Abstract
Fibroblast growth factor 9 (FGF9) is an autocrine/paracrine growth factor that plays vital roles in many physiologic processes including embryonic development. Aberrant expression of FGF9 causes human diseases and thus it highlights the importance of controlling FGF9 expression; however, the mechanism responsible for regulation of FGF9 expression is largely unknown. Here, we show the crucial role of an AU-rich element (ARE) in FGF9 3′-untranslated region (UTR) on controlling FGF9 expression. Our data demonstrated that AUF1 binds to this ARE to regulate FGF9 mRNA stability. Overexpression of each isoform of AUF1 (p37, p40, p42 and p45) showed that only the p42 isoform reduced the steady-state FGF9 mRNA. Also, knockdown of p42AUF1 prolonged the half-life of FGF9 mRNA. The induction of FGF9 mRNA in prostaglandin (PG) E2-treated human endometrial stromal cells was accompanied with declined cytoplasmic AUF1. Nevertheless, ablation of AUF1 led to sustained elevation of FGF9 expression in these cells. Our study demonstrated that p42AUF1 regulates both steady-state and PGE2-induced FGF9 mRNA stability through ARE-mediated mRNA degradation. Since almost half of the FGF family members are ARE-containing genes, our findings also suggest that ARE-mediated mRNA decay is a common pathway to control FGFs expression, and it represents a novel RNA regulon to coordinate FGFs homeostasis in various physiological conditions.
INTRODUCTION
The mammalian fibroblast growth factor (FGF) family includes 18 distinct members and are grouped into six subfamilies based on the differences in sequence homology and phylogeny [review see (1)]. Many of these FGFs are expressed abundantly in a specific spatial and temporal pattern and considered to play substantial roles in development (2), angiogenesis (3), hematopoiesis (4) and tumorigenesis (5). Human fibroblast growth factor 9 (FGF9; MIN# 600921) shares >93% sequence identity with Xenopus, mice and rats (6,7), suggesting that FGF9 is important and may have similar functions across species. Previous studies suggested that FGF9 functions as an autocrine/paracrine growth factor for neuron cells (8), fibroblasts (9) and endometrial stromal cells (10), and as an oncogene for NIH3T3 fibroblast cells (11).
FGF9 mRNA is ubiquitously expressed in the embryo but is restricted to kidney, brain and uterus in the adult (10–12). Furthermore, abnormal expression of FGF9 is involved in several human diseases, including cancer (13,14), endometriosis (15), neuron degeneration (16), male-to-female sex reversal (17) and male infertility (18). Nevertheless, upon stimulation by factors like prostaglandin (PG) E2 (19), estrogen (10), androgen (20) and retinoic acid (21), the mRNA of FGF9 is upregulated but rapidly returns to basal levels. These studies implied that the expression of FGF9 needs to be strictly controlled. While the functions of FGF9 in physiological and pathological processes are well studied, mechanisms that control steady-state and stimuli-induced FGF9 mRNA levels to maintain FGF9 homeostasis remain largely unknown.
Regulation of the rate of mRNA decay is an important mechanism to control gene expression at the post-transcriptional level (22). The interaction between cis-acting elements in the transcripts and sequence-specific RNA-binding proteins permits precise control of the mRNA level and, subsequently, the protein level (23). Adenylate/uridylate-rich elements (AREs) are regulatory elements present in the 3′-untranslated region (UTR) of certain mRNAs that have been implicated in post-transcriptional gene regulation by mediating rapid degradation of target mRNA (24,25). AREs are generally found in transcripts that need to be precisely controlled, such as genes that function as growth factors, proto-oncogenes and cytokines (23,26,27). At least 17 trans-acting factors have been identified (28) to interact with mRNA containing AREs. Most of these proteins, such as the AU-rich element-binding protein 1 (AUF1) family (29), the tristetraprolin (TTP) family (30) and the KH-type splicing regulatory protein (KSRP) (31) reduce the target mRNA level. In contrast, factors like human antigen R (HuR) positively regulate target gene expression (32).
We previously reported that a microsatellite motif in the 3′-UTR of the FGF9 gene is important for controlling the mRNA stability of FGF9 (17). The work demonstrated a new post-transcriptional regulation of FGF9 expression and suggested the expression of FGF9 may be controlled by a network that involves multiple layers of regulations. In this study, we set out to investigate other novel mechanisms responsible for the tight regulation of FGF9 mRNA stability.
MATERIALS AND METHODS
Cell culture
A human embryonic kidney cell line HEK293 expressing endogenous FGF9 was grown in minimum essential medium (MEM) (Gibco, Carlsbad, CA, USA) with 10% heat-inactivated horse serum and 1.0 mM sodium pyruvate. Stromal cells were cultured in DMEM/F12 with 10% fetal bovine serum (FBS). In both cases, media were supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA, USA). Cells were routinely maintained at 37°C in a humidified 5% CO2 incubator. For PGE2 treatment (Cayman Chemical, Ann Arbor, MI, USA), phenol red-free DMEM/F12 medium was supplemented with 1% charcoal-stripped FBS and 1μM PGE2. Cells infected by shAUF1 and null virus were exposed to 1 μM PGE2 for the indicated durations. In parallel, the same amount of ethanol was used as control.
Plasmids
The entire FGF9 3′-UTR sequence was amplified using forward primer (FGF9 3′-FUTR-F: 5′-GCTCTAGAGCGGACAAAGACAGTTTCTTCACT-3′) and reverse primer (FGF9 3′-FUTR-R1: 5′-GCTCTAGAGCTTTGGAATTTCTATAAATAAATTTAAC-3′) anchored with XbaI restriction enzyme cutting sites. These XbaI polymerase chain reaction (PCR) products were ultimately subcloned into the XbaI site of the pGL3-promoter (pGL3-p) vector containing a SV40 promoter upstream of the firefly luciferase gene (Promega, Madison, WI, USA). The deletion and mutation constructs were generated by the same strategy with different reverse primers. FGF9 3′-UTR 549R (5′-GCTCTAGACTAAGAGGTCTTTGCTTTAAG-3′) and FGF9 3′-FUTR-mut-R1 (5′-GCTCTAGAGCTTTGGAATTTCTATCCCTCCCTTTAAC-3′) were used for the delARE and mutARE constructs, respectively. All constructs were confirmed by direct sequencing.
For AUF1 overexpression, the cDNA of AUF1 was amplified with AUF1CDS-1 (5′-TTTTTAAGCTTTGCTGCTAGTTTCGGTTCG) and AUF1CDS-2 (CCCGCTCGAGGTATGGTTTGTAGCTATTTTG) primers anchored with restriction enzyme cutting sites HindIII and XhoI (indicated by underline) on each 5′-end, respectively. The PCR products were digested by HindIII and XhoI enzymes and cloned into pcDNA3.1/Myc_His(+)A vector (Invitrogen).
Transient transfection and luciferase reporter assay
HEK293 cells were seeded on each well of a 24-well tissue culture plate (2.5 × 105 cells/well) (TPP AG, Trasadingen, Switzerland) and transfected with each plasmid construct (0.5 μg) plus plasmids carrying renilla luciferase as an internal control. The transfection was carried out with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were incubated for 24 h and harvested by adding 100 μl of reporter lysis buffer (Luciferase Assay System; Promega). The activity of luciferase was measured using a luminometer (Lumat LB 9507; EG&G Berthold, Bad Wildbad, Germany). Firefly luminescence was normalized to the protein concentration. Results are presented relative to normalized luminescence driven from pGL3-promoter and reported as relative luciferase units (RLU). All experiments were done in triplicate and independently performed at least three times.
mRNA isolation and half-life measurement
Total RNA from cells was isolated with the RNeasy mini kit (Qiagen, Hilden, Germany). For RNA half-life measurements of luciferase mRNA, HEK293 cells were transfected with 2.5 µg plasmid/106 cells in 6-well plates using Lipofectamine 2000. Twelve hours later, the cells were treated with 5 μg/ml actinomycin D (Act D) (Sigma, St Louis, MO, USA). RNA was extracted from cells treated with Act D at 0, 2, 4, 6 and 8 h for RNA half-life measurement. As for FGF9 mRNA half-life measurement, HEK293 cells were transfected with 40 nM siRNA/106 cells in 6-well plates using Lipofectamine 2000. Act D was added and RNA was isolated at 0, 10, 20, 30, 60 and 120 min after treatment. Complementary DNA was synthesized from 2 µg of DNase-treated total RNA using random primers and an ABI RT kit (Apply Biosystems, Forster city, CA, USA).
Real-time PCR measurements were carried out with the Applied Biosystem GeneAmp 7900. This assay was performed with 40 ng of RNA equivalent cDNA in triplicate on an ABI 7900HT Sequence Detection System and analyzed with SDS2.1 software (Apply Biosystems). TaqMan formulations were used and modified by validating the efficiency between target and reference genes according to the manufacturer’s protocols (Apply Biosystems). Luciferase TaqMan assays were performed with the primer set pGL3-Luc-1512F (5′-GCGTTATTTATCGGAGTTGCAGTTG-3′)/pGL3-Luc-1512R (5′-CATACTGTTGAGCAATTCACGTTCA-3′) and detected with pGL3-Luc-1512-Probe (5′-(FAM) CCGCGAACGACATTT (TAMRA)-3′) from the Assays-by-DesignSM service (Apply Biosystems). FGF9 TaqMan assays were performed with primer set (HS00181829-ml FGF9). Ribosomal 18s (4319413E) (Apply Biosystems) was used as endogenous control. The comparative CT method was used to calculate the relative amount of each sample according to the previous description. Finally, the mRNA half-life was determined by linear regression analysis.
UV crosslink and biotin pull-down assay
RNA–protein-binding reactions were performed using 3–5 μg of cytoplasmic extract, 5 μg of recombinant AUF1 (p45 isoform) purchased from USBiological (Swampscott, MA, USA), 10 ng biotin-labeled RNA and 2 μg tRNA, in a final volume of 20 μl using Binding Buffer A [20 mM HEPES-KOH at pH 7.5, 2.5 mM magnesium chloride (MgCl2), 100 mM potassium chloride (KCl), 20% glycerol, 0.5 mM dithiothreitol and protease inhibitor tablets]. Heparin was added at 0.25 μg/μl. Addition of heparin up to 0.75 mg/ml did not have any significant influence on the pattern of UV cross-linking. Reaction mixtures were incubated for 1 h at 37°C and UV irradiated at 254 nm for 20 min (Stratagene, La JoUa, CA, USA) on ice. The samples were analyzed by 10% SDS–PAGE and detected by western analysis.
For biotin pull-down analysis, 200 µg of whole-cell lysates were incubated with 2 μg purified biotinylated riboprobes (FGF9 ARE: Biotin-AAAUUUAUUUAUA; FGF9 mutant ARE: Biotin-AAAGGGAGGGAUA; Dharmacon Research) for 1 h at 25°C. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Invitrogen), the unbound proteins were removed, and after extensive washes with the binding buffer, the proteins were step-eluted with protein sample buffer. The eluted proteins were then analyzed by western blot analysis.
Western blot analysis
Whole-cell lysates were obtained by RIPA lysis buffer [50 mM KCl, 5% (v/v) glycerol, 0.1% (v/v) Nonidet P-40, 1 mM MgCl2, 1 mM dithiothreitol, 10 mM Tris/HCl pH 8.0]. Nucleus and cytoplasmic fraction were isolated by NE-PER kit (Thermo, Waltham, MA, USA). Protein concentration was determined by the Bradford assay (Bio-rad, Hercules, CA, USA). After protein quantification, equal amounts of whole-cell lysates were boiled in 4% sodium dodecyl sulfate (SDS) sample buffer (250 mM Tris–HCl, 20% 2-mercaptoethanol, 8% SDS, 40% glycerol and 0.02% bromophenol blue) and subjected to SDS-PAGE separation. Proteins were transferred onto polyvinylidene difluoride membrane (Amersham, Buckinghamshire, UK) and detected by an enhanced chemiluminescence (ECL) kit (Amersham). Endogenous AUF1 was detected by polyclonal rabbit anti-mouse AUF1 antiserum (Upstate Biotechnology, Charlottesville, VA, USA) at 1:4000 dilutions. Various recombinant AUF1 isoforms were detected by rabbit monoclonal anti-myc antibody at 1:2000 dilutions. α-Tubulin was used as an internal control and detected by rabbit monoclonal anti-α-tubulin antibody (Cell Signaling Technologies, Beverly, MA, USA) at 1:2000 dilutions. Lamin A/C was used as loading control for nucleus fraction and detected by mouse monoclonal anti-lamin A/C antibody at 1:200 dilutions. (Santa Cruz Biotech, Santa Cruz, CA, USA).
RNA interference
Small interfering RNAs (siRNA) targeting AUF1 and scrambled control duplexes (Invitrogen) were transfected into HEK293 cells with Lipofectamine 2000 (Invitrogen). After 48 h, total protein was isolated and analyzed by western blot analysis. The knockdown efficiency was calculated by normalizing to the signal intensity first to α-tubulin, and then divided by the value of scramble control. For knockdown of AUF1 in stromal cells, plasmids encoding shRNA that targeted all four AUF1 isoforms were transfected into 293T cells to generate shAUF1 lentivirus. Plasmid without any shRNA (Null) was used as a mock control in this experiment. The lentiviral shRNA clones were from the National RNAi Core Facility of Academic Sinica, Taiwan. Stromal cells were infected with Null (pLK0.1) lentivirus, or AUF1 shRNA lentivirus.
Computational analysis of FGF9 3′-UTR sequences
Sequences of FGF9 from the species Homo sapiens (Human; NM_002010), Canis lupus familiaris (dog; CF406880), Bos taurus (cow; DT899064.1), Sus scrofa (swine; EW180881), Taeniopygia guttata (bird; EH120653), Mus musculus (mouse; AA623627) and Rattus norvegicus (rat; DV717281) were downloaded from NCB1 CoreNucleotide and aligned with a multiple sequence alignment program (http://searchlauncher.bcm.tmc.edu/ multi-align/multi-align.html). Images of multiple sequence alignment were output by BOXSHADE (http://www.ch.embnet.org/software /BOX_form.html). In addition, conserved cis-elements of FGF9 3′-UTR were screened by UTRdb (33) and UTRscan (34).
Statistical analysis
All the data sets were analyzed using GraphPad Prism 4.0 (GraphPad Software, Inc., San Diego, CA, USA) and presented as mean ± SEM. Results were further analyzed using one-way analysis of variance (ANOVA), and post-test was processed using Tukey’s multiple comparison tests or the Student’s t-test.
RESULTS
The 3′-UTR of FGF9 mRNA contains AU-rich elements
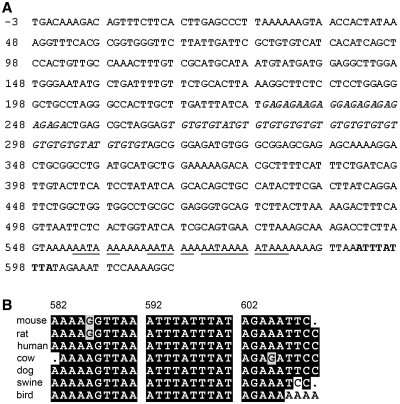
To identify potential factors that regulate FGF9 gene expression post-transcriptionally, we used bioinformatic tools to search for putative regulatory elements in the human FGF9 3′-UTR. Beside the (GA)n and (TG)n microsatellite motifs we previously reported (17), an ARE located at 593–601 bp downstream of the stop codon was identified (Figure 1A). Similar ARE motifs were also identified in FGF9 of mouse, rat, cow, dog, swine and distantly related vertebrate, bird. When these ARE sequences were aligned with that found in the human, the result showed 79% and 100% identities in the AU-rich region and the core nonamer, respectively (Figure 1B). These results demonstrated that the ARE in the 3′-UTR of human FGF9 is evolutionarily conserved and suggested that it may play a functional role in controlling the level of FGF9 mRNA.
Figure 1.
Sequence features of the human FGF9 3′-UTR. (A) The sequence corresponding to the 3′-UTR of FGF9 is depicted. The initial TGA nucleotide sequence (−3 to −1) corresponds to the translation stop codon. The GA and TG microsatellite motifs are marked in italic type. The ARE sequence (ATTTATTTA) is marked in bold and the underlines mark the poly-A signals (AATAAA). (B) Cross-species alignment of mouse, rat, human, cow, dog, swine and bird showed an overall 79% identity in the AU-rich element region.
ARE is a destabilization element
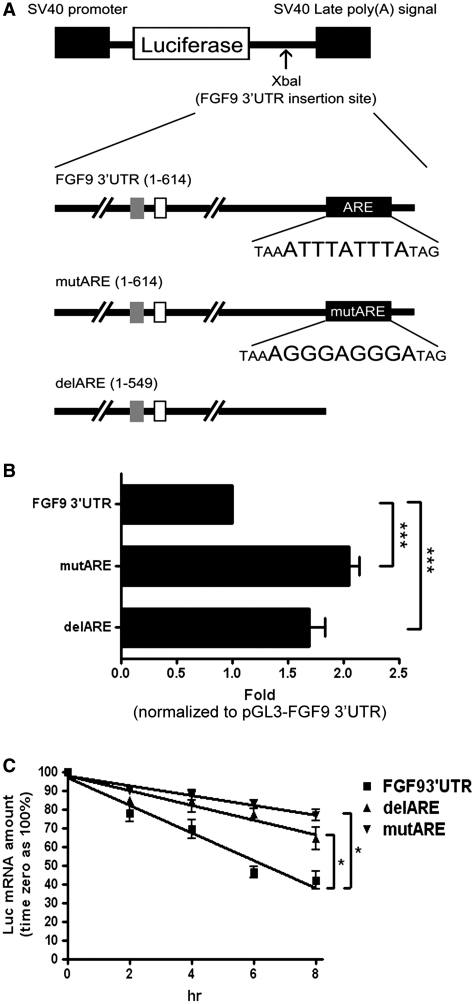
To functionally characterize the effects of FGF9 3′-UTR ARE on regulating FGF9 expression, sequences containing full-length FGF9 3′-UTR (FGF93′-UTR), FGF9 3′-UTR with ARE deleted (delARE) and full length with site-directed mutated ARE (mutARE) were cloned into the 3′-UTR of a firefly luciferase reporter construct driven by an SV40 promoter (Figure 2A). Results from the reporter assays showed that the luciferase activities of delARE and mutARE constructs were increased ∼1.8- (P < 0.001) and 2-fold (P < 0.001), respectively, compared with the construct having full-length FGF9 3′-UTR (Figure 2B). This result indicates that the ARE is a functional cis-element of FGF9 mRNA and its presence may cause mRNA degradation.
Figure 2.
FGF9 ARE decreases gene expression in HEK293 cells. (A) Diagrams showing the firefly luciferase reporter gene construct (top diagram), full-length FGF9 3′-UTR cloned into the 3′-end of the firefly luciferase reporter gene (second diagram), and constructs containing site-direct mutation of ARE (mutARE) (third diagram) and deletion of ARE (delARE) (fourth diagram). Empty and gray squares represent TG and GA dinucleotide repeats, respectively. The black square represents ARE motif. (B) Reporter gene activity shown as relative luciferase activity (fold) normalized to FGF9 3′-UTR. (C) Effect of FGF9 ARE on mRNA stability detected by real-time RT-qPCR. FGF9 3′-UTR (filled square), delARE (Filled triangle) or mutARE (filled inverted triangle) was transfected into HEK293 cells, which were treated with Act D (5 μg/ml) to inhibit transcription of the luciferase reporter gene. Results are normalized to time zero for each construct. All data are shown as mean ± SEM of 4–6 independent experiments. *P < 0.05; ***P < 0.001.
Next, we tested whether ARE affects the mRNA stability by transfecting FGF93′-UTR, delARE and mutARE constructs into HEK293 cells and treating them with 5 μg/ml actinomycin D to inhibit nascent RNA transcription. After quantification by reverse transcription real-time PCR (RT-qPCR), the half-life of each fusion mRNA was determined by linear regression analysis. Our data showed that the fusion mRNA with full-length FGF9 3′-UTR has the shortest half-life (t1/2) of 6.98 ± 0.10 h (Figure 2C). In contrast, fusion mRNAs with either deleted or mutated FGF9 ARE significantly prolong the mRNA stability to 9.94 ± 0.13 (P < 0.05) or 16.34 ± 0.05 h (P < 0.05; Figure 2C). These results demonstrated that FGF9 ARE is required to reduce the half-life of fusion transcripts.
AUF1 specifically binds to FGF9 ARE
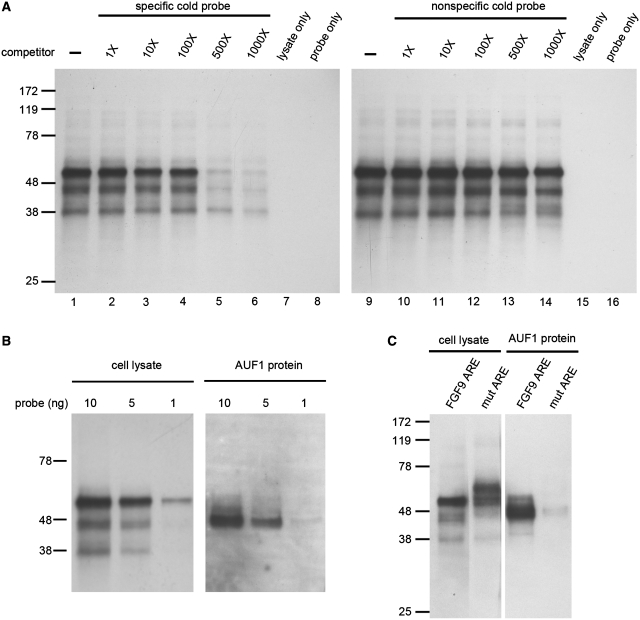
To determine whether FGF9 ARE can form protein–RNA complex, we performed UV cross-linking assays and detected three major bands corresponding to molecular masses of ∼37, 45 and 55 kDa in whole-cell extracts (Figure 3A). Competition assays showed that these three interacting complexes were quenched with specific cold probes (Figure 3A, left), but not with non-specific ones (Figure 3A, right). These data indicated the proteins are specifically binding to FGF9 ARE.
Figure 3.
AUF1 specifically binds to FGF9 3′-UTR ARE. (A) UV cross-linking to biotin-labeled riboprobe from whole-cell lysate of HEK293 cells showed three protein–RNA complexes. Competition assays were performed by adding extra non-biotin-label (cold) riboprobe as competitor with labeled probe. Left panel, specific cold probe; right panel, non-specific cold probe. (B) Dose-dependence of proteins in cell lysate (left panel) and AUF1 (right panel) binding to FGF9 ARE. (C) UV cross-linking in cell lysate (left panel) and AUF1 protein (right panel) showing linkage to proteins in the wild-type FGF9 ARE but not to the mutant ARE.
Since the molecular weights of the proteins binding to FGF9 ARE are similar to those of AUF1, we set out experiments to test whether AUF1 is the binding protein of FGF9 ARE. Commercially purified AUF1 protein was used in parallel with cell lysate to interact with FGF9 RNA probe. The AUF1–FGF9 complex showed a migration rate similar to the 45-kDa complex protein and the protein–RNA binding seems to be dose dependent (Figure 3B). Furthermore, using the mutant probe with a sequence change from AUUUAUUUA to AGGGAGGGA, the protein–RNA complexes located at 37–45 kDa disappeared, while a new complex with molecular mass of ∼60 kDa was shown (Figure 3C, left). Interestingly, the AUF1–FGF9 complex showed a migration rate similar to the 45-kDa complex protein and the protein–RNA binding seems to be abolished when the mutant probe was applied in the reaction (Figure 3B, right). Taken together, these data demonstrated that AUF1 specifically binds to FGF9 ARE and the molecular weight of AUF1–protein complex is ∼45 kDa.
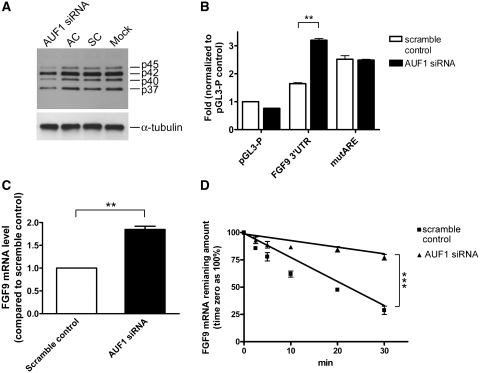
AUF1 knockdown increases FGF9 mRNA level
To determine the effect of AUF1 on regulation of FGF9 mRNA, synthetic siRNAs were used to knockdown the endogenous AUF1 in HEK293 cells. Among three exon-specific siRNAs tested, the use of AUF1 exon 1-specific siRNA resulted in the best knockdown effect (Supplementary Figure S1) and was applied in the experiments thereafter. As shown in Figure 4A, compare to the scramble (SC) and AUF1-specific siRNA controls (AC), the AUF1 exon 1-specific siRNA commonly knockdown all 4 AUF1 isoforms. Knocking down AUF1 showed a 2.5-fold increase in luciferase reporter activity in the presence of FGF9 ARE (P < 0.01; Figure 4B). In contrast, the mutARE reporter construct was not affected by AUF1 knockdown indicating the specificity of the AUF1 effect (Figure 4B). Furthermore, knocking down AUF1 also significantly increased endogenous FGF9 mRNA level to almost 2-fold (P < 0.01; Figure 4C). Using RNA stability assays, our data demonstrated that the half-life of endogenous FGF9 mRNA indeed extended from 19.5 to 60.2 min in AUF1-depleted cells (P < 0.001; Figure 4D). Collectively, these data strongly suggest that AUF1 is involved in the destabilization of human FGF9 transcripts.
Figure 4.
AUF1 controls FGF9 mRNA stability. (A) Western blots showing AUF1-specific siRNA, but not the scramble control (SC) or AUF1-specific siRNA control (AC), knocks down AUF1 protein. Mock is HEK293 cells treated with transfecting reagent only and α-tubulin was used as a control. (B) Luciferase reporter activities in empty vector (pGL3-P), full length (FGF9 3′-UTR), and mutant (mutARE) after AUF1 or scramble siRNA knockdown were shown. (C) Endogenous FGF9 mRNA levels in HEK293 cells following AUF1 or scramble siRNA knockdown. (D) Real-time RT-qPCR measurements showing the half-life of endogenous FGF9 mRNA with or without siRNA knockdown. Results are normalized to time zero for each construct. All data are shown as mean ± SEM of 4–6 independent experiments. **P < 0.01; ***P < 0.001.
p42AUF1 overexpression decreases FGF9 mRNA level
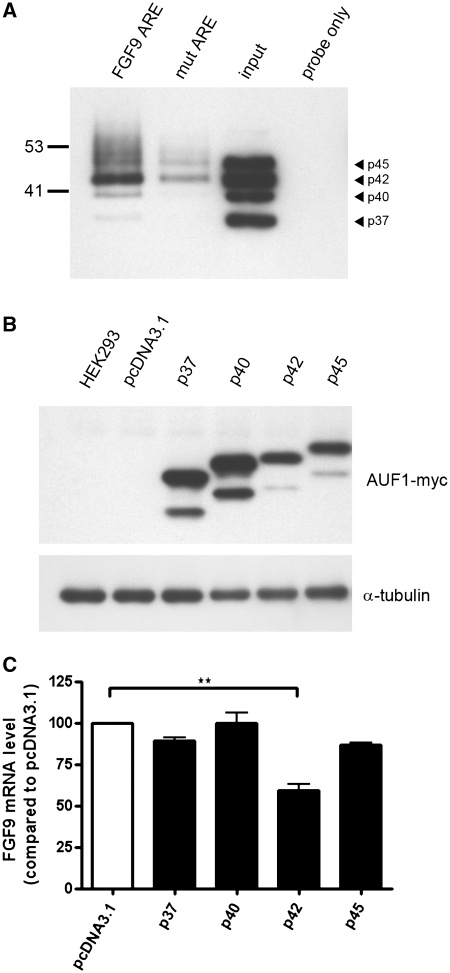
Finally, we determined whether overexpression of AUF1 has the opposite effect on FGF9 mRNA stability. A biotin pull-down assay was carried out to test the interaction between FGF9 ARE and AUF1. As expected, AUF1 was specifically interacted with FGF9 ARE sequence, especially the p42 isoform. Moreover, mutation of the core sequence from U to G resulted in a decrease of the binding affinity to <20% (Figure 5A). To study the isoform-specific interaction between FGF9 ARE and AUF1 proteins, four AUF1 isoforms were cloned and overexpressed in HEK293 cells. The expression of each isoform was confirmed by western blot analysis with anti-myc antibody (Figure 5B). Followed by RNA isolation and RT-qPCR analysis, our data showed a 50% decrease of FGF9 mRNA in cells overexpressing p42AUF1 (P < 0.01), while the other AUF1 isoforms (p37, p40 and p45) only had minor and non-significant effects on the FGF9 mRNA level (Figure 5C). As we have demonstrated, AUF1 is involved in destabilization of human FGF9 transcripts (Figure 4) and these data suggested that p42AUF1 has the ability to selectively downregulate FGF9 expression by binding to 3′-UTR ARE and destabilizing the FGF9 transcripts.
Figure 5.
AUF1 p42 isoform selectively downregulates FGF9 mRNA expression. (A) Pull-down assay showing that p42AUF1 is the major isoform pulled down by biotin-labeled FGF9 ARE probe. Arrowhead indicates the position of each AUF1 isoform. Note that the mutant is markedly weaker than the wild type. Input and probe only indicate positive and negative controls. (B) Western blots using anti-Myc antibody showing the expression of indicated recombinant AUF1 proteins. HEK293 and pcDNA3.1 refer to cells without transfection and cells transfected with empty vector, respectively. (C) Endogenous FGF9 mRNA levels relative to control (pcDNA3.1) in cells overexpressing the AUF1 isoforms p37, p40, p42 and p45. **P < 0.01.
AUF1 contributes to the regulation of PGE2-mediated induction of FGF9 mRNA
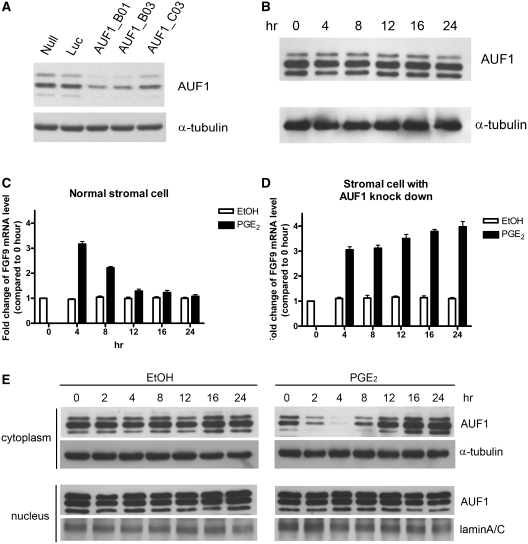
We previously showed that PGE2 induces an acute and transient increase of FGF9 mRNA in human primary endometrial stromal cells (19). We hypothesized that AUF1 is involved in the reduction of FGF9 mRNA in stromal cells. To test this hypothesis, we knocked down AUF1 by shRNA interference in primary endometrial stromal cells. Western blot analysis revealed that the AUF1 abundance was reduced by 60–90% in primary stromal cells infected with AUF1 shRNA (Figure 6A). We then determined whether knockdown of AUF1 affects FGF9 mRNA expression in non-stimulated cells or cells exposed to PGE2. In agreement with previous report, administration of 1 μM PGE2 enhanced FGF9 mRNA by 3-fold; the level peaked at 4 h and then declined toward the basal level by 12 h (P < 0.001; Figure 6B). Interestingly, AUF1 knockdown caused a prolonged elevation of PGE2-induced FGF9 mRNA level (P < 0.001; Figure 6C). The FGF9 mRNA remained elevated even at 24 h after PGE2 treatment.
Figure 6.
AUF1 knockdown sustains the PGE2-medicated induction of FGF9 mRNA in human primary endometrial stromal cells. (A) Western blots showing successful AUF1 knockdown by AUF1-specific shRNA (B01, B03 and C03), but not the Luc control shRNA. The Null indicates cells without shRNA treatment. (B) Western blots showing AUF1 protein expression in whole-cell lysates isolated from stromal cells after PGE2 treatment. (C) FGF9 mRNA levels measured at indicated times after PGE2 treatment in stromal cells. Ethanol (EtOH) was used as control treatment. (D) Experiment similar to that shown in C, but with AUF1 knockdown by AUF1_B03 shRNA. All data are shown as mean ± SEM of 4–6 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001. (E) Western blots showing the distribution pattern of AUF1 protein in stromal cells after PGE2 treatment. α-Tubulin and LaminA/C used as loading controls for proteins isolated from cytoplasmic and nuclear fractions, respectively.
To reveal the mechanism underlying this regulation, we measured the expression of AUF1 after PGE2 treatment in whole-cell lysates, and found that PGE2 had no effect on AUF1 expression in primary stromal cells (Figure 6D). This suggested that other mechanisms, for example, decreased amounts of cytoplasmic AUF1 (35) or phosphorylation of AUF1 proteins (36), are involved in the regulation of PGE2-induced excess of FGF9 mRNA. Further examination revealed that the levels of AUF1 phosphorylation were not changed in non-stimulated or PGE2-treated cells (Supplementary Figure S2). Nevertheless, western blot data showed that PGE2 significantly reduce AUF1 amount in the cytoplasmic fraction while no effect was detected in the control experiments (Figure 6E). Moreover, the time-dependent, PGE2-mediated decline of cytoplasmic AUF1 showed perfect correlation with the reduction of FGF9 mRNA during the time course of this stimulation (Figure 6B and E). These results demonstrated that AUF1, through the change of its cellular distribution, plays a vital role in controlling FGF9 mRNA level in PGE2-induced FGF9 transcription.
DISCUSSION
Fibroblast growth factor 9 plays numerous physiological and pathological roles in many species (37). Considering that FGF9 controls many vital functions in developmental, physiological and pathological processes, it is important to study the molecular mechanisms responsible for regulation of FGF9 gene expression. Previously, we reported that PGE2, via its EP2 receptor-mediated signaling pathway, transcriptionally upregulates FGF9 gene expression (19). Although the data provide valuable information for understanding the regulation of FGF9 gene expression, it cannot answer the question of why upregulation of FGF9 is transient. In this study, we showed that a nonamer AU-rich element (AUUUAUUUA) within the 3′-UTR of the FGF9 transcript is involved in rapid mRNA turnover. RNA–protein interaction and overexpression studies revealed that p42AUF1 is the major isoform of the AUF1 protein family that binds to FGF9 ARE to selectively regulate the levels of FGF9 mRNA. Our data provided direct evidence to show that AUF1 acts as a negative regulator for FGF9 expression post-transcriptionally. To our knowledge, this is the first report to demonstrate a post-transcriptional regulation of FGF9 in any species. Our results provide the molecular and cellular mechanism underlying the homeostasis of FGF9 mRNA level in cells, an important balance between the normal physiological function and abnormal pathological process.
We determined that the half-life of FGF9 mRNA is <20 min, a typical characteristic of the mRNAs of tightly regulated genes, which indicates that a post-transcriptional regulatory mechanism exists to control the lifespan of FGF9 mRNA. In addition, FGF9 is a potent peptide growth factor that stimulates the proliferation of many cells including endometrial epithelium and stroma, prostate cancer cells and neuronal cells (19). However, these effects must be controlled during a short period of time to avoid overstimulation due to sustained elevation of FGF9. PGE2 is a versatile eicosanoid that regulates key responses in numerous physiological and pathological processes (38). Results from the present study showed that AUF1 is able to bind and reduce FGF9 mRNA to the basal level once it is transcriptionally activated by PGE2. And AUF1 knockdown leads to maintained FGF9 mRNA levels after activation. These findings indicate that AUF1 stops FGF9 signaling by destabilizing FGF9 mRNA and thus reducing the amount of FGF9 protein synthesis. Taken together, these data also suggest that AUF1 precisely regulates FGF9 signaling by terminating the effect of environmental stimuli on FGF9. This is a very important safeguarding system to control FGF9 signaling within a precise time window or at a certain developmental stage. Therefore, loss of this control may prolong the effect of FGF9 and contribute to disease processes. Reports of other AUF1 target mRNAs from other groups are consistent with this theory. For instance, alteration of the U-rich region in the 3′-UTR of Bcl-2 mRNA increases mRNA stability and contributes to the overproduction of the anti-apoptotic Bcl-2 protein that is responsible for the transformation of follicular B-cell lymphoma (39,40). In addition, AUF1 knockout mice display symptoms of severe endotoxic shock, including vascular hemorrhage, intravascular coagulation and high mortality, upon endotoxin challenge (41) suggesