Abstract
Integrons are genetic elements that incorporate mobile gene cassettes by site-specific recombination and express them as an operon from a promoter (Pc) located upstream of the cassette insertion site. Most gene cassettes found in integrons contain only one gene followed by an attC recombination site. We have recently shown that a specific lineage of group IIC introns, named group IIC-attC introns, inserts into the bottom strand sequence of attC sites. Here, we show that S.ma.I2, a group IIC-attC intron inserted in an integron cassette array of Serratia marcescens, impedes transcription from Pc while allowing expression of the following antibiotic resistance cassette using an internal outward-oriented promoter (Pout). Bioinformatic analyses indicate that one or two putative Pout, which have sequence similarities with the Escherichia coli consensus promoters, are conserved in most group IIC-attC intron sequences. We show that Pout with different versions of the −35 and −10 sequences are functionally active in expressing a promoterless chloramphenicol acetyltransferase (cat) reporter gene in E. coli. Pout in group IIC-attC introns may therefore play a role in the expression of one or more gene cassettes whose transcription from Pc would otherwise be impeded by insertion of the intron.
INTRODUCTION
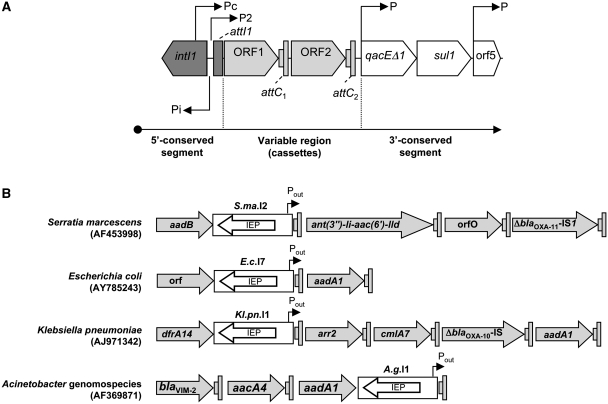
Integrons are genetic elements that capture gene cassettes using a site-specific tyrosine recombinase (called an integron integrase) and promote their co-expression by supplying a unique functional promoter, Pc, divergent to the integrase gene (1–3). Most gene cassettes are composed of a single structural gene followed by a short recombination site designated attC (or 59-base element), that is specifically recognized by integron integrases (4). Integrons are found on chromosomes and on diverse mobile elements, such as plasmids and transposons, and play a major role in lateral gene transfer in gram-negative bacteria (5,6). Distinct classes of mobile integrons, corresponding to their integrase genes, have been reported in the literature (6). Mobile class 1 integrons are the most widespread among multi-drug resistant bacteria and are often associated with transposons from the Tn21 family (7). The class 1 integron platform is composed of two conserved segments, the 5′-conserved (5′-CS) and 3′-conserved (3′-CS) regions, and one variable region (Figure 1A). The 5′-CS segment contains the integrase gene (intI1), two divergent promoter regions (called Pi for the integrase gene and Pc for gene cassettes), and a recombination site (attI1) into which cassettes are integrated. The 3′-CS segment usually contains a partially functional intercalating dyes/quaternary ammonium compound resistance gene (qacEΔ1) and most also contain a sulfonamide resistance gene (sul1), and an open reading frame (ORF5), whose product has some similarity to puromycin acetyltransferase (8,9). Between the two conserved segments, the variable region usually includes a short array of gene cassettes coding for various antibiotic resistance mechanisms or ORFs whose products have no known function (10–12). Almost all gene cassettes are promoterless structures that depend on the Pc promoter to express their genes. Among class 1 integrons, several Pc variants (the most prevalent being Pcweak, Pchybrid 1, Pcstrong and Pchybrid 2, respectively) and a second cassette promoter region, P2 (almost exclusively associated with the Pcweak variant), have been described in the literature with different versions of the −35 and −10 sequences for each promoter (1,3,13–16). Therefore, expression of gene cassettes is potentially influenced by the genomic localization of the integron (i.e. on a multicopy plasmid versus on the chromosome) and mutation of the transcription and translation initiation signals (1,13,14,17). Moreover, if several cassettes are inserted in the variable region, additional factors, such as premature transcription termination within attC sites (13), a cassette with its own promoter (18,19), or insertion of mobile genetic elements in attC sites (e.g. insertion sequences (IS) or group II introns) (20,21), may also influence expression of gene cassettes.
Figure 1.
Class 1 integron and cassette arrays. (A) Schematic diagrams of the general structure of a class 1 integron. P, promoters; intI1, integrase gene; qacEΔ1, antiseptic resistance gene; sul1, sulfonamide resistance gene; orf5, gene of unknown function. (B) Schematic diagrams of the variable region (gene cassettes) of class 1 integrons found in S. marcescens SCH909, E. coli 702, K. pneumoniae and Acinetobacter genomospecies genomes. The gray arrows indicate cassette ORFs; the gray boxes indicate cassette attC sites; the white rectangles and arrows indicate group IIC-attC introns with their intron encoded proteins (IEP); and Pout indicates a putative outward-oriented promoter within the intron.
Group II introns, together with LINEs and SINEs, are mobile elements from among non-LTR retrotransposons (22). They are found in bacteria (23), Archaea (24) and in organelle genes of plants, fungi and yeast (25). Group II intron RNAs are characterized by a conserved secondary structure organized into six domains (DI–DVI) (26). They fold into active ribozymes that catalyze their excision (from precursor RNAs) and invade new genomic locations, aided by the intron-encoded protein (IEP) (27). Eight lineages of group II introns, termed bacterial classes A-F, ML (mitochondrial-like) and CL (chloroplast-like), have been established according to phylogenetic analysis of their IEP sequences (28–30). Group IIC introns are of special interest because they are found in intergenic regions, usually after palindromic sequences (23,31–33), and have unique RNA structure and self-splicing properties (34,35). Phylogenetic analyses of intron IEP sequences has shown that introns found in attC sites constitute a monophyletic subset of group IIC, named group IIC-attC introns (32,36). Group IIC introns found in integrons are specifically inserted into the bottom strand sequence of gene cassettes and consequently are oriented opposite to the transcription of the adjacent genes. While most introns found in integrons are in the last cassettes of the variable region (37), those found in the Serratia marcescens SCH909 (accession no. AF453998), Escherichia coli 702 (AY785243), and Klebsiella pneumoniae (AJ971342) integrons, are in the first cassette and potentially influence the expression of the following gene cassettes (Figure 1B).
In this study, we first show that S.ma.I2, a group IIC-attC intron inserted in an integron cassette array of S. marcescens, impedes transcription from Pcweak-P2 promoters located within the 5′-CS region, while allowing expression of the following antibiotic resistance cassette using an internal outward-oriented promoter (Pout). Then, we performed bioinformatic analyses of all group II-attC intron sequences available in databases in order to determine the prevalence of Pout. We found that one or two putative Pout, which have sequence similarities with the E. coli consensus promoters, are conserved in several group IIC-attC introns. We show that Pout with different versions of the −35 and −10 sequences from various group IIC-attC introns are functionally active in expressing a promoterless chloramphenicol acetyltransferase (cat) reporter gene in E. coli.
