated or incubated with DpnI, then amplified by PCR using primers that spanned 1632 bp and seven DpnI sites. The restriction enzyme DpnI specifically recognizes methylated GATC sites. In the absence of NaCl, the enzyme efficiently cleaves fully methylated GATC sites and slowly cleaves hemi-methylated sites. In the presence of 200 mM NaCl, DpnI effectively cleaves fully methylated DNA at a reduced rate but no longer cleaves hemi-methylated DNA (32). Hence, in the presence of 200 mM NaCl, plasmid DNA that only replicated once, resulting in hemi-methylated DNA, would be resistant to DpnI cleavage and subsequently amplified by PCR.
Fully methylated plasmid DNA isolated from dam+/dcm+ DH5α bacteria was amplified to a small extent following digestion (Figure 2, Lanes A2 and A3). This was likely due to residual, partially digested plasmid DNA, leaving intact amplicon fragments (33). When exonuclease III was used to digest linear DNA following incubation with DpnI, no amplification product was observed, indicating that all the plasmid DNA had been cut at least once by DpnI, making it susceptible to exonuclease digestion (Figure 2, Lanes A5 and A6). In contrast, unmethylated plasmid DNA isolated from dam−/dcm− bacteria, which should not be a substrate for DpnI, was amplifed as expected following DpnI and exonuclease III incubation, showing that unmethylated, closed-circular plasmid was not subject to DpnI digestion (Figure 2, Lanes B1–B6). Plasmid DNA isolated from HEK293 cells behaved identically to methylated control DNA, indicating that the plasmids used in this work were not replicated following transfection (Figure 2, Lanes C1−C6 for transcriptionally active plasmids; the replication assay for transcriptionally silent plasmids looked identical).
Figure 2.
pRFPm is not replicated in human cells. Template DNA for Lanes A1–A6, was isolated from dam(+) bacteria. Template DNA for Lanes B1–B6 was isolated from dam/dcm(−) bacteria. Template DNA for Lanes C1–C6 was isolated from HEK293 cells 48 h after transfection in the absence of dox. Lanes A1, A4, B1, B4, C1 and C4 are positive controls for PCR. Lane (−) is a negative control with no template DNA in the PCR reaction. See text for details.
O6-MeG induced transcriptional mutagenesis in HEK293 cells
To test for transcriptional mutagenesis by O6-meG, HEK293 Tet-Off cells were transfected with the vectors described in Figure 1. In some instances, cells were treated with O6-benzylguanine, eliminating AGT activity that could repair the methylated base (38,39). Transcription was induced by excluding the tetracycline analog doxycycline (dox) from the growth medium, and RNA was recovered from the cells 24 h after transfection. The RFP mRNA was converted to DNA using RT–PCR, and restriction enzyme sensitivity and sequencing were used to characterize the products.
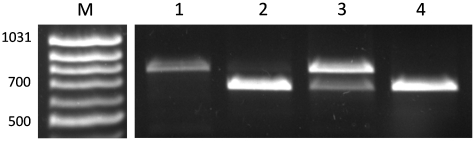
As in the assay to detect the presence of O6-meG in the modified vector, PspOMI was used to test for transcriptional mutagenesis. PCR was used to amplify RFP cDNA, using primers that spanned the PspOMI site present in the RFPm gene. cDNA generated from RNA following transcription of the pRFPwt RFP gene does not contain a PspOMI restriction site, leading to the formation of a band of DNA corresponding to the expected 751 bp after incubation with PspOMI (Figure 3, Lane 1). cDNA generated from cells transfected with pRFPm contained one PspOMI restriction site, leading to the formation of a band of DNA at 623 bp (and a band at 128 bp, not shown) after incubation with PspOMI (Figure 3, Lane 4). As predicted, RNA from cells transfected with pRFPm-O6meG in the absence or presence of inhibitor resulted in a mixture of PspOMI-resistant and PspOMI-sensitive cDNA due to the incorporation of uridine or cytidine opposite O6-meG during transcription, respectively (Figure 3, Lanes 2 and 3).
Figure 3.
PspOMI digestion of RT–PCR products from cells reveals misincorporation opposite O6meG. RT–PCR products from cells transfected with the following plasmids were digested with PspOMI. 1: pRFPwt. 2: pRFPm-O6meG. 3: pRFPm-O6meG in the presence of 10 µM O6-benzylguanine. 4: pRFPm.
Band intensities from the PspOMI digest assay were used to estimate the relative base incorporation during transcription past O6-meG in HEK293 cells. In the absence of O6-benzylguanine, 3.0 ± 0.5% [standard error of the mean (SEM) n = 3] of the transcripts contained a misincorporation at the position opposite the O6-meG. However, RFP transcripts from cells in which AGT was inhibited with either 10 or 50 μM O6-benzylguanine prior to transfection resulted in 64 ± 9% (SEM, n = 2) and 65 ± 9% (SEM, n = 4) misincorporation, respectively.
RT–PCR was used to produce RFP cDNA for sequencing from cells. Twenty individual cDNAs were sequenced from four independent experiments each giving a total of 80 sequences for each experimental condition. The results from cells pretreated with 10 or 50 μM O6-benzylguanine (three experiments using 10 μM O6-benzylguanine and one experiment using 50 μM O6-benzylguanine) and transfected with pRFPm-O6meG confirmed that uridine and cytidine were principally incorporated opposite the lesion. The percentage of uridine misincorporation opposite O6-meG was 58 ± 7% (SEM, n = 4), which was in agreement with the results from the restriction digest assay. Additionally, one transcript from cells treated with 10 μM O6-benzylguanine contained a 10-base deletion, and a second contained a 1-base deletion located 2–4 bases from the lesion site coincident with misincorporation opposite the lesion (Supplementary Table SII). The deletions in the RNA are reminiscent of those observed by Marietta and Brooks during transcription past a cyclopyrimidine dimer on the transcribed strand (8). No aberrant sequences were observed in cells transfected with pRFPwt or pRFPm in the presence or absence of O6-benzylguanine, or with pRFPm-O6meG in the absence of inhibitor. These data indicate that O6-meG, if left unrepaired, is highly mutagenic at the level of transcription in human cells, inducing uridine incorporation in approximately two-thirds of the transcripts produced.
Transcriptional mutagenesis by O6-MeG induced changes in RFP activity
To determine if transcriptional mutagenesis by O6-meG results in changes in protein function following translation, recovery of RFP fluorescence was measured in cells transfected with the vector pRFPm-O6meG. Changes in fluorescence were assessed qualitatively with fluorescence microscopy and measured quantitatively with flow cytometry.
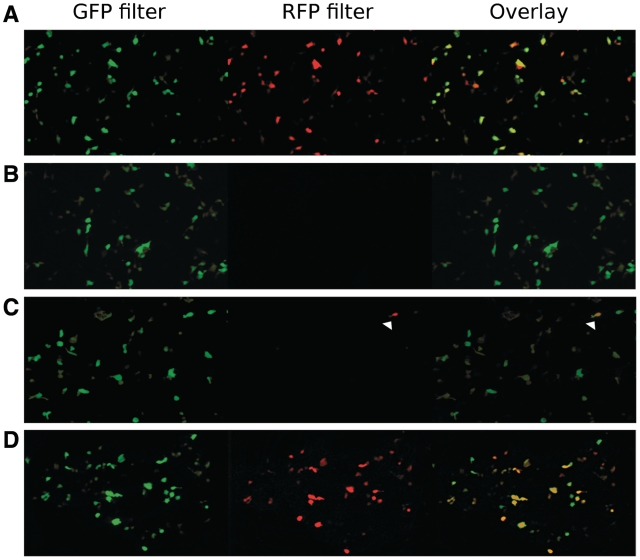
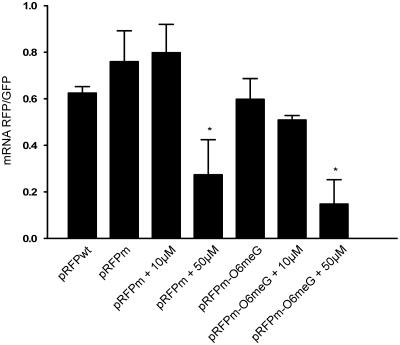
Cells transfected with the control vector pRFPwt exhibited both red and green fluorescence in the absence of dox (Figure 4A). Cells transfected with the control vector pRFPm exhibited only green fluorescence as expected, since the S21P mutation eliminated the RFP activity (Figure 4B); however, the presence of RFPm mRNA was confirmed by quantitative real-time RT–PCR (Figure 5). Interestingly, mRNA quantification revealed a significant effect of 50 μM O6-benzyguanine on the RFP/GFP mRNA ratio in cells transfected with pRFPm and pRFPm-O6meG. This was not observed in the presence of 10 μM O6-benzylguanine. An effect of 50 μM O6-benzylguanine alone on transcript levels has been observed before (40,41). It is possible that, while not cytotoxic, 50 μM O6-benzylguanine exerts an effect on the transcript levels of certain specific genes or on transcription in general.
Figure 4.
Fluorescence microscopy showing transcriptional mutagenesis. HEK293 cells were transfected with a specific vector as shown. In all cases, images were obtained 24 h after transfection. Row A: cells were transfected with plasmid pRFPwt and showed strong GFP and RFP fluorescence. Row B: cells were transfected with pRFPm, which expresses GFP and a non-fluorescent variant of RFP. No red fluorescence was observed. Row C: cells were transfected with pRFPm-O6meG. A small percentage of the cells exhibited red fluorescence, showing that a small quantity of mutant mRNA was formed following transcription past the adduct (white arrowhead). Row D: cells were transfected with pRFPm-O6meG in the presence of 10 µM O6-benzylguanine. These cells exhibited strong red fluorescence, indicating an elevated level of transcriptional mutagenesis due to the extended presence of O6-meG.
Figure 5.
qRT2-PCR indicates presence of RFPm mRNA in the absence of RFP fluorescence. Although there is no red fluorescence in cells transfected with pRFPm, RNA quantification shows there is no significant difference between RFP mRNA levels in cells transfected with pRFPwt, pRFPm, pRFPm-O6meG and pRFPm-O6meG plus 10 µM O6-benzylguanine. Additionally, treatment with 50 µM O6-benzylguanine significantly affected mRNA levels in cells transfected with pRFPm, and pRFPm-O6meG. Significance was determined by a two-sample Student’s t-test which compared the RFP–GFP mRNA abundance ratio from untreated pRwt transfected cells to the ratio observed under each of the experimental conditions in at least three independent experiments (*P < 0.05).
In contrast to cells transfected with undamaged pRFPm, cells pre-incubated with 10 μM O6-benzylguanine and transfected with pRFPm-O6meG exhibited robust green and red fluorescence, indicating that uridine incorporation opposite O6-meG during transcription reactivated RFP (Figure 4D). Furthermore, cells transfected with pRFPm-O6meG in the absence of inhibitor exhibited green fluorescence with ∼1% RFP-positive cells, showing that in some cells, the damaged base was sufficiently long lived, even in the presence of active AGT, to produce observable amounts of altered RFP mRNA (Figure 4C).
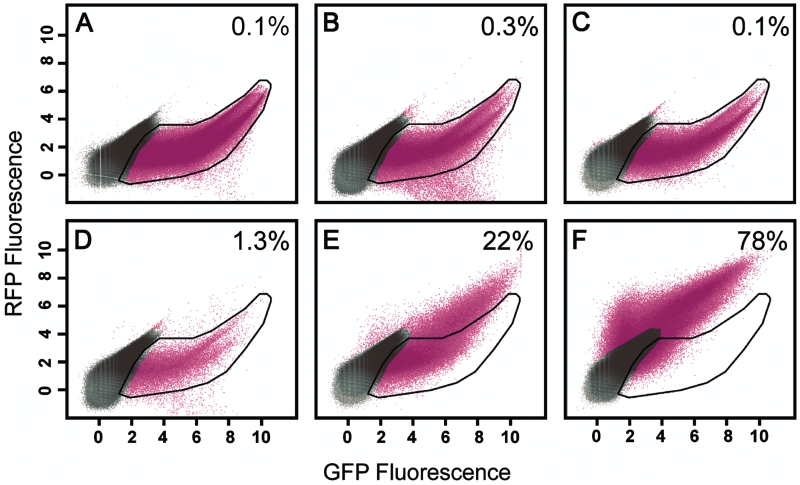
Flow cytometry showed a strong recovery of red fluorescence when O6-meG was positioned on the transcribed strand of DNA, especially in the presence of O6-benzylguanine. In RFP–GFP fluorescence scatterplots, recovery of red fluorescence was observed as a shift in the distribution of cells along the y-axis (RFP fluorescence) with no corresponding difference in the distribution along the x-axis (GFP fluorescence). Red and green fluorescence values from cells transfected with pRFPm fell within the space defined by cells transfected with pGFP, a single reporter plasmid expressing GFP (black polygon, Figure 6A), indicating that GFP fluorescence was dominant as expected. Note that the results for pRFPm RFP–GFP fluorescence were not influenced by the absence or presence of O6-benzylguanine (Figure 6B and C). Cells transfected with the plasmid pRFPm-O6meG in the presence of O6-benzylguanine exhibited a clear shift toward higher values of RFP fluorescence (Figure 6E, 22% of cells lie outside the black polygon), while cells transfected with the same plasmid in the absence of inhibitor fell primarily within the GFP region (Figure 6D). However, some scatter toward the RFP-positive region was observed (1.3% of cells lie outside the black polygon). Cells transfected with pRFPwt expressed high levels of both RFP and GFP (Figure 6F). Additional statistical analysis of the flow cytometry data can be found in Supplementary Figure S2. Overall, these data indicate that the presence of O6-meG in DNA can produce altered RNA that results in changes to protein function via transcriptional mutagenesis.
