Abstract
Most organisms, from Escherichia coli to humans, use the ‘universal’ genetic code, which have been unchanged or ‘frozen’ for billions of years. It has been argued that codon reassignment causes mistranslation of genetic information, and must be lethal. In this study, we successfully reassigned the UAG triplet from a stop to a sense codon in the E. coli genome, by eliminating the UAG-recognizing release factor, an essential cellular component, from the bacterium. Only a few genetic modifications of E. coli were needed to circumvent the lethality of codon reassignment; erasing all UAG triplets from the genome was unnecessary. Thus, UAG was assigned unambiguously to a natural or non-natural amino acid, according to the specificity of the UAG-decoding tRNA. The result reveals the unexpected flexibility of the genetic code.
INTRODUCTION
The genetic code defines the relationship between the genetic information in DNA and the amino acid sequences of proteins, with the assignment of the 64 codons to amino acids or the stop signal. These assignments have remained unchanged or ‘frozen’, with only rare exceptions (1–3), since they were presumably established in the common ancestor of all organisms. It has been argued that codon reassignment would alter the amino acid sequences of most proteins simultaneously, and thus have a destructive impact on the organism (4). This lethal effect might be avoided, if the usage of the codon to be redefined is minimized throughout the genome, prior to the codon redefinition (5), or is gradually adapted to the new assignment (6). However, each of these changes involves the accumulation of a number of mutations in the genome, which is considered to be the major constraint ‘freezing’ the genetic code. In the present study, we aimed to define the conditions required for the reassignment of a codon, and found that only a few mutations are needed to prepare Escherichia coli for the reassignment of the amber UAG triplet from a stop signal to a sense codon.
In the E. coli genome, the amber UAG triplet occurs at the ends of about 300 open reading frames (ORFs) (Profiling of E. coli Chromosome, http://www.shigen.nig.ac.jp/ecoli/pecplus/index.jsp). Release factor 1 (RF-1), encoded by the prfA gene, is the only molecule that recognizes UAG and terminates protein synthesis (7). Amber suppressor tRNAs naturally occur in E. coli and translate UAG to amino acids (8,9). As a consequence of amber suppression, UAG is recognized ambiguously in two manners, as a sense codon and a stop signal, in the presence of competing RF-1 (Figure 1). On the other hand, the complete reassignment of UAG requires the elimination of RF-1 from the cell, but the knock-out of the prfA gene, encoding RF-1, from the E. coli genome is reportedly lethal (7).
Figure 1.
Amber suppression and the complete reassignment of the UAG triplet from a stop to a sense codon by eliminating RF-1.
MATERIALS AND METHODS
Strains, plasmids, non-natural amino acid and mass spectrometry
The E. coli K12 strains, TOP10 and HST08, were purchased from Invitrogen and Takara Bio (Japan), respectively. The hemA and hemK genes, but not the intervening prfA gene, were cloned in the vector pAp102 (10), together with the transcriptional promoter upstream of hemA. The prfA gene, with an upstream lacZ promoter, was cloned in pACYC184, to create pLacPrfA. The amber suppressor tRNAGln and tRNATyr genes, each under the control of the E. coli tyrT promoter, were cloned in the vector pACYC184, to create pKS3supE and pKS3supF, respectively. The plasmids pKSsupF-kan and piodoTyrRS-MJR1-kan are derivatives of the plasmid pKS3supF and TyrRS-MJR1 (11), respectively. The chloramphenicol acetyltransferase (cat) gene within pACYC184-kan, a kanamycin-resistant pACYC184 derivative, was engineered to contain 3 and 10 UAG triplets, to create cat(3Am) and cat(10Am), respectively. The gst(Am25) gene, described previously (11), was engineered to have six UAG triplets near the N-terminus, and was then placed under the control of the tac promoter, to create the plasmid pTacGST(6Am). 3-Iodo-l-tyrosine was purchased from Sigma-Aldrich, and was added to the growth media at a concentration of 0.1 g/l. Mass spectrometric analyses were commercially performed by Shimadzu Biotech (Japan).
Construction of BAC7
The miniF replicon with the kanamycin resistance (kan) gene was amplified from the AcNPV bacmid (Invitrogen) by PCR with the F1 and F2 primers, as described previously (12), using PrimeStar GXL DNA polymerase (Takara Bio), to generate the vector BAC-kan. DNA fragments carrying seven essential genes (coaD, hda, hemA, mreC, murF, lolA and lpxK) were obtained by PCR with BL21(DE3) genomic DNA as the template, prepared using Dr GenTLE (Takara Bio) yeast genome extraction kit. The TAG stop triplets of these seven genes were mutagenized to TAA, by PCR amplification with mutagenic primers. The waaA-coaD operon, followed by the ftsI-murE-murF genes from the mra cluster, was cloned in the BAC-kan vector, to create the plasmid BAC-murF-coaD. Actually, the ftsI-murE-murF genes do not comprise an entire operon, since they lack a transcriptional promoter. The essential murF gene is probably expressed from the promoter of the upstream waaA-coaD operon. The kan gene in the BAC-murF-coaD plasmid was then replaced by the hda and cat genes, using the RT/ET kit, to create the BAC-murF-coaD-had plasmid. Meanwhile, the mreBCD and ycaI-msbA-lpxK operons were cloned together in the vector BAC-kan, to create the BAC-mreC-lpxK plasmid. The DNA fragment carrying the mreC-lpxK operons, prepared from this plasmid, was substituted for the cat gene in the BAC-murF-coaD-hda plasmid, to create BAC-murF-coaD-hda-mreC-lpxK. The prfA gene was removed from the hemA-prfA-hemK operon, to create the hemAK pair of genes. The downstream rarA was removed from the lolA-rarA operon, to leave a lolA gene with the native promoter. The hemAK and lolA genes were connected with the gentamicin resistance (gent) gene from the vector pFastBac1 (Invitrogen), under the control of the kan promoter, to create a DNA fragment consisting of the hemA-lolA-gent genes. The kan gene in the plasmid BAC-murF-coaD-hda-mreC-lpxK was replaced by this DNA fragment, to create the BAC7gent plasmid. The gent gene was replaced by the Sh ble Zeocin resistance (zeo) gene from the vector pcDNA3.1/Zeo(+) (Invitrogen), to create BAC7. BAC0 was an ‘empty’ BAC vector containing only the zeo gene. The essential genes in BAC7 were separately knocked out by the cat gene, to create the BAC6 plasmids.
Chromosomal engineering
Chromosomal engineering was performed using an RT/ET kit (Gene Bridges). To create a prfA conditional mutant, the DNA encoding the ribosome-binding site, the araBAD promoter and the araC gene (13) was introduced, together with the kan gene, into the chromosome of strain TOP10, upstream of the hemA-prfA-hemK operon. To knock-out prfA, the 50-bp N-terminal fragment and 225-bp C-terminal fragment of the prfA-coding sequence were linked to the 5′ and 3′ sides, respectively, of the zeo gene, and then introduced into the chromosome. The supE44 (or glnX44) gene of HST08 was disrupted by the cat gene, linked to the left arm sequence (CGTACCCCAGCCAATTTATTCAAGACGCTTACCTTGTAAGTGCACCCAGT) and the right arm sequence (ATTAAAAAAGCTCGCTTCGGCGAGCTTTTTGCTTTTCTGCGTTCATTCA) at the 5′- and 3′-ends, respectively.
RESULTS
Elimination of the prfA gene from the E. coli genome
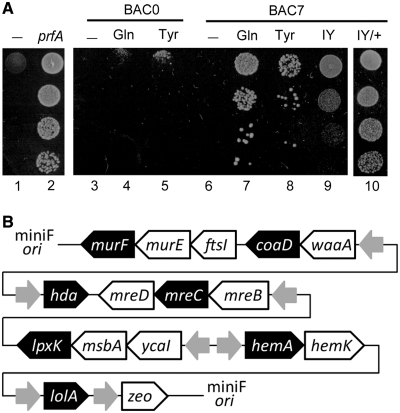
To determine the requirements for avoiding the lethal effect of the prfA disruption, we created a conditional prfA mutant, which repressed the expression of the essential prfA gene under a restrictive condition. The arabinose promoter, together with the araC gene encoding the regulator, was introduced into the E. coli chromosome upstream of the hemA-prfA-hemK operon. Thus, the expression of this operon was induced in the presence of l-arabinose and repressed by the addition of d-glucose to the growth media. To make only the prfA expression conditional, the constitutively expressed hemA and hemK genes were cloned on a plasmid, and were then introduced into the cell. The resulting cell was a conditional prfA mutant that did not grow in the presence of d-glucose, and was complemented by the introduction of another plasmid constitutively expressing the prfA gene (Figure 2A, Lanes 1 and 2).
Figure 2.