MATERIALS AND METHODS
Recombinant plasmids, bacterial strains and growth conditions
Plasmids are described in Table 1. The pKK-InΔSmaI2 clone was obtained from pKK-In by PCR amplification using primer pair Sm909-3947.for and Sm909-1507.rev to remove the group II intron S.ma.I2. The PCR reaction mixture was digested with DpnI (in order to remove the methylated pKK-In template) and ethanol precipitated. Then, the recovered unmethylated 4396-bp PCR product (i.e. pKK-InΔSmaI2) was ligated with T4 DNA ligase (400 U; NEB) and transformed into E. coli DH5-α competent cells with ampicillin selection. Serratia marcescens SCH909, Shewanella baltica OS155 and E. coli DH5-α (supE44 ΔlacU169 (ϕ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1) were grown in Luria–Bertani (LB) broth (5 g NaCl, 10 g tryptone, 5 g yeast extract) supplemented with 1 g glucose at 37°C. When necessary, antibiotics were used at the following concentrations: ampicillin (Ap), 100 µg/ml; and chloramphenicol (Cm), 34 µg/ml. Nitrosomonas europaea was cultured as previously described (38). Geobacter sulfurreducens genomic DNA and the S. baltica OS155 strain were kindly provided by The Institute for Genomic Research and by the DOE Joint Genome Institute, respectively. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added at 1 mM final concentration for induction of the tac promoter in pLQ880. Total DNA was isolated using a phenol–chloroform purification method as described by Sambrook and Russell (39).
Table 1.
Plasmids used in this study
| Plasmids | Description or relevant characteristics | Reference |
|---|---|---|
| pKK232-8 | Cloning vector with a promoterless cat used for promoter selection. | (53) |
| pKK-IntI1 | 349-bp SalI-HindIII PCR fragment amplified from S. marcescens SCH909 genomic DNA (primer pair IntI1-SalI and aadB-HindIII) containing part of intI1 and the nucleotide sequence up to the initiation codon of the aadB::S.ma.I2 gene cassette and cloned into pKK232-8 digested by SalI-HindIII. | This study |
| pKK-SmaI2 | 2095-bp SalI-HindIII PCR fragment amplified from S. marcescens SCH909 genomic DNA [primer pair SmaI2-SalI and ant(3″)Ii-HindIII] containing the entire group II intron S.ma.I2 (1971 bp) and the nucleotide sequence up to the initiation codon of the ant(3″)-Ii-aac(6′)-IId gene cassette and cloned into pKK232-8 digested by SalI-HindIII. | This study |
| pKK-In | 2900-bp SalI-HindIII PCR fragment amplified from S. marcescens SCH909 genomic DNA [primer pair IntI1-SalI and ant(3″)Ii-HindIII] containing part of intI1, the aadB::S.ma.I2 gene cassette and the nucleotide sequence up to the initiation codon of the ant(3″)Ii-aac(6′)-IId gene cassette and cloned into pKK232-8 digested by SalI-HindIII. | This study |
| pKK-InΔSmaI2 | Clone derived from pKK-In by PCR (primer pair Sm909–3947.for and Sm909–1507.rev) to remove the group II intron S.ma.I2 (‘Materials and Methods’ section). | This study |
| pKK-NeI1-P1out | 200-bp PCR fragment amplified from N. europaea genomic DNA (primer pair NeI1-prom.for and NeI1-prom.rev) containing part of the group II intron N.e.I1 (base positions 1–200 in N.e.I1) and cloned into pKK232-8 digested by SmaI. | This study |
| pKK-GsI1-P1out | 200-bp PCR fragment amplified from G. sulfurreducens genomic DNA (primer pair GsI1-prom.for and GsI1-prom.rev) containing part of the group II intron G.s.I1 (base positions 1–200 in G.s.I1) and cloned into pKK232-8 digested by SmaI. | This study |
| pKK-ShbaI2-P1out | 200-bp PCR fragment amplified from S. baltica genomic DNA (primer pair ShbaI2-prom.for and ShbaI2-prom.rev) containing part of the group II intron Sh.ba.I2 (base positions 1–200 in Sh.ba.I2) and cloned into pKK232-8 digested by SmaI. | This study |
| pKK-SmaI2-P2out | 383-bp SspI-BglII restriction fragment digested from the pUCSmI plasmid (36) containing part of the group IIC intron S.ma.I2 (base positions 288–675 in S.ma.I2) and cloned into pKK232-8 digested by SmaI by the TA-cloning method. | This study |
| pLQ872 | Weak Pc promoter from integron In0 (pVS1) cloned in pKK232-8. | (1) |
| pLQ876 | Strong Pc promoter from integron In4 (Tn1696) cloned in pKK232-8. | (1) |
| pLQ880 | 96-bp HindIII-BamHI fragment of tac promoter cloned in pKK232-8. | (1) |
Polymerase chain reaction procedures and primers
We used the Phusion DNA polymerase (Finnzymes) for plasmid assembly and the Biotools DNA polymerase (Biotools) for the 5′-RACE, according to the manufacturer’s instructions. PCR primers IntI-SalI (5′-CGCACACCGTCGACACGGATGAAG), aadB-HindIII (5′-CTGCCGCAGCTAGAAGCTTGTGTATCAATG), SmaI2-SalI (5′-CCGCTTTCAGGTCGACATATGCGG), ant (3″) Ii-HindIII (5′-AGCTGTACCGAAGCTTCGGCGGGTAC), Sm909–3497.for (5′-ACAATTCATTCAAGCCGAACCC), Sm909–1507.rev (5′-TAGGCCGCATATCGCGACC), NeI1-prom.for (5′-GTGCGCCCAGCATGGGCGCG), NeI1-prom.rev (5′-AGCTCGCCTCGCCTGCCTCG), GsI1-prom.for (5′-GTACGCCCGGCATGGGCGTG), GsI1-prom.rev (5′-CTGACTTGCCCGGACACCCC), ShbaI2-prom.for (5′-GTACGCCCAGCATGGGCATG), ShbaI2-prom.rev (5′-ATGAACTTTCTTTGCACTGC), PKKL311 (5′-TTCTTTACGATGCGATTG) and POLY(C) (5′-CCCCCCCCCCCCCCC) were synthesized using an ABI-3900 DNA Synthesizer from Applied Biosystems Inc. (Foster City, CA, USA).
Genome project database searches for group IIC-attC introns
A protein–protein Basic Local Alignment Search Tool (BLASTP) search was performed on the entire GenBank non redundant protein sequences (nr) using as a query the IEP peptide sequence of group IIC-attC intron S.ma.I2 from S. marcescens (accession no. AF453998).
Multiple sequence alignments and phylogenetic tree
Phylogenetic analysis was based on intron RT subdomains and X domains. Bacterial class C IEP sequences from Azotobacter vinelandii (accession no. CP001157), Bacillus halodurans (BA000004), Bacteroides thetaiotaomicron (AE015928), Burkholderia cenocepacia (CP000959), Clostridium acetobutylicum (AE001437), Lactobacillus reuteri (AY911856), Microscilla sp. (AF339846), Oceanobacillus iheyensis (BA000028), Pseudomonas alcaligenes (U77945), Pseudomonas syringae pv. tomato (AE016853), Streptococcus agalactiae (AJ292930), Streptococcus pneumoniae (AF030367) and Symbiobacterium thermophilum (AP006840) were retrieved from the Mobile group II intron web site (40). The tree was rooted with IEP sequences from the Lactococcus lactis Ll.LtrB (mitochondrial-like; accession no. U50902) and Sinorhizobium meliloti RmInt1 (bacterial class D; accession no. Y11597) introns. The compiled IEP peptide sequences were aligned using CLUSTAL W (41). The resulting multiple sequence alignments were subjected to analyses using the neighbor-joining algorithm, with the Poisson correction distance method, of the Molecular Evolutionary Genetics Analysis (MEGA) package version 4.0 (42). One thousand bootstrap analyses were performed to estimate the robustness of the phylogenetic inference.
Bioinformatic predictions of internal outward-oriented promoters (Pout) in group IIC-attC introns
We searched for Pout in intron sequences, ranging from the 5′-end of the intron to the nucleotide opposite the start codon of the ORF encoding the IEP on the bottom strand, using the Neural Network for Promoter Prediction (NNPP) version 2.2 (Berkeley Drosophila Genome Project, http://www.fruitfly.org/index.html) and BPROM (SoftBerry, http://linux1.softberry.com/berry.phtml) programs.
5′-rapid amplification of cDNA end
Transcription initiation sites from the putative Pout were determined using the 5′-rapid amplification of cDNA ends (5′-RACE) method as described by Sambrook and Russell (39). Escherichia coli DH5-α competent cells were transformed with the indicated pKK232-8 clone and subjected to Ap selection. One colony of each transformant was cultured in LB medium containing both Ap and Cm at 37°C until the optical density at 600 nm was 0.7. Total RNA was purified using the RNeasy Mini Kit (Qiagen). cDNA synthesis was done using the Superscript III reverse transcriptase (200 U; Invitrogen) according to the manufacturer’s instructions and the PKKL311 primer (10 µM; reverse primer within cat) and incubated for 60 min at 50°C. RNase H (5 U; NEB) was added to the RT reactions and incubated for 30 min at 37°C. cDNA transcripts were purified using the QIAquick PCR Purification Kit (Qiagen). A dG-tail was added to the purified cDNA transcripts using dGTP (100 mM; Amersham Biosciences) and terminal transferase (20 U; NEB) according to the manufacturer’s instructions. The tailed cDNA transcripts were purified using the QIAquick PCR Purification Kit. PCR amplification of the tailed cDNA was conducted with the PKKL311 and POLY(C) primer pair (10 µM each) using Biotools DNA polymerase (2.5 U; Biotools) according to the manufacturer’s instructions. In order to find transcription start sites, the PCR products were purified and sequenced using the PKKL311 primer.
CAT assay
CAT assays were performed as described by Levesque and collaborators (1). CAT activity was assayed on crude cell extracts, from E. coli DH5-α cells carrying one of the pKK232-8 clones, prepared by sonication in Tris–HCl (1 mM [pH 7.6]). For each assay a 150 µl reaction mix containing 9.6 µl of [14C]Cm (0.05 mCi/ml; PerkinElmer), 24 µl of acetyl-coenzyme A (4 mM, resuspended in 20 mM sodium phosphate buffer [pH 7.0]), 39 µl of Tris (1 M [pH 7.5]) and 83.4 µl of deionized water was prepared. The CAT assay was started by adding 20 µl of total protein (1 ng/µl) to 130 µl of the reaction mix. After 60 min incubation at 37°C, the reactions were stopped using 1 ml of ethyl acetate and dried. The samples (resuspended in 20 µl of ethyl acetate) were spotted onto thin-layer chromatography sheets of silica gel H (Analtech) and run in a chromatography chamber with chloroform:methanol (95:5 v/v) for 60 min. Once dry, the silica plate was covered with plastic wrap and processed for phosphorimaging. CAT activity was calculated as the count of acetylated Cm (i.e. the total count of 1-acetoxy-Cm and 3-acetoxy-Cm divided by the sum of acetylated and non-acetylated Cm). We used as negative controls either 20 µl of Tris–HCl (1 mM [pH 7.6]) or 20 µl of crude cell extract of E. coli DH5-α competent cells transformed with the pKK232-8 plasmid.
RESULTS
Insertion of S.ma.I2 into integron #2 of Serratia marcescens SCH909 affects the expression of the following gene cassette
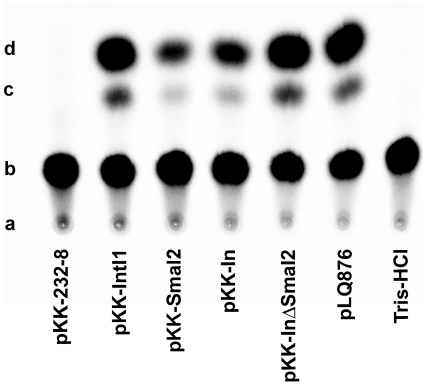
The integron #2 of S. marcescens SCH909 (AF453998) is one of three class 1 multiresistance integrons located on a 60-kb conjugative plasmid (20). The first cassette contains the aadB [also called ant(2″)-Ia] aminoglycoside resistance gene, separated from its attC site by S.ma.I2 that inserted into the bottom strand sequence (Figure 1B). The attC site is followed by the ant(3″)-Ii-aac(6′)-IId aminoglycoside resistance gene cassette. This cassette is followed by an unknown ORF with an attC site and a partial gene composed of the beginning of the blaOXA-10 cassette interrupted by IS1. The sequence downstream of IS1 revealed that the blaOXA-10 gene cassette is incomplete and that the 3′-CS segment of this integron is absent. Sequencing of the 5′-CS region showed that integron #2 harbors the Pcweak-P2 combination of promoters (data not shown). Previous studies showed that in the Pcweak-P2 combination, Pcweak does not contribute significantly to the expression of gene cassettes, which is mainly driven by P2 (13,14). In order to estimate whether insertion of S.ma.I2 affects the expression of the following ant(3″)-Ii-aac(6′)-IId gene cassette, we cloned various DNA fragments from integron #2 into the pKK232-8 plasmid upstream of a cat reporter gene. The resulting plasmids pKK-IntI1, pKK-SmaI2, pKK-In and pKK-InΔSmaI2 (see Table 1 for plasmid descriptions) were used in a quantitative CAT assay to examine expression of cat in E. coli DH5-α (‘Materials and Methods’ section). Figure 2 shows the separation by thin-layer-chromatography of Cm from its derivatives, 1-acetoxy-Cm and 3-acetoxy-Cm, in a 60 min assay at 37°C (1,3-diacetoxy-Cm was not detected). In our experimental conditions, we found that expression of cat from the clone pKK-InΔSmaI2 (i.e. in absence of S.ma.I2) was about 3.5-fold higher than with the clone pKK-In (27.3 ± 1.6% and 7.5 ± 0.5%, respectively) (Table 2). We also found that expression of cat from the clone pKK-SmaI2 (i.e. cloned S.ma.I2 sequence only) was slightly lower than with the clone pKK-In (6.7 ± 1.7% and 7.5 ± 0.5%, respectively). Therefore, our data suggest that insertion of S.ma.I2 in integron #2 of S. marcescens potentially results in a 72% decrease of expression of the following ant(3″)-Ii-aac(6′)-IId gene cassette. Moreover, a 0.89 relative ratio of acetylated Cm between the pKK-SmaI2 and pKK-In clones suggests that most of ant(3″)-Ii-aac(6′)-IId transcripts comes from a putative outward-oriented promoter (Pout) within S.ma.I2, and that S.ma.I2 disrupts transcripts from the Pcweak-P2 promoters. Nevertheless, a reverse trancriptase-(RT) assay showed that a small amount of transcription of cat from the Pcweak-P2 promoters occurs in the presence of S.ma.I2 (data not shown). On the other hand, a similar cat activity of the pKK-IntI1 and pKK-InΔSmaI2 clones (28.1 ± 5.2% and 27.3 ± 1.6%, respectively) suggests that, unlike S.ma.I2, the aadB gene cassette does not impede transcription from Pcweak-P2.
Figure 2.
Thin-layer chromatography of the [14C]chloramphenicol (Cm) CAT assay products in order to determine the effect of S.ma.I2 insertion in the integron #2 from S. marcescens. The pKK232 clones used for this assay are described in Table 1. The TLC plate was exposed to a Kodak BioMax MR film to obtain this image. CAT activity was assayed as described in ‘Materials and Methods’ section. (a) indicates the origin; (b) indicates non-acetylated Cm; (c) indicates 1-acetoxy-Cm; (d) indicates 3-acetoxy-Cm.
Table 2.
Expression of cat reporter gene in E. coli from promoter sequences within various cloned DNA fragments from S. marcescens SCH909 integron #2
| Clonea | DNA fragments from integron #2 cloned in pKK232-8 | Promoter | Cm acetylated (%)b | Ratio relative to pKK-In |
|---|---|---|---|---|
| Nonec | NA | NA | 0.3 ± 0.1 | 0.04 |
| pKK232-8 | none | None | 0.4 ± 0.1 | 0.05 |
| pKK-IntI1 | Partial 5′-CS | Pcweak-P2 in 5′-CS | 28.1 ± 5.2 | 3.75 |
| pKK-InΔSmaI2 | Partial 5′-CS + aadB-attC | Pcweak-P2 in 5′-CS | 27.3 ± 1.6 | 3.64 |
| pKK-In | Partial 5′-CS + aadB::S.ma.I2-attC | Pcweak-P2 in 5′-CS + putative Pout in S.ma.I2 | 7.5 ± 0.5 | 1.00 |
| pKK-SmaI2 | S.ma.I2-attC | Putative Pout in S.ma.I2 | 6.7 ± 1.7 | 0.89 |
aFor detailed information about these clones see Table 1.
bMeans ± SD of three independent experiments.
cTris–HCl (1 mM [pH 7.6]) was added to the reaction mix instead of crude cell extracts.
NA, not applicable
Bioinformatic analyses indicate that putative outward-oriented promoters (Pout) are found in several group IIC-attC introns
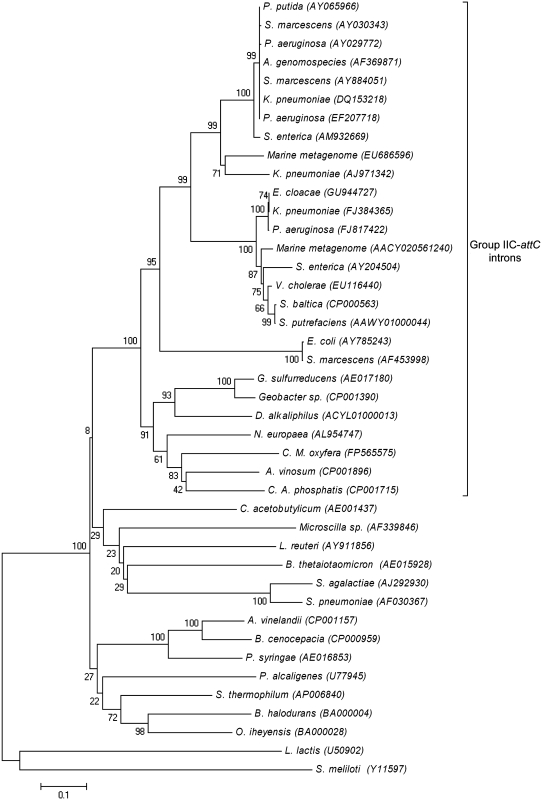
We wished to determine whether Pout also occurs in the E.c.I7 (99.8% sequence identity with S.ma.I2) and Kl.pn.I1 introns that are inserted into the first attC site of integron cassette arrays (i.e. between the first cassette and the following cassettes, potentially affecting the expression of the latter) from E. coli (accession no. AY785243) and K. pneumoniae (AJ971342), respectively (Figure 1B). We were also interested in knowing whether or not Pout is a conserved feature within the group IIC-attC intron lineage (32). Therefore, we first identified and analyzed several full length group IIC-attC introns distributed among 25 distinct bacterial genomes and two marine metagenome projects (‘Materials and Methods’ section and Table 3). Phylogenetic analysis of intron IEPs confirmed that the introns belonged to bacterial class IIC (Figure 3). Nodes of 100% bootstrap support define the bases of both bacterial IIC and IIC-attC lineages. Then, we used the BPROM and NNPP prediction programs for bacterial promoters in order to find putative Pout (‘Materials and Methods’ section). Table 3 shows a compilation of putative Pout (−35 region, −10 region and the spacing between these regions) that obtained the highest scores from both programs. One or two putative promoters, designated P1out and P2out, were predicted for most introns, except for the Desulfurivibrio alkaliphilus (accession no. ACYL01000013) and Allochromatium vinosum (CP001896) introns, for which no promoter was predicted. The putative promoter sequences are generally similar to the E. coli consensus promoter, TTGACA-N16–18-TATAAT (43). Table 3 also shows the positions of P1out and P2out among the introns. Interestingly, the putative P1out and P2out are precisely positioned within the ribozyme portion of the introns corresponding to domain I (DI) and domain II (DII), respectively. One exception was observed for the putative P2out from the Candidatus Accumulibacter phosphatis (accession no. CP001715) intron, which is positioned within domain III (DIII). These results suggest that outward-oriented promoters are general features of group IIC-attC introns, rather than being present only in S.ma.I2.
Table 3.
Bioinformatic analysis for putative outward-oriented promoters (Pout) in group IIC-attC introns
| Host organisma | Accession no. - nucleotide | Inb | Accession no. - protein (IEP) | 5′ exon genec | Putative Poutd |
||||
|---|---|---|---|---|---|---|---|---|---|
| Name | −35 region | Spacing | −10 region | Positions | |||||
| Pseudomonas putida | AY065966 | Y | AAL47550 | qacEΔ1 | P1out | TTGCCA | 17 nt | TCTAAT | 81–109 (DI) |
| Serratia marcescens | AY030343 | Y | AAK40354 | qacEΔ1 | P2out | TTGCCT | 17 nt | TTGCAT | 387–415 (DII) |
| Pseudomanas aeruginosa | AY029772 | Y | AAK50439 | qacEΔ1 | |||||
| Acinetobacter genomospecies | AF369871 | Y | AAK54203 | qacEΔ1 | |||||
| Serratia marcescens | AY884051 | Y | AAX16009 | NDe | |||||
| Klebsiella pneumoniae | DQ153218 | Y | AAZ82494 | qacEΔ1 | |||||
| Pseudomanas aeruginosa | EF207718 | Y | ABN10344 | qacEΔ1 | |||||
| Salmonella enterica | AM932669 | Y | CAP69662 | qacEΔ1 | P1out | TTGCCA | 17 nt | TCTAAT | 81–109 (DI) |
| P2out | TTGCCT | 17 nt | TTGCAT | 387–415 (DII) | |||||
| Marine metagenome | EU686596 | Y | NDe | orf (hypothetical prot.) | P1out | TTGCCA | 17 nt | TCTAAT | 80–108 (DI) |
| P2out | TTACCC | 17 nt | TCTCAT | 384–412 (DII) | |||||
| Klebsiella pneumoniae | AJ971342 | Y | CAJ29542 | arr2 | P1out | TTGCCA | 17 nt | TTGAAT | 76–104 (DI) |
| P2out | TTGCAT | 17 nt | GATGAT | 359–387 (DII) | |||||
| Enterobacter cloacae | GU944727 | Y | ADF59072 | NDe | P1out | TTGCCA | 17 nt | TTTAAT | 76–104 (DI) |
| Klebsiella pneumoniae | FJ384365 | Y | ACJ76645 | qacEΔ1 | P2out | TTGCCC | 17 nt | TTTCAT | 381–409 (DII) |
| Pseudomonas aeruginosa | FJ817422 | Y | ACO53361 | NDe | |||||
| Marine metagenome | AACY020561240 | N | NDe | orf (hypothetical prot.) | P1out | TTGCCA | 17 nt | TTTAAT | 76–104 (DI) |
| P2out | TTGCCC | 17 nt | TTTCAT | 382–410 (DII) | |||||
| Salmonella enterica | AY204504 | Y | AAO46869 | NDe | P1out | TTGCCA | 17 nt | TTTAAT | 91–119 (DI) |
| Vibrio cholerae | EU116440 | Y | ABV21790 | NDe | P1out | TTGCCA | 17 nt | TTTAAT | 76–104 (DI) |
| P2out | TTGCCC | 17 nt | TTTCAT | 381–409 (DII) | |||||
| Shewanella baltica | CP000563 | N | YP_001050216 | Transcriptional regulator | P1out | TTGCCA | 17 nt | TTTAAT | 76–104 (DI) |
| Shewanella putrefaciens | AAWY01000044 | Y | ZP_01707545 | Second group II intron | P2out | TTACCC | 17 nt | TTTCAT | 382–410 (DII) |
| Escherichia coli | AY785243 | Y | AAV34200 | aadA1 | P1out | TTGCCA | 17 nt | TTTAAT | 77–105 (DI) |
| Serratia marcescens | AF453998 | Y | AAL51020 | ant(3″)-Ii- aac(6′)-IId | P2out | TTGAAC | 17 nt | TAATCT | 322–350 (DII) |
| Geobacter sulfurreducens | AE017180 | Y | NP_953517 | vapC (NA) | P1out | TTGCCC | 16 nt | TATGCT | 168–195 (DI) |
| Geobacter sp. | CP001390 | Y | YP_002536457 | orf (hypothetical prot.) | P1out | TTGCCT | 17 nt | TACGCT | 74–102 (DI) |
| Desulfurivibrio alkaliphilus | ACYL01000013 | Y | ZP_05710592 | NADH:flavin oxidoreductase/NADH oxidase | none predicted | ||||
| Nitrosomonas europaea | AL954747 | Y | NP_842195 | ampG (NA) | P1out | TTGCCC | 18 nt | TATACT | 77–106 (DI) |
| P2out | TTGCCA | 16 nt | TCTGAT | 409–435 (DII) | |||||
| Candidatus Methylomirabilis oxyfera | FP565575 | N | CBE67152 | orf (hypothetical prot.) | P1out | TTGCCT | 17 nt | TCACAT | 66–94 (DI) |
| Allochromatium vinosum | CP001896 | N | YP_003442808 | orf (hypothetical prot.) | none predicted | ||||
| Candidatus Accumulibacter phosphatis | CP001715 | Y | ACV35120 | orf (hypothetical prot.) | P1out | TTGCCC | 18 nt | TATCAT | 77–106 (DI) |
| P2out | TTCGCG | 17 nt | TACTAT | 468–496 (DIII) | |||||
aHost organisms with identical introns were grouped together according to phylogenetic analysis (Figure 3).
bThis column indicates which introns are inserted (Y) or not inserted (N) into an integron cassette array.
cGene or gene cassette divergent with the intron IEP, and downstream of putative Pout. NA, (not applicable) was indicated when the 5′-exon ORF is convergent with the intron sequence.
dOutward-oriented promoters predicted using the BPROM and NNPP programs. Positions of promoter extremities (beginning of the −35 hexamer sequence—end of the −10 hexamer sequence) are indicated for the complementary strand. Domain I (DI), domain II (DII), or domain III (DIII) was indicated based on secondary structure analysis of intron RNA (data not shown) using the MFOLD program (54) and the consensus RNA secondary structure for group IIC introns (35).
eND, not defined in databases.
Figure 3.
Phylogenetic tree for group IIC-attC intron IEP amino acid sequences from various organisms. Evolutionary distances were computed using the neighbor-joining algorithm of the MEGA4 software (‘Materials and Methods’ section).
In order to see whether putative Pout with different versions of the −35 and −10 sequences are functionally active in vivo, the P1out sequences from the Nitrosomonas europaea intron N.e.I1 (accession no. AL954747), the Geobacter sulfurreducens intron G.s.I1 (AE017180) and the Shewanella baltica intron Sh.ba.I2 (CP000563) and the P2out sequence from S.ma.I2 were cloned upstream of a promoterless cat reporter gene in the pKK232-8 plasmid (Table 1). For most introns, we cloned P1out rather than P2out because they are closer to exon 1 and are more similar to the consensus sequence. However, for S.ma.I2, we cloned P2out since it is closer to the consensus promoter and potentially involved in expression of the following gene cassettes. Transformation of any of the four pKK232-8 clones (i.e. pKK-NeI1-P1out, pKK-GsI1-P1out, pKK-ShbaI2-P1out or pKK-SmaI2-P2out) into competent E. coli DH5-α cells conferred resistance to Cm (32 µg/ml) because of CAT activity, whereas E. coli DH5-α cells transformed with the original pKK232-8 plasmid were sensitive to the same concentration of Cm (data not shown). Therefore, transcription of cat occurred from a promoter within the cloned intron sequence that is functionally active in E. coli.
Identification of promoter elements and transcription start sites of cat within the pKK232-8-based clones
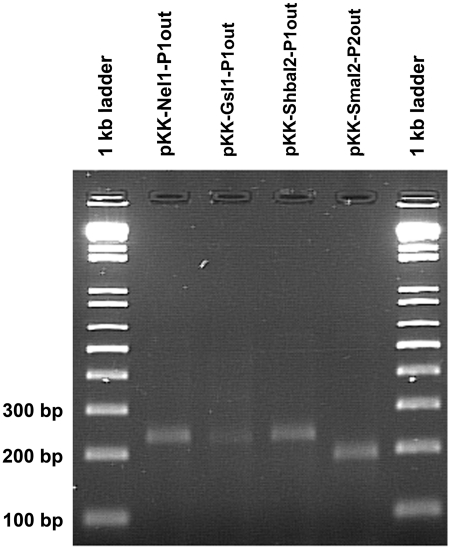
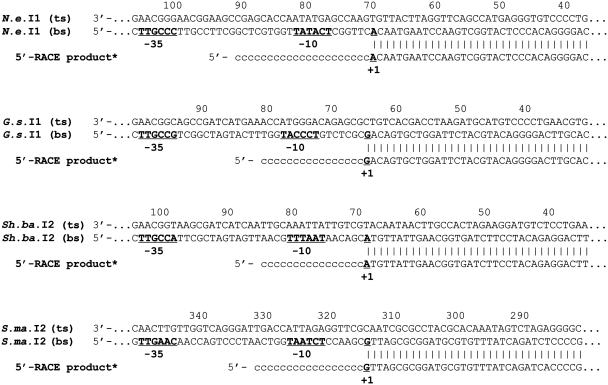
In order to determine whether transcription of cat originated from the predicted Pout, we used the 5′-RACE method (‘Materials and Methods’ section). Figure 4 shows an agarose gel containing specific 5′-RACE–PCR products for the identified clones using the PKKL311 (antisense primer in the cat gene) and POLY(C) primers. In order to find the transcription start sites, the four 5′-RACE–PCR products were sequenced using the PKKL311 primer. Figure 5 shows an alignment of the sequenced 5′-RACE–PCR products (reversed and complemented) with the cloned intron sequences. Each alignment shows the −35 and −10 regions of a promoter in the intron bottom-strand sequence and the transcription start site. Comparison of the −35 and −10 regions from the 5′-RACE data to those predicted using the BPROM and NNPP programs showed that both programs successfully identified either P1out or P2out, except for the G.s.I1 intron (Table 3). In fact, according to the 5′-RACE data the functionally active P1out sequence in G.s.I1 is TTGCCG-N16-TACCCT (positions 73–100 on the complementary strand). This sequence is located within the average range (i.e. 77–105 ± 5) for putative P1out predicted in other intron sequences. We also show that P1out sequences found in both Sh.ba.I2 and S.ma.I2 introns (64% sequence identity) and the P2out found in S.ma.I2 (cloned in pKK-ShbaI2-P1out and pKK-SmaI2-P2out, respectively) are functionally active in E. coli. Therefore, in our experimental conditions, S.ma.I2 contains two functionally active Pout.
Figure 4.
Agarose gel (2%) of the 5′-RACE-PCR products. 5′-RACE assays were performed as described in ‘Materials and Methods’ section in order to find the transcription initiation site of cat in the indicated pKK232 clones (see Table 1 for description).
Figure 5.
Alignments of the 5′-RACE product sequences with their corresponding intron DNA sequences. ts and bs indicate the top strand and bottom strand sequences; −35 and −10, components of the promoter. +1, transcription initiation site. Reversed and complemented 5′-RACE sequences are indicated (asterisks).
Analysis of Pout activity by comparison with that of the tac and integron promoters
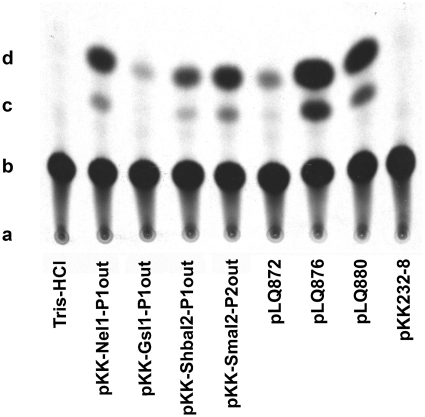
In order to determine the relative strength of the four functionally active Pout identified using the 5′-RACE method, we used a quantitative CAT assay. We compared their relative efficiency to that of the tac promoter (TTGACA-N16-TATAAT) and the weak and strong versions of integron Pc promoter (Pcweak TGGACA-N17-TAAGCT and Pcstrong TTGACA-N17-TAAACT, respectively) (1). Figure 6 shows the separation by thin-layer-chromatography of Cm from it derivatives, 1-acetoxy-Cm and 3-acetoxy-Cm, from a 60 min assay at 37°C (1,3-diacetoxy-Cm was not detected). Table 4 shows the percentage of acetylated Cm for each version of Pout. The most active intron promoters are S.ma.I2 P2out and N.e.I1 P1out. Transcription of cat using these promoters resulted in 6.65 ± 0.68% and 6.15 ± 0.88% of acetylated Cm, respectively (Table 3). The percentages of Cm acetylated using the Pcstrong (36.83 ± 4.42%) and tac (15.41 ± 1.74%) promoters are respectively >5-fold and 2-fold higher than using either S.ma.I2 P2out or N.e.I1 P1out. However, expression of CAT using either S.ma.I2 P2out or N.e.I1 P1out was >5-fold higher than using the Pcweak (1.10 ± 0.09%) promoter. In the same experimental conditions, expression of CAT using the Sh.ba.I2 P1out (identical to the S.ma.I2 P1out) is less than half that of S.ma.I2 P2out, but more than twice that of Pcweak. Finally, expression of CAT using the G.s.I1 P1out promoter resulted in the weakest level of acetylated Cm, 0.58 ± 0.06%, which is less than half that of the Pcweak promoter.
Figure 6.
Thin-layer chromatography of the [14C]chloramphenicol (Cm) CAT assay products for determination of the relative strengths of group IIC-attC intron promoters (Pout). The TLC plate was exposed to a Kodak BioMax MR film to obtain this image. CAT activity was assayed as described in ‘Materials and Methods’ section. The pKK232 clones used for this assay are described in Table 1. pLQ872 and pLQ876 contain the weak and strong versions of integron Pc promoters (respectively) cloned into pKK232-8 (1). (a) indicates the origin; (b) indicates non-acetylated Cm; (c) indicates 1-acetoxy-Cm; (d) indicates 3-acetoxy-Cm.
Table 4.
Relative strengths of group IIC-attC intron promoters (Pout) compared with the tac and the weak and strong versions of integron Pc promoters
| Clonea | Promoter | Cm acetylated (%)b | Ratio relative to tac |
|---|---|---|---|
| –c | NAd | 0.22 ± 0.06 | 0.01 |
| pKK232-8 | none | 0.26 ± 0.07 | 0.02 |
| pKK-SmaI2-P2out | P2oute | 6.65 ± 0.68 | 0.43 |
| pKK-NeI1-P1out | P1oute | 6.15 ± 0.88 | 0.40 |
| pKK-ShbaI2-P1out | P1oute | 2.99 ± 0.51 | 0.19 |
| pKK-GsI1-P1out | P1oute | 0.58 ± 0.06 | 0.04 |
| pLQ872 | Pcweak | 1.10 ± 0.09 | 0.07 |
| pLQ876 | Pcstrong | 36.83 ± 4.42 | 2.40 |
| pLQ880 | tac | 15.41 ± 1.74 | 1.00 |
DISCUSSION
In this study, we show that S.ma.I2, a group IIC-attC intron inserted in an integron cassette array of S. marcescens, impedes transcription from the Pcweak-P2 promoters located within the 5′-CS region, while allowing expression of the following antibiotic resistance cassette using two internal outward-oriented promoters (Pout). Despite these promoters, insertion of S.ma.I2 into integron #2 of S. marcescens potentially results in a 72% decrease of expression of the following ant(3″)-Ii-aac(6′)-IId gene cassette. Bioinformatic analyses of group IIC-attC introns from 25 distinct bacterial genomes and two marine metagenome projects indicate that one or two putative Pout are also found in other introns. These promoters, designated P1out and P2out, are located at similar distances from their exon 1 in RNA domain I and domain II, respectively. Comparison of promoter sequences with the consensus RNA structure/sequence for group IIC introns (35) showed that Pout are located within the region of variable sequences (data not shown). Our data suggest that Pout are conserved features of the group IIC-attC lineage. A distinct inward-directed internal promoter within the Lactococcus lactis intron, Ll.ltrB, was identified upstream of the gene for the IEP (LtrA) (44). Mutation of this promoter reduced the steady-state level of ltrA mRNA, LtrA, intron splicing and conjugation in L. lactis. A functional inward-directed promoter (tested in E. coli) was also found in S.ma.I2, CCTACA-N16-TAAACA (positions 375–402 in S.ma.I2), upstream of the gene for the IEP (Smtr) (data not shown). We show that Pout with different versions of the −35 and −10 sequences are functionally active in expressing a promoterless cat reporter gene in E. coli. These results are consistent considering that a consensus sequence of all the putative Pout has a strong similarity with the E. coli consensus promoter (43). The quantitative data obtained for the tested Pout sequences indicate that, despite their heterologous origins, these promoters work well in E. coli. On the other hand, GsI1-P1out and ShbaI2-P1out, which showed weak activity in E. coli, may have greater activities in their respective hosts, i.e. G. sulfurreducens (Delta-proteobacteria) and S. baltica (Gamma-proteobacteria), respectively.
Integrons can express multiple gene cassettes via read-through transcription from Pc to at least some extent (13,45). While the aadB gene cassette does not block transcription from Pcweak-P2, we showed that S.ma.I2 impedes transcription, most probably due to secondary structure within the intron. Pout may therefore confer a selective advantage to inserted group IIC-attC introns by ensuring transcription of following gene cassettes. For instance, Pout in the S.ma.I2 and E.c.I7 introns may play a role in the expression of the following ant(3″)-Ii-aac(6′)-IId and aadA1 resistance genes whose transcription from Pc would be reduced by insertion of the intron (Figure 1B). A Pout in group IIC-attC introns may also ensure maintenance of the introns in integrons and their dissemination to other organisms.
Despite the potential selective advantages conferred by Pout and specificity for attC site motifs, it is perplexing that only a few introns are found in either mobile or chromosomal integrons (32). We have previously demonstrated that the S.ma.I2 intron is not transcribed in the S. marcescens strain (36), suggesting that the insertion of group IIC-attC introns into the antisense strand relative to cassette transcription limits mobility of the intron to other attC sites.
The 3′-CS segment of class 1 integrons usually contains a partially functional intercalating dyes/quaternary ammonium compound resistance gene (qacEΔ1) and most also contain a sulfonamide resistance gene (sul1) (Figure 1A). Although transcription of both genes, from either the Pc promoter or a promoter of their own, was shown (46,47), we suggest that bacteria with class 1 integrons may use an additional source of transcription for the qacEΔ1 and sul1 genes as a selective advantage in order to survive in the presence of intercalating dyes, low levels of quaternary ammonium compounds or sulfonamide. In this regard, the Pout of group IIC-attC introns that are inserted into the last attC site of cassette arrays may contribute to the survival of the strain by potentially ensuring an enhanced transcription to qacEΔ1 and sul1 genes. It has been shown that selection by quaternary ammonium compounds and sulfonamide in natural or clinical environments has the potential to coselect for multidrug resistance (9,48–51).
Mobile IS from the IS1111 family, named ISPa21 and ISPst6, also target the attC sites of integron cassette arrays (21,52). Phylogenetic analyses of transposase sequences has revealed that ISPa21, ISPst6 and ISPst6-related sequences constitute a monophyletic subset within the IS1111 family, which is associated with attC sites (i.e. the IS1111-attC subgroup) (52). Interestingly, as with group IIC-attC introns, IS elements found in integrons are inserted near the 5′-end of distinct attC site motifs and into the antisense strand with respect to the gene cassette array transcription. A putative Pout was also suggested in both ISPa21 and ISPst6 elements. However, activity of such a promoter was either not reported (for ISPa21) or negative (for ISPst6) (21,52). Therefore, unlike IS elements, insertion of group IIC-attC into gene cassettes is more likely advantageous.
Analysis of the unique mobility pathway and distribution of group IIC-attC introns has shown that several factors potentially influence their presence and dissemination in bacterial genomes. The exact role of group IIC-attC introns in bacteria and especially in integrons remains undetermined. However, the unique features of integron cassettes suggest that these introns may play a role in cassette formation by recruiting and then joining genes and attC sites (20,37).
FUNDING
Fellowship from Canadian Institutes of Health Research (CIHR); Strategic Training Initiatives in Health Research (STIHR) grant STP-59324 (to G.L.); C.O.N.I.C.E.T. fellowship (to C.Q.); Canadian Institutes of Health Research grant MT-13564 (to P.H.R.). Funding for open access charges: Personal Funds.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
D.C. is a member of the Carrera del Investigador Científico, Consejo Nacional de Investigaciones Científicas y Técnicas (C.O.N.I.C.E.T.), Argentina.
REFERENCES
- 1.Lévesque C, Brassard S, Lapointe J, Roy PH. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene. 1994;142:49–54. doi: 10.1016/0378-1119(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 2.Ouellette M, Bissonnette L, Roy PH. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 beta-lactamase gene. Proc. Natl Acad. Sci. USA. 1987;84:7378–7382. doi: 10.1073/pnas.84.21.7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 4.Hall RM, Brookes DE, Stokes HW. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol. Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 5.Boucher Y, Labbate M, Koenig JE, Stokes HW. Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol. 2007;15:301–309. doi: 10.1016/j.tim.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Mazel D. Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 2006;4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 7.Hall RM. Mobile gene cassettes and integrons: moving antibiotic resistance genes in gram-negative bacteria. Ciba Found. Symp. 1997;207:192–202. doi: 10.1002/9780470515358.ch12. [DOI] [PubMed] [Google Scholar]
- 8.Bissonnette L, Roy PH. Characterization of In0 of Pseudomonas aeruginosa plasmid pVS1, an ancestor of integrons of multiresistance plasmids and transposons of gram-negative bacteria. J. Bacteriol. 1992;174:1248–1257. doi: 10.1128/jb.174.4.1248-1257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulsen IT, Littlejohn TG, Rådström P, Sundström L, Sköld O, Swedberg G, Skurray RA. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob. Agents Chemother. 1993;37:761–768. doi: 10.1128/aac.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fluit AC, Schmitz FJ. Class 1 integrons, gene cassettes, mobility, and epidemiology. Eur. J. Clin. Microbiol. Infect. Dis. 1999;18:761–770. doi: 10.1007/s100960050398. [DOI] [PubMed] [Google Scholar]
- 11.Fluit AC, Schmitz FJ. Resistance integrons and super-integrons. Clin. Microbiol. Infect. 2004;10:272–288. doi: 10.1111/j.1198-743X.2004.00858.x. [DOI] [PubMed] [Google Scholar]
- 12.Partridge SR, Tsafnat G, Coiera E, Iredell JR. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 2009;33:757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 13.Collis CM, Hall RM. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 1995;39:155–162. doi: 10.1128/aac.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jové T, Da Re S, Denis F, Mazel D, Ploy MC. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet. 2010;6:e1000793. doi: 10.1371/journal.pgen.1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt FR, Nücken EJ, Henschke RB. Nucleotide sequence analysis of 2′′-aminoglycoside nucleotidyl-transferase ANT(2′′) from Tn4000: its relationship with AAD(3′′) and impact on Tn21 evolution. Mol. Microbiol. 1988;2:709–717. doi: 10.1111/j.1365-2958.1988.tb00081.x. [DOI] [PubMed] [Google Scholar]
- 16.Tenover FC, Filpula D, Phillips KL, Plorde JJ. Cloning and sequencing of a gene encoding an aminoglycoside 6′-N-acetyltransferase from an R factor of Citrobacter diversus. J Bacteriol. 1988;170:471–473. doi: 10.1128/jb.170.1.471-473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papagiannitsis CC, Tzouvelekis LS, Miriagou V. Relative strengths of the class 1 integron promoter hybrid 2 and the combinations of strong and hybrid 1 with an active P2 promoter. Antimicrob Agents Chemother. 2009;53:277–280. doi: 10.1128/AAC.00912-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bissonnette L, Champetier S, Buisson JP, Roy PH. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J. Bacteriol. 1991;173:4493–4502. doi: 10.1128/jb.173.14.4493-4502.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stokes HW, Hall RM. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation. Plasmid. 1991;26:10–19. doi: 10.1016/0147-619x(91)90032-r. [DOI] [PubMed] [Google Scholar]
- 20.Centrón D, Roy PH. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob. Agents Chemother. 2002;46:1402–1409. doi: 10.1128/AAC.46.5.1402-1409.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poirel L, Brinas L, Fortineau N, Nordmann P. Integron-encoded GES-type extended-spectrum beta-lactamase with increased activity toward aztreonam in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005;49:3593–3597. doi: 10.1128/AAC.49.8.3593-3597.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu. Rev. Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai L, Zimmerly S. Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior. Nucleic Acids Res. 2002;30:1091–1102. doi: 10.1093/nar/30.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toro N. Bacteria and Archaea group II introns: additional mobile genetic elements in the environment. Environ. Microbiol. 2003;5:143–151. doi: 10.1046/j.1462-2920.2003.00398.x. [DOI] [PubMed] [Google Scholar]
- 25.Bonen L, Vogel J. The ins and outs of group II introns. Trends Genet. 2001;17:322–331. doi: 10.1016/s0168-9525(01)02324-1. [DOI] [PubMed] [Google Scholar]
- 26.Michel F, Ferat JL. Structure and activities of group II introns. Annu. Rev. Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 27.Pyle AM, Lambowitz AM. Group II introns: ribozymes that splice RNA and invade DNA. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 469–505. [Google Scholar]
- 28.Simon DM, Clarke NA, McNeil BA, Johnson I, Pantuso D, Dai L, Chai D, Zimmerly S. Group II introns in Eubacteria and Archaea: ORF-less introns and new varieties. RNA. 2008;14:1704–1713. doi: 10.1261/rna.1056108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon DM, Kelchner SA, Zimmerly S. A broadscale phylogenetic analysis of group II intron RNAs and intron-encoded reverse transcriptases. Mol. Biol. Evol. 2009;26:2795–2808. doi: 10.1093/molbev/msp193. [DOI] [PubMed] [Google Scholar]
- 30.Zimmerly S, Hausner G, Wu X. Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res. 2001;29:1238–1250. doi: 10.1093/nar/29.5.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Granlund M, Michel F, Norgren M. Mutually exclusive distribution of IS1548 and GBSi1, an active group II intron identified in human isolates of group B streptococci. J. Bacteriol. 2001;183:2560–2569. doi: 10.1128/JB.183.8.2560-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quiroga C, Centrón D. Using genomic data to determine the diversity and distribution of target site motifs recognized by class C-attC group II introns. J. Mol. Evol. 2009;68:539–549. doi: 10.1007/s00239-009-9228-3. [DOI] [PubMed] [Google Scholar]
- 33.Robart AR, Seo W, Zimmerly S. Insertion of group II intron retroelements after intrinsic transcriptional terminators. Proc. Natl Acad. Sci. USA. 2007;104:6620–6625. doi: 10.1073/pnas.0700561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Podar M, Chu VT, Pyle AM, Perlman PS. Group II intron splicing in vivo by first-step hydrolysis. Nature. 1998;391:915–918. doi: 10.1038/36142. [DOI] [PubMed] [Google Scholar]
- 35.Toor N, Hausner G, Zimmerly S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA. 2001;7:1142–1152. doi: 10.1017/s1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quiroga C, Roy PH, Centrón D. The S.ma.I2 class C group II intron inserts at integron attC sites. Microbiology. 2008;154:1341–1353. doi: 10.1099/mic.0.2007/016360-0. [DOI] [PubMed] [Google Scholar]
- 37.Léon G, Roy PH. Potential role of group IIC-attC introns in integron cassette formation. J Bacteriol. 2009;191:6040–6051. doi: 10.1128/JB.00674-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Léon G, Roy PH. Excision and integration of cassettes by an integron integrase of Nitrosomonas europaea. J. Bacteriol. 2003;185:2036–2041. doi: 10.1128/JB.185.6.2036-2041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 40.Dai L, Toor N, Olson R, Keeping A, Zimmerly S. Database for mobile group II introns. Nucleic Acids Res. 2003;31:424–426. doi: 10.1093/nar/gkg049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 43.Harley CB, Reynolds RP. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou L, Manias DA, Dunny GM. Regulation of intron function: efficient splicing in vivo of a bacterial group II intron requires a functional promoter within the intron. Mol. Microbiol. 2000;37:639–651. doi: 10.1046/j.1365-2958.2000.02033.x. [DOI] [PubMed] [Google Scholar]
- 45.Jacquier H, Zaoui C, Sanson-le Pors MJ, Mazel D, Bercot B. Translation regulation of integrons gene cassette expression by the attC sites. Mol. Microbiol. 2009;72:1475–1486. doi: 10.1111/j.1365-2958.2009.06736.x. [DOI] [PubMed] [Google Scholar]
- 46.Sundström L, Rådström P, Swedberg G, Sköld O. Site-specific recombination promotes linkage between trimethoprim- and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol Gen Genet. 1988;213:191–201. doi: 10.1007/BF00339581. [DOI] [PubMed] [Google Scholar]
- 47.Guerineau F, Brooks L, Mullineaux P. Expression of the sulfonamide resistance gene from plasmid R46. Plasmid. 1990;23:35–41. doi: 10.1016/0147-619x(90)90042-b. [DOI] [PubMed] [Google Scholar]
- 48.Antunes P, Machado J, Sousa JC, Peixe L. Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob. Agents Chemother. 2005;49:836–839. doi: 10.1128/AAC.49.2.836-839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaze WH, Abdouslam N, Hawkey PM, Wellington EM. Incidence of class 1 integrons in a quaternary ammonium compound-polluted environment. Antimicrob. Agents Chemother. 2005;49:1802–1807. doi: 10.1128/AAC.49.5.1802-1807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kazama H, Hamashima H, Sasatsu M, Arai T. Distribution of the antiseptic-resistance genes qacE and qacEΔ1 in gram-negative bacteria. FEMS Microbiol. Lett. 1998;159:173–178. doi: 10.1111/j.1574-6968.1998.tb12857.x. [DOI] [PubMed] [Google Scholar]
- 51.Rådström P, Swedberg G, Sköld O. Genetic analyses of sulfonamide resistance and its dissemination in gram-negative bacteria illustrate new aspects of R plasmid evolution. Antimicrob. Agents Chemother. 1991;35:1840–1848. doi: 10.1128/aac.35.9.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tetu SG, Holmes AJ. A family of insertion sequences that impacts integrons by specific targeting of gene cassette recombination sites, the IS1111-attC group. J. Bacteriol. 2008;190:4959–4970. doi: 10.1128/JB.00229-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brosius J. Plasmid vectors for the selection of promoters. Gene. 1984;27:151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]
- 54.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]