Figure 6.
Representative scatter plots of 293 cells show changes in protein function due to transcriptional mutatgenesis. (A) pGFP plasmid with CMV promoter driving expression of GFP only. (B) pRFPm. (C) pRFPm + 10 μM BzG. (D) pRFPm-O6meG. (E) pRFPm-O6meG + 10 μM BzG. (F) pRFPwt. The polygon represents extent of red and green signal in (A) (pGFP). The number in the upper right of each panel is the percentage of cells that fall outside of the GFP only polygon. Magenta color represents cells gated as significantly fluorescent compared with non-fluorescent controls. The data are plotted on a natural log scale.
DISCUSSION
The results presented here show that O6-meG located on the transcribed strand of an active gene is mutagenic at the level of transcription in human cells. These findings are in agreement with biochemical data demonstrating that this adduct induces base misincorporation events during transcription by human RNA polymerase II, resulting in significant insertion of uridine opposite the lesion (28). Furthermore, the production of altered mRNA following transcription past O6-meG is consistent with the recovery of RFP activity in HEK293 cells, showing that mutant transcripts do indeed change the primary amino acid sequence of proteins and that such change is reflected as altered protein function in cells.
DNA repair affects the magnitude of base changes to RNA that are induced during transcriptional mutagenesis (42–46). In the presence of functional AGT, ∼3% of the transcripts were mutated following HEK293 transfection with a plasmid containing O6-meG, and only an occasional RFP-positive cell was observed. In contrast, inhibition of AGT by the drug O6-benzylguanine resulted in virtually all cells exhibiting RFP fluorescence, with two-thirds of the transcripts containing a mutation. Hence, inhibiting repair of O6-meG led to increased transcriptional mutagenesis. Thus, the consequences of O6-meG formation in cellular DNA are not limited to S phase, when it can induce mutations during replication, but are also observed during transcription. Hence, AGT may well protect all cells from the harmful effects of alkylation at the O6-position of guanine, including those that are terminally differentiated and not replicating (10).
For human RNA polymerase II, the extent of uridine misincorporation during transcription past O6-meG in cells differs from results using nuclear extracts as the source of the transcription machinery. When HeLa nuclear extract was the source of RNA polymerase II, misincorporation of uridine opposite O6-meG occurred 25% of the time (28). This is in contrast to the 58% uridine misincorporation reported here for transcription past the lesion in cells when AGT was inhibited with O6-benzylguanine. In vitro transcription experiments with the local sequence used in this study revealed a misincorporation ratio similar to the results obtained in cells suggesting possible sequence context effects rather than differences between the two assays (Supplementary Figure S3). A comparison between O6-meG-induced transcriptional mutagenesis and replicative mutagenesis reveals some distinct similarities. Replicative DNA polymerases incorporate thymidine or cytidine opposite O6-meG, with reported T : C ratios ranging from 1 : 1 to 7 : 1, depending on the precise polymerase and DNA sequence studied (19–21). Other replication products have been observed but to a much lesser extent, including incorporation of adenine and small deletions (44). For DNA polymerases, these data are consistent with the GC → AT transition mutations that have been reported in a variety of assays used to examine replication past O6-meG in cells. The mutational events reported for DNA polymerases correspond to those observed for human RNA polymerase II, suggesting that the underlying mechanism governing base misincorporation opposite O6-meG during transcription and replication are similar (47).
DNA repair modulates the consequences of DNA damage during replication, often suppressing mutagenesis by removing the DNA lesions prior to S phase. Hence, in the absence of AGT, the O6-meG mutation frequency escalates during replication, reaching 75–90% in human 293 cells and 90–99% in Escherichia coli (42,48–50). This is in marked contrast to the much lower mutation frequencies of 5–25% observed when AGT is active. Interestingly, the O6-meG-induced mutation frequency in cells is virtually independent of DNA sequence, which is in contrast to biochemical data. For example, primer extension studies with E. coli DNA polymerase showed differences in mutation frequency depending on sequence context, but this result was not reproduced in cells (42).
Methylation damage to DNA poses a threat to cell viability. Exogenous and endogenous methylating agents are ubiquitous and contribute to the production of methylation damage in DNA, including the formation of O6-meG. It is estimated that S-adenosylmethionine alone may react with DNA in a typical mammalian cell to produce 10–30 O6-meG adducts per cell per day (51). Actual measurements of O6-meG in DNA from human tissues suggest that the lesion persists with steady-state values that, while widely variable, amount to 1.4–220 lesions per 109 nt, with lower values measured in blood and the higher levels found in lung tissue from smokers (11,52,53). While the O6-meG levels are low relative to the total number of bases found in DNA, there is the possibility that the lesion could be present within a transcription unit in a few cells per day, contributing to transcriptional mutagenesis. The majority of such mutations to RNA may well be transient and perhaps even benign due to RNA turnover; however, based on the evidence presented here and elsewhere, it is possible that transcriptional mutagenesis, rather than replication-induced mutations, may be among the principal inducers of many human diseases, ranging from cancer to prion activation, though there is no direct evidence that this is the case (45).
The drug O6-benzylguanine, which was used to inhibit AGT activity in this study, has been used as a co-chemotherapeutic agent in clinical trials to treat tumors of the central nervous system. By inhibiting AGT, O6-benzylguanine sensitizes cells to alkylating agents such as BCNU that form adducts at the O6-position of guanine. Inhibiting AGT prevents the repair of the O6-alkyl lesion, potentially making the alkyating agent more effective as an anti-tumorigenic compound. But the data presented here show that inhibiting the action of AGT induces transcriptional mutagenesis. In fact, the maximum concentration of O6-benzylguanine measured in patient blood has been reported as 8–11 μM, which is consistent with the concentration used in the growth medium in experiments reported here, suggesting that the drug could induce transcriptional mutations in patients (54).
The studies described here used an RFP reporter that permitted the analysis of transcriptional mutagenesis in individual cells, both at the level of the RNA and the subsequent effects on protein function. While RFP alterations do not affect cell physiology, the reporter-based system does allow visualization of transcriptional mutagenesis in single cells, providing insight into the distribution in mutation frequencies across the population of cells rather than just observing averages. This information will be important in considering what is happening in a population of cells where the damaged gene cannot be visualized directly. The work of Saxowsky et al. (46) has been able to show that mutations at the level of transcription can affect cellular processes. They examined the effect of 8-oxoguanine in inducing mutations in Ras protein at the level of transcriptional mutagenesis. They discovered that a population of constitutively active Ras proteins was produced via transcriptional mutagenesis, which led to downstream events in the Ras pathway. Furthermore, these same events were induced by mutations to the ras gene, showing that transcriptional mutations, while transient in nature, can significantly affect cells, possibly leading to oncogene activation (46). The work reported here concerning O6-meG indicates that this lesion can also induce transcriptional mutagenesis, and its effect could be serious since it exhibits a high mutation frequency in the absence of repair. An active gene containing O6-meG in a position sensitive to a T to C transition could transiently produce as much as 66% altered transcripts from that copy of the gene, resulting in a reduction of wt protein. This would be detrimental during development and could affect overall cell viability.
FUNDING
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Environmental Health Science or the National Institutes of Health. Funding for open access charge: National Institute of Health (ES010581 to D.A.S.).
Conflict of interest statement. None declared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Professor Alexandra Dimitri for her critical reading of the manuscript and helpful discussions.
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. Washington: ASM Press; 1995. [Google Scholar]
- 2.Kunkel TA, Bebenek K. DNA replication fidelity. Annu. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 3.Walker GC. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984;48:60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viswanathan A, You HJ, Doetsch PW. Phenotypic change caused by transcriptional bypass of uracil in nondividing cells. Science. 1999;284:159–162. doi: 10.1126/science.284.5411.159. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Zhou W, Doetsch PW. RNA polymerase bypass at sites of dihydrouracil: implications for transcriptional mutagenesis. Mol. Cell. Biol. 1995;15:6729–6735. doi: 10.1128/mcb.15.12.6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bregeon D, Peignon PA, Sarasin A. Transcriptional mutagenesis induced by 8-oxoguanine in mammalian cells. PLoS Genet. 2009;5:e1000577. doi: 10.1371/journal.pgen.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clauson CL, Oestreich KJ, Austin JW, Doetsch PW. Abasic sites and strand breaks in DNA cause transcriptional mutagenesis in Escherichia coli. Proc. Natl Acad. Sci. USA. 107:3657–3662. doi: 10.1073/pnas.0913191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marietta C, Brooks PJ. Transcriptional bypass of bulky DNA lesions causes new mutant RNA transcripts in human cells. EMBO Rep. 2007;8:388–393. doi: 10.1038/sj.embor.7400932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith JM, Koopman PA. The ins and outs of transcriptional control: nucleocytoplasmic shuttling in development and disease. Trends Genet. 2004;20:4–8. doi: 10.1016/j.tig.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Kisby GE, Olivas A, Park T, Churchwell M, Doerge D, Samson LD, Gerson SL, Turker MS. DNA repair modulates the vulnerability of the developing brain to alkylating agents. DNA Repair. 2009;8:400–412. doi: 10.1016/j.dnarep.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 12.Wurdeman RL, Douskey MC, Gold B. DNA methylation by N-methyl-N-nitrosourea: methylation pattern changes in single- and double-stranded DNA, and in DNA with mismatched or bulged guanines. Nucleic Acids Res. 1993;21:4975–4980. doi: 10.1093/nar/21.21.4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irving CC, Cox R, Murphy WM. Influence of dose of N-methyl-N-nitrosourea on the induction of urinary bladder cancer in rats. Cancer Lett. 1979;8:3–7. doi: 10.1016/0304-3835(79)90016-8. [DOI] [PubMed] [Google Scholar]
- 14.Cox R, Irving CC. O6-methylguanine accumulates in DNA of mammary glands after administration of N-methyl-N-nitrosourea to rats. Cancer Lett. 1979;6:273–278. doi: 10.1016/s0304-3835(79)80045-2. [DOI] [PubMed] [Google Scholar]
- 15.Dolan ME, Pegg AE, Moschel RC, Grindey GB. Effect of O6-benzylguanine on the sensitivity of human colon tumor xenografts to 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU) Biochem. Pharmacol. 1993;46:285–290. doi: 10.1016/0006-2952(93)90416-t. [DOI] [PubMed] [Google Scholar]
- 16.Tong WP, Kirk MC, Ludlum DB. Formation of the cross-link 1-[N3-deoxycytidyl),2-[N1-deoxyguanosinyl]ethane in DNA treated with N, N'-bis(2-chloroethyl)-N-nitrosourea. Cancer Res. 1982;42:3102–3105. [PubMed] [Google Scholar]
- 17.Kohn KW. Interstrand cross-linking of DNA by 1,3-bis(2-chloroethyl)-1-nitrosourea and other 1-(2-haloethyl)-1-nitrosoureas. Cancer Res. 1977;37:1450–1454. [PubMed] [Google Scholar]
- 18.Tisdale MJ. Antitumor imidazotetrazines–XV. Role of guanine O6 alkylation in the mechanism of cytotoxicity of imidazotetrazinones. Biochem. Pharmacol. 1987;36:457–462. doi: 10.1016/0006-2952(87)90351-0. [DOI] [PubMed] [Google Scholar]
- 19.Singer B, Chavez F, Goodman MF, Essigmann JM, Dosanjh MK. Effect of 3′ flanking neighbors on kinetics of pairing of dCTP or dTTP opposite O6-methylguanine in a defined primed oligonucleotide when Escherichia coli DNA polymerase I is used. Proc. Natl Acad. Sci. USA. 1989;86:8271–8274. doi: 10.1073/pnas.86.21.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dosanjh MK, Galeros G, Goodman MF, Singer B. Kinetics of extension of O6-methylguanine paired with cytosine or thymine in defined oligonucleotide sequences. Biochemistry. 1991;30:11595–11599. doi: 10.1021/bi00113a015. [DOI] [PubMed] [Google Scholar]
- 21.Shibutani S. Quantitation of base substitutions and deletions induced by chemical mutagens during DNA synthesis in vitro. Chem. Res. Toxicol. 1993;6:625–629. doi: 10.1021/tx00035a006. [DOI] [PubMed] [Google Scholar]
- 22.Mojas N, Lopes M, Jiricny J. Mismatch repair-dependent processing of methylation damage gives rise to persistent single-stranded gaps in newly replicated DNA. Genes Dev. 2007;21:3342–3355. doi: 10.1101/gad.455407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pegg AE, Dolan ME, Scicchitano D, Morimoto K. Studies of the repair of O6-alkylguanine and O4-alkylthymine in DNA by alkyltransferases from mammalian cells and bacteria. Environ. Health Perspect. 1985;62:109–114. doi: 10.1289/ehp.8562109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu J, Ma A, Dinner AR. A two-step nucleotide-flipping mechanism enables kinetic discrimination of DNA lesions by AGT. Proc. Natl Acad. Sci. USA. 2008;105:4615–4620. doi: 10.1073/pnas.0708058105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wibley JE, Pegg AE, Moody PC. Crystal structure of the human O(6)-alkylguanine-DNA alkyltransferase. Nucleic Acids Res. 2000;28:393–401. doi: 10.1093/nar/28.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dolan ME, Norbeck L, Clyde C, Hora NK, Erickson LC, Pegg AE. Expression of mammalian O6-alkylguanine-DNA alkyltransferase in a cell line sensitive to alkylating agents. Carcinogenesis. 1989;10:1613–1619. doi: 10.1093/carcin/10.9.1613. [DOI] [PubMed] [Google Scholar]
- 27.Gerson SL. Clinical relevance of MGMT in the treatment of cancer. J. Clin. Oncol. 2002;20:2388–2399. doi: 10.1200/JCO.2002.06.110. [DOI] [PubMed] [Google Scholar]
- 28.Dimitri A, Burns JA, Broyde S, Scicchitano DA. Transcription elongation past O6-methylguanine by human RNA polymerase II and bacteriophage T7 RNA polymerase. Nucleic Acids Res. 2008;36:6459–6471. doi: 10.1093/nar/gkn657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viswanathan A, Doetsch PW. Effects of nonbulky DNA base damages on Escherichia coli RNA polymerase-mediated elongation and promoter clearance. J. Biol. Chem. 1998;273:21276–21281. doi: 10.1074/jbc.273.33.21276. [DOI] [PubMed] [Google Scholar]
- 30.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 31.Jensch F, Kemper B. Endonuclease VII resolves Y-junctions in branched DNA in vitro. EMBO J. 1986;5:181–189. doi: 10.1002/j.1460-2075.1986.tb04194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez JA, Marek D, Wangh LJ. The efficiency and timing of plasmid DNA replication in Xenopus eggs: correlations to the extent of prior chromatin assembly. J. Cell Sci. 1992;103(Pt 4):907–918. doi: 10.1242/jcs.103.4.907. [DOI] [PubMed] [Google Scholar]
- 33.Taylor ER, Morgan IM. A novel technique with enhanced detection and quantitation of HPV-16 E1- and E2-mediated DNA replication. Virology. 2003;315:103–109. doi: 10.1016/s0042-6822(03)00588-9. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng LZ, Workman JL, Kingston RE, Kelly TJ. Regulation of DNA replication in vitro by the transcriptional activation domain of GAL4-VP16. Proc. Natl Acad. Sci. USA. 1992;89:589–593. doi: 10.1073/pnas.89.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herbig U, Weisshart K, Taneja P, Fanning E. Interaction of the transcription factor TFIID with simian virus 40 (SV40) large T antigen interferes with replication of SV40 DNA in vitro. J. Virol. 1999;73:1099–1107. doi: 10.1128/jvi.73.2.1099-1107.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dolan ME, Pegg AE, Dumenco LL, Moschel RC, Gerson SL. Comparison of the inactivation of mammalian and bacterial O6-alkylguanine-DNA alkyltransferases by O6-benzylguanine and O6-methylguanine. Carcinogenesis. 1991;12:2305–2309. doi: 10.1093/carcin/12.12.2305. [DOI] [PubMed] [Google Scholar]
- 39.Chen JM, Zhang YP, Moschel RC, Ikenaga M. Depletion of O6-methylguanine-DNA methyltransferase and potentiation of 1,3-bis(2-chloroethyl)-1-nitrosourea antitumor activity by O6-benzylguanine in vitro. Carcinogenesis. 1993;14:1057–1060. doi: 10.1093/carcin/14.5.1057. [DOI] [PubMed] [Google Scholar]
- 40.Fishel ML, Rabik CA, Bleibel WK, Li X, Moschel RC, Dolan ME. Role of GADD34 in modulation of cisplatin cytotoxicity. Biochem. Pharmacol. 2006;71:239–247. doi: 10.1016/j.bcp.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 41.Rabik CA, Fishel ML, Holleran JL, Kasza K, Kelley MR, Egorin MJ, Dolan ME. Enhancement of cisplatin [cis-diammine dichloroplatinum (II)] cytotoxicity by O6-benzylguanine involves endoplasmic reticulum stress. J. Pharmacol. Exp. Ther. 2008;327:442–452. doi: 10.1124/jpet.108.141291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delaney JC, Essigmann JM. Effect of sequence context on O(6)-methylguanine repair and replication in vivo. Biochemistry. 2001;40:14968–14975. doi: 10.1021/bi015578f. [DOI] [PubMed] [Google Scholar]
- 43.Halford S, Rowan A, Sawyer E, Talbot I, Tomlinson I. O(6)-methylguanine methyltransferase in colorectal cancers: detection of mutations, loss of expression, and weak association with G:C>A:T transitions. Gut. 2005;54:797–802. doi: 10.1136/gut.2004.059535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pletsa V, Gentil A, Margot A, Armier J, Kyrtopoulos SA, Sarasin A. Mutagenesis by O6 meG residues within codon 12 of the human Ha-ras proto-oncogene in monkey cells. Nucleic Acids Res. 1992;20:4897–4901. doi: 10.1093/nar/20.18.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doetsch PW. Translesion synthesis by RNA polymerases: occurrence and biological implications for transcriptional mutagenesis. Mutat. Res. 2002;510:131–140. doi: 10.1016/s0027-5107(02)00258-0. [DOI] [PubMed] [Google Scholar]
- 46.Saxowsky TT, Meadows KL, Klungland A, Doetsch PW. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc. Natl Acad. Sci. USA. 2008;105:18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spratt TE. Klenow fragment-DNA interaction required for the incorporation of nucleotides opposite guanine and O6-methylguanine. Biochemistry. 1997;36:13292–13297. doi: 10.1021/bi971253g. [DOI] [PubMed] [Google Scholar]
- 48.Pauly GT, Moschel RC. Mutagenesis by O(6)-methyl-, O(6)-ethyl-, and O(6)-benzylguanine and O(4)-methylthymine in human cells: effects of O(6)-alkylguanine-DNA alkyltransferase and mismatch repair. Chem. Res. Toxicol. 2001;14:894–900. doi: 10.1021/tx010032f. [DOI] [PubMed] [Google Scholar]
- 49.Pauly GT, Hughes SH, Moschel RC. Mutagenesis in Escherichia coli by three O6-substituted guanines in double-stranded or gapped plasmids. Biochemistry. 1995;34:8924–8930. doi: 10.1021/bi00027a045. [DOI] [PubMed] [Google Scholar]
- 50.Pauly GT, Hughes SH, Moschel RC. Comparison of mutagenesis by O6-methyl- and O6-ethylguanine and O4-methylthymine in Escherichia coli using double-stranded and gapped plasmids. Carcinogenesis. 1998;19:457–461. doi: 10.1093/carcin/19.3.457. [DOI] [PubMed] [Google Scholar]
- 51.Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang H, Konishi C, Kuroki T, Huh N. Detection of O6-methylguanine, O4-methylthymine and O4-ethylthymine in human liver and peripheral blood leukocyte DNA. Carcinogenesis. 1995;16:1277–1280. doi: 10.1093/carcin/16.6.1277. [DOI] [PubMed] [Google Scholar]
- 53.Shields PG, Povey AC, Wilson VL, Weston A, Harris CC. Combined high-performance liquid chromatography/32P-postlabeling assay of N7-methyldeoxyguanosine. Cancer Res. 1990;50:6580–6584. [PubMed] [Google Scholar]
- 54.Adams DM, Zhou T, Berg SL, Bernstein M, Neville K, Blaney SM. Phase 1 trial of O6-benzylguanine and BCNU in children with CNS tumors: a Children's Oncology Group study. Pediatr. Blood Cancer. 2008;50:549–553. doi: 10.1002/pbc.21362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.