Complementation experiments with the conditional prfA mutant. (A) Cell cultures, diluted from top to bottom, were spotted on growth media containing 0.4% glucose (w/v). The pACYC184 vector was introduced into the mutant (Lanes 1, 3 and 6). The prfA gene (Lane 2), the suppressor tRNAGln (Lanes 4 and 7) and tRNATyr (Lanes 5 and 8) were introduced, alone (Lane 2) or together with the BAC0 ‘empty’ plasmid (Lanes 4 and 5) or BAC7 (Lanes 7 and 8). BAC0 carried only the zeo gene. The iodotyrosine-incorporating plasmid was introduced, together with BAC7 (Lanes 9 and 10), in the absence (Lane 9) or presence (Lane 10) of 3-iodo-l-tyrosine in the medium. (B) Gene arrangement in BAC7. The gray arrows show the direction of the promoters. The seven essential genes are shown by white letters on the black background.
A recent report described the individual knock-outs of most of the E. coli genes, and revealed which genes cannot be disrupted, because of lethality (PEC, http://www.shigen.nig.ac.jp/ecoli/pecplus/index.jsp). Among these essential genes, we found that only seven essential ORFs (coaD, hda, hemA, mreC, murF, lolA and lpxK) end with UAG. We then engineered these seven ORFs to end with UAA, another stop triplet, to allow their expression even in the absence of RF-1. These seven modified ORFs were cloned, in a bacterial artificial chromosome (BAC), to create the BAC7 plasmid (Figure 2B). For the genes that are part of a transcriptional unit, entire or large part of the operon was cloned in the plasmid. However, BAC7 did not support cell growth under the restrictive condition (Figure 2A, Lane 6). This result suggested that the functions of additional genes, other than the seven essential genes, were also necessary for cell growth. These additionally required ORFs ending with UAG were probably prevented from being expressed, due to the absence of RF-1.
Instead of engineering more ORFs to end with UAA, we introduced the E. coli amber suppressor tRNAGln or tRNATyr into the prfA mutant. In the absence of RF-1, UAG was to be recognized by these tRNAs, and the ORFs ending with UAG were to be extended by an in-frame UAA or UAG in the 3′ region, to produce only aberrant proteins with unnecessary peptide tails. The introduction of the suppressor tRNAs, together with BAC7, successfully complemented the prfA mutant, whereas the tRNA genes alone showed no complementing activity (Figure 2A, Lanes 4–8). This result implied that many of the aberrant proteins with peptide tails retained their activities and were able to support cell growth, when these activities were expressed along with those of the seven essential genes.
We then examined if the E. coli suppressor tRNAs could be replaced with the archaeal pair of a UAG-decoding tRNA and an engineered tyrosyl-tRNA synthetase, for the complementation. This enzyme, designated as iodoTyrRS-mj, specifically charges the tRNA with a non-natural amino acid, 3-iodo-l-tyrosine, and incorporates 3-iodo-l-tyrosine into proteins in response to UAG in E. coli cells, when this amino acid is supplemented in the growth media (11). This archaeal pair exhibited complementing activity, depending on the presence of 3-iodo-l-tyrosine in the growth medium (Figure 2A, Lanes 9 and 10). These observations suggested that the expression of a tRNA molecule able to translate UAG, in combination with the engineering of only 2% of the ORFs ending with UAG, would suffice to avoid the lethal effect of RF-1 elimination.
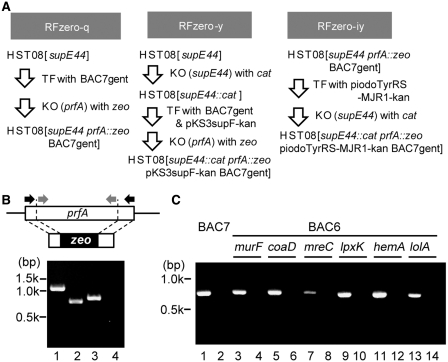
We applied these parameters to achieve the knock-out of prfA and redefinition of UAG in E. coli cells. The E. coli K-12 HST08 strain has the endogenous supE44 gene encoding the amber suppressor tRNAGln, and was transformed with BAC7gent, a gentamicin-resistant derivative of BAC7 (Figure 3A). The prfA gene (1080 bp) was then knocked out from the chromosome by replacing a major part (51–858 bp) with the zeocin resistance (zeo) gene. The replacement in the resulting strain, HST08[supE44 prfA::zeo BAC7gent] or RFzero-q, was confirmed by sequencing the prfA locus, while the absence of the sequence in the genome was confirmed by PCR (Figure 3B). The growth rate of RFzero-q was determined as 0.9 h−1, by monitoring the change in the natural logarithm of the optical density at 600 nm of the culture per hour; this rate differed little from the 1.3 h−1 rate of the parent HST08(BAC7gent).
Figure 3.
Construction of the RFzero strains. (A) Genetic modifications to create the RFzero strains. TF and KO mean transformation and knock-out, respectively. (B) PCR amplification was performed, using primers (black arrows) to amplify the entire prfA locus (Lanes 1 and 2) and primers (gray arrows) to amplify the prfA sequence inside the knocked out region (Lanes 3 and 4), with the genomic DNAs of HST08 (Lanes 1 and 3) and RFzero-q (Lanes 2 and 4) as the template. (C) PCR amplification of the prfA sequence inside the knocked out region, with the genomic DNAs of the HST08 strain (Lanes 1, 3, 5, 7, 9, 11 and 13) and the RFzero-q strain (Lanes 2, 4, 6, 8, 10, 12 and 14), each with BAC7 (Lanes 1 and 2), and BAC6 plasmids (Lanes 3–14) lacking the genes indicated above the lanes.
Finally, we determined which of the seven essential ORFs needed to be engineered to end with UAA, for the successful knock-out of the prfA gene. Each of the seven engineered ORFs was removed from BAC7, to create seven BAC6 plasmids. Six plasmids, except for the plasmid lacking the hda gene, allowed us to eliminate the prfA gene from the HST08 chromosome (Figure 3C), indicating that at least hda must be engineered. We then found that engineering only hda was not sufficient for a successful prfA knock-out. The viability of the cell was impaired severely, when a few or more of the six essential ORFs were left ending with UAG. Therefore, BAC7, with all seven of the essential ORFs engineered, was used for further experiments.
Reassignment of the UAG triplet as a sense codon
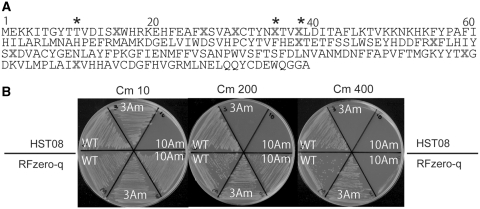
To prove the complete reassignment of UAG from a stop signal to glutamine, RFzero-q was transformed with a mutant chloramphenicol acetyltransferase (cat) gene, cat(3Am), containing three UAG triplets within the ORF (Figure 4A). The transformants with the wild-type cat and cat(3Am) genes showed similar levels of chloramphenicol (Cm) resistance, up to a concentration of 400 µg/ml of Cm in the growth media (Figure 4B). Furthermore, another cat mutant gene, with UAG in place of 10 glutamine codons, cat(10Am), conferred only slightly lower resistance (up to Cm 200 µg/ml) to RFzero-q than the wild-type cat gene (Figure 4B). This finding showed that 10 UAG triplets were translated with efficiency comparable with that of the glutamine codons. The difference in the resistance level is probably caused by the fact that the UAG-reading tRNA, a mutant tRNAGln, recognizes the UAG triplet with a lower efficiency than the wild-type tRNAGln reads the CAA and CAG glutamine codons. The parent HST08, also expressing the suppressor tRNAGln, was transformed with cat(10Am), and the transformant was not resistant to Cm at ≥10 µg/ml concentrations (Figure 4B). The absence of RF-1 in RFzero-q was also shown by the drastic increase in the efficiency of translating UAG as glutamine in RFzero-q, as compared with that of the nonsense suppression in competition with RF-1 in the parent HST08.
Figure 4.
Cm resistance of RFzero-q with the wild-type cat gene and the cat(3Am) and cat(10Am) mutant genes. (A) The amino acid sequence of the cat(10Am) gene is shown with X’s representing the positions of UAG. The asterisks show the UAG positions in cat(3Am). (B) Each third of the upper and lower halves of each plate shows the growth of the HST08 and RFzero-q strains, respectively, transformed with the indicated cat genes. Cm concentrations are indicated above the plates.
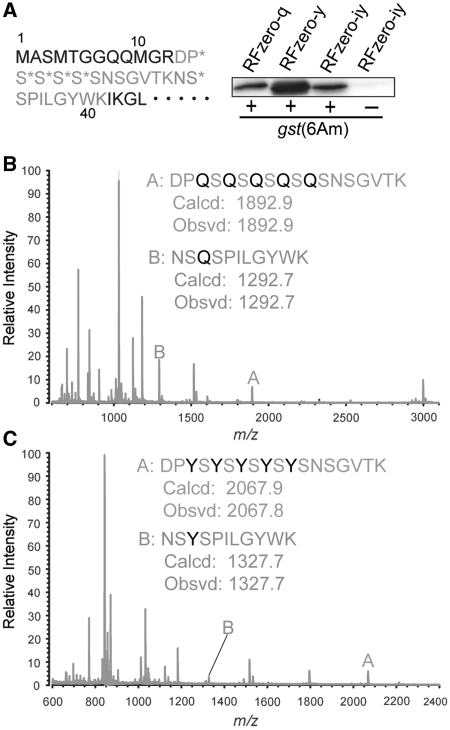
To confirm the identity of the amino acid incorporated at UAG in the RFzero-q strain, six UAG triplets were introduced near the N-terminus of the glutathione S-transferase (GST) gene (Figure 5A). This mutant gene, gst(6Am), produced the full-length GST in RFzero-q (Figure 5A), and the product was then subjected to a mass spectrometric analysis. After trypsin digestion, the gst(6Am) product from RFzero-q generated a peptide with an average mass corresponding to the theoretical value (m/z = 1892.9) for residues 13–30 including five glutamines, and a peptide with an average mass corresponding to the theoretical value (m/z = 1292.7) for residues 31–41 including one glutamine (Figure 5B). This result indicated that UAG was translated as glutamine in the RFzero-q strain.
Figure 5.
Expression of the gst(6Am) gene in the RFzero strains. (A) The N-terminal 45 residues of gst(6Am) are shown, where the asterisks indicate the positions of UAG triplets. The gst(6Am) products from RFzero-q, -y and -iy cells each with gst(6Am), and RFzero-iy cells without gst(6Am) (30-μl culture each) were subjected to SDS–PAGE and western blotting using an anti-GST antibody. Mass spectrometric analyses of the trypsin digests of the gst(6Am) products from RFzero-q (B) and RFzero-y (C). Theoretical (Calcd) and observed (Obsvd) values of the average masses for peptides A and B are shown.
Next, the UAG triplet was reassigned to a tyrosine codon. The supE44 gene in the HST08 genome was disrupted, prior to the knock-out of prfA (Figure 3A). The resulting strain was transformed with BAC7gent and a plasmid expressing the amber suppressor tRNATyr (pKS3supF-kan). The prfA gene was successfully disrupted in the transformant, to create HST08[supE44::cat prfA::zeo BAC7gent pKS3supF-kan] or RFzero-y. To confirm the identity of the amino acid incorporated at UAG, the gst(6Am) mutant was introduced into RFzero-y. The full-length GST was produced (Figure 5A), and was then subjected to a mass spectrometric analysis. The gst(6Am) product from RFzero-y generated a peptide with an average mass corresponding to the theoretical value (m/z = 2067.9) for residues 13–30 including five tyrosines, and a peptide with an average mass corresponding to the theoretical value (m/z = 1327.7) for residues 31–41 including one tyrosine (Figure 5C). This result indicated that UAG was translated as tyrosine in RFzero-y. Together with the result obtained with RFzero-q, our findings show that UAG can be reassigned to a natural amino acid, according to the specificity of the UAG-decoding tRNA.
Reassignment of the UAG triplet to a non-natural amino acid
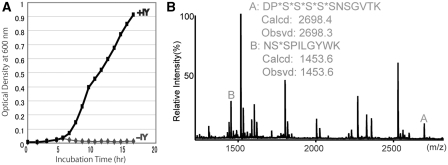
Finally, we examined the feasibility of genetically encoding a non-natural amino acid by codon reassignment. The RFzero-q strain was transformed with a plasmid (piodoTyrRS-MJR1-kan) expressing the iodotyrosine-incorporating tRNA–iodoTyrRS-mj pair (Figure 3A). In this transformed strain and in the presence of 3-iodo-l-tyrosine, the supE44 gene was successfully disrupted in the genome, to create HST08[supE44::cat prfA::zeo BAC7gent piodoTyrRS-MJR1-kan] or RFzero-iy. A PCR analysis of the genomic DNA of this strain confirmed that the prfA gene was absent, and that the supE44 gene was replaced by the marker cat gene (data not shown). The growth of RFzero-iy depended on the presence of 3-iodo-l-tyrosine in the growth media (Figure 6A), which was consistent with the observation that the iodotyrosine-incorporating tRNA–iodoTyrRS-mj pair complemented the prfA conditional mutant, depending on the presence of 3-iodo-l-tyrosine.
Figure 6.
UAG reassignment as a sense codon specifying a non-natural amino acid. (A) Growth of RFzero-iy cells in the presence (+IY) and absence (−IY) of 3-iodo-l-tyrosine (IY) supplemented in the growth media. (B) Mass spectrometric analysis of the trypsin digest of the gst(6Am) product from RFzero-iy. Theoretical (Calcd) and observed (Obsvd) values of the average masses are shown for peptides A and B.
To identify the amino acid incorporated at UAG, the gst(6Am) gene was introduced into RFzero-iy. The full-length GST product was produced from this mutant gene in the RFzero-iy strain (Figure 5A). In a mass spectrometric analysis (Figure 6B), the product generated a peptide with an average mass corresponding to the theoretical value (m/z = 2698.4) for residues 13–30 including five iodotyrosines (DP*S*S*S*S*SNSGVTK, where the asterisk indicates the position of UAG triplets), and a peptide with an average mass corresponding to the theoretical value (m/z = 1453.6) for residues 31–41 including one iodotyrosine (NS*SPILGYWK). This result showed that all of the six UAG triplets were translated as iodotyrosine. Thus, UAG can be completely reassigned to a novel repertoire of amino acids in the bacterial genome.
DISCUSSION
The ‘universal’ genetic code assigns three triplets (UAG, UAA and UGA) as stop signals. Each of the eukaryal and archaeal RFs recognizes all three of these triplets (14,15), while there are two types of RFs in bacterial cells: RF-1 for UAG and UAA, and RF-2 for UAA and UGA (16). The disruption of the gene encoding a RF is lethal (7), probably because the absence of the factor causes ribosome stalling at the stop triplets recognized by the factor. Although this harmful effect is reportedly suppressed, to a certain extent, by mRNA cleavage and tmRNA-mediated tagging (17), this mechanism leads to the degradation of the tagged product, and thus prevents the expression of ORFs ending with the stop triplet. When the RF-1-encoding prfA gene was disrupted in E. coli, both UAA and UGA are available as stop signals, and the UAG-ending ORFs were thus extended by an in-frame UAA or UGA triplet in the 3′ region in the presence of UAG-decoding tRNA. Although amber suppression also causes this extended translation, the UAG-ending ORFs are normally translated as well, due to the presence of RF-1. On the other hand, by the present codon reassignment, involving the knock-out of prfA, the UAG-ending ORFs only produced aberrant proteins with unnecessary peptide tails, in the presence of UAG-decoding tRNA.
In addition to the engineering of the seven essential ORFs, the expression of a UAG-decoding tRNA was required to avoid the lethality of RF-1 elimination. This tRNA expression not only solves the potential problem of ribosome stalling at UAG codons, but also allows the non-essential ORFs ending with UAG to be expressed, although their products have C-terminal peptide tails. A non-essential gene is defined as a gene that can be disrupted separately, and the disruption of a few or more non-essential genes at a time may impair the viability of the cell. Thus, E. coli growth should require the functions of many other genes, in addition to the essential genes, which account for <10% of the genome (PEC, http://www.shigen.nig.ac.jp/ecoli/pecplus/index.jsp). In fact, the reduction of the E. coli chromosome to 70% of the original size reportedly has detrimental effects on the cell (18). In light of this knowledge, a significant proportion of the aberrant proteins, expressed from the extended ORFs, should retain their activities necessary for cell growth. We found that none of the six essential ORFs ending with UAG, except for hda, needed to be engineered to end with UAA, to create an RF-1-lacking strain. This finding revealed that the aberrant proteins, generated from the extension of these ORFs, retained their essential function for cell growth, suggesting that the E. coli proteome largely tolerates the C-terminal extension.
The second stop triplet, to terminate the translation extended by UAG readthrough, may not occur at random. In the E. coli genome, 42% of the UAG triplets are accompanied by a second stop triplet, UAA or UGA, occurring within 10 triplets after UAG. In contrast, a significantly smaller proportion of UAG triplets (27%) would have the second stop triplet within 10 triplets, if it were present randomly. This comparison suggests that the average aberrant protein, produced by UAG readthrough, has a shorter peptide tail than that expected from the random occurrence of the second stop triplet. This tendency of ‘doubling’ stop signal has also been observed in the genomes of yeasts and ciliates (19,20), and might have evolved, because the unusual proteins with long C-terminal tails, produced by nonsense suppression or other events, were harmful to the organism. The double-stop-codon system, which makes the tails shorter, might have alleviated some of the detrimental effects, and as a result, facilitated the reassignment of stop codons, as naturally established in the nuclear genomes of certain organisms, including ciliates (1).
The ‘ambiguous intermediate’ theory has been proposed for explaining the molecular mechanism underlying the natural occurrence of codon reassignment, including sense codon reassignment (6). This theory assumes that the codon to be reassigned retains two different meanings during the period when its usage in the genome gradually adapts to the second meaning. This scenario proved valid for the UAG reassignment. Prior to the RF-1 elimination, the expression of UAG-decoding tRNA conferred double meaning on UAG, which either terminated translation or specified an amino acid. Then, the required number of UAG triplets was replaced with UAA, to make the UAG reassignment tolerable for E. coli. Although only a small number of UAG replacements prepared E. coli for the UAG reassignment, the number of genomic mutations required to achieve a sense codon reassignment may be much larger, but may also depend on the tolerance of the proteome to the event.
For the present UAG reassignment, two mutations to create a UAG-reading tRNA and to inactivate RF-1, in addition to the UAG-to-UAA synonymous changes in the seven ORFs, are required for the UAG reassignment. Since suppressor tRNAs can naturally occur in E. coli (8,9), these nine mutations can be accumulated in the bacterial genome, with the lethal RF-1 mutation being the last. Assuming a mutation rate of 10−9 per base, an E. coli cell with all of these mutations could emerge within 270 generations starting from a single wild-type cell. Therefore, the flexibility of the genetic code is remarkably high. Nevertheless, the reassignment of UAG to a canonical amino acid has no apparent selective advantage, and would not help the cell to prevail in a mixed population. On the other hand, the reassignment to a novel amino acid could be advantageous to the organism, which would help to establish this change in the genome. A potential complexity is that more genetic changes are required, since an aminoacyl-tRNA synthetase gene must be duplicated, with the second gene being modified to create a synthetase that attaches the novel amino acid to tRNA. This is probably the case for pyrrolysine, a non-canonical amino acid, recognized by a specific synthetase and encoded by UAG in methane-generating Archaea (21). Pyrrolysine confers reproductive advantage to the organisms, by playing a catalytic role in enzymes involved in methanogenesis (22).
A novel repertoire of amino acids has been incorporated into proteins in E. coli, yeast, insect and mammalian cells, and has already contributed to protein science and technology (23–26). The previous approaches, however, relied on nonsense or frameshift suppression, with the inherent drawback of low product yields due to the competition between endogenous factors and suppressor tRNA molecules. Although an ‘orthogonal’ ribosome exhibiting increased suppression efficiency was recently developed (27), the competition with RF-1 still limited product yields. By eliminating the competing factor, our approach promises to drastically increase the yield of proteins containing non-natural amino acids. Furthermore, the codon reassignment allows the incorporation of these novel building blocks at multiple sites in a single protein, and will enhance the creation of proteins with novel structures and functions, as well as the preparation of proteins with naturally occurring modifications.
FUNDING
The Ministry of Education, Culture, Sports, Science and Technology of Japan (Targeted Proteins Research Program, and a Grant-in-Aid for Scientific Research (B) 19380195 to K.S. in part); RIKEN (Structural Genomics/Proteomics Initiative in the National Project on Protein Structural and Functional Analyses). Funding for open access charge: RIKEN.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr S. Kira for valuable discussions, Dr T. Hohsaka for critical reading of the manuscript, and Ms A. Ishii and Ms T. Nakayama for clerical assistance.
REFERENCES
- 1.Osawa S, Jukes TH, Watanabe K, Muto A. Recent evidence for evolution of the genetic code. Microbiol. Rev. 1992;56:229–264. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knight RD, Freeland SJ, Landweber LF. Rewiring the keyboard: evolvability of the genetic code. Nat. Rev. Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- 3.Santos MAS, Moura G, Massey SE, Tuite MF. Driving change: the evolution of alternative genetic codes. Trends Genet. 2004;20:95–102. doi: 10.1016/j.tig.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Crick FH. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 5.Osawa S, Jukes TH. Codon reassignment (codon capture) in evolution. J. Mol. Evol. 1989;28:271–278. doi: 10.1007/BF02103422. [DOI] [PubMed] [Google Scholar]
- 6.Schultz DW, Yarus M. Transfer RNA mutation and the malleability of the genetic code. J. Mol. Biol. 1994;235:1377–1380. doi: 10.1006/jmbi.1994.1094. [DOI] [PubMed] [Google Scholar]
- 7.Rydén SM, Isaksson LA. A temperature-sensitive mutant of Escherichia coli that shows enhanced misreading of UAG/A and increased efficiency for some tRNA nonsense suppressors. Mol. Gen. Genet. 1984;193:38–45. doi: 10.1007/BF00327411. [DOI] [PubMed] [Google Scholar]
- 8.Benzer S, Champe SP. A change from nonsense to sense in the genetic code. Proc. Natl Acad. Sci. USA. 1962;48:1114–1121. doi: 10.1073/pnas.48.7.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garen A, Siddiqi O. Suppression of mutations in the alkaline phosphatase structural cistron of E. coli. Proc. Natl Acad. Sci. USA. 1962;48:1121–1127. doi: 10.1073/pnas.48.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakamoto K, Ishimaru S, Kobayashi T, Walker JR, Yokoyama S. The Escherichia coli argU10(Ts) phenotype is caused by a reduction in the cellular level of the argU tRNA for the rare codons AGA and AGG. J. Bacteriol. 2004;186:5899–5905. doi: 10.1128/JB.186.17.5899-5905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakamoto K, Murayama K, Oki K, Iraha F, Kato-Murayama M, Takahashi M, Ohtake K, Kobayashi T, Kuramitsu S, Shirouzu M, et al. Genetic encoding of 3-iodo-L-tyrosine in Escherichia coli for single-wavelength anomalous dispersion phasing in protein crystallography. Structure. 2009;17:335–344. doi: 10.1016/j.str.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Motohashi T, Shimojima T, Fukagawa T, Maenaka K, Park EY. Efficient large-scale protein production of larvae and pupae of silkworm by Bombyx mori nuclear polyhedrosis virus bacmid system. Biochem. Biophys. Res. Commun. 2005;326:564–569. doi: 10.1016/j.bbrc.2004.11.060. [DOI] [PubMed] [Google Scholar]
- 13.Iraha F, Oki K, Kobayashi T, Ohno S, Yokogawa T, Nishikawa K, Yokoyama S, Sakamoto K. Functional replacement of the endogenous tyrosyl-tRNA synthetase-tRNATyr pair by the archaeal tyrosine pair in Escherichia coli for genetic code expansion. Nucleic Acids Res. 2010;38:3682–3691. doi: 10.1093/nar/gkq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konecki DS, Aune KS, Tate WP, Caskey CT. Characterization of reticulocyte release factor. J. Biol. Chem. 1977;252:4514–4520. [PubMed] [Google Scholar]
- 15.Dontsova M, Frolova L, Vassilieva J, Piendl W, Kisselev L, Garber M. Translation termination factor aRF1 from the archaeon Methanococcus jannaschii is active with eukaryotic ribosomes. FEBS Lett. 2000;472:213–216. doi: 10.1016/s0014-5793(00)01466-6. [DOI] [PubMed] [Google Scholar]
- 16.Scolnick E, Tompkins R, Caskey T, Nirenberg M. Release factors differing in specificity for terminator codons. Proc. Natl Acad. Sci. USA. 1968;61:768–774. doi: 10.1073/pnas.61.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Yokota T, Ito K, Nakamura Y, Aiba H. Reduced action of polypeptide release factors induces mRNA cleavage and tmRNA tagging at stop codons in Escherichia coli. Mol. Microbiol. 2007;63:116–126. doi: 10.1111/j.1365-2958.2006.05498.x. [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto M, Ichimura T, Mizoguchi H, Tanaka K, Fujimitsu K, Keyamura K, Ote T, Yamakawa T, Yamazaki Y, Mori H, et al. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol. Microbiol. 2005;55:137–149. doi: 10.1111/j.1365-2958.2004.04386.x. [DOI] [PubMed] [Google Scholar]
- 19.Liang H, Cavalcanti AR, Landweber LF. Conservation of tandem stop codons in yeasts. Genome Biol. 2005;6:R31. doi: 10.1186/gb-2005-6-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adachi M, Cavalcanti AR. Tandem stop codons in ciliates that reassign stop codons. J. Mol. Evol. 2009;68:424–431. doi: 10.1007/s00239-009-9220-y. [DOI] [PubMed] [Google Scholar]
- 21.Alkalaeva E, Eliseev B, Ambrogelly A, Vlasov P, Kondrashov FA, Gundllapalli S, Frolova L, Söll D, Kisselev L. Translation termination in pyrrolysine-utilizing archaea. FEBS Lett. 2009;583:3455–3460. doi: 10.1016/j.febslet.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao B, Gong W, Ferguson TK, James CM, Krzycki JA, Chan MK. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 24.Sakamoto K, Hayashi A, Sakamoto A, Kiga D, Nakayama H, Soma A, Kobayashi T, Kitabatake M, Takio T, Saito K, et al. Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 26.Mukai T, Wakiyama M, Sakamoto K, Yokoyama S. Genetic encoding of non-natural amino acids in Drosophila melanogaster Schneider 2 cells. Protein Sci. 2010;19:440–448. doi: 10.1002/pro.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Neumann H, Peak-Chew SY, Chin JW. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat. Biotechnol. 2007;25:770–777. doi: 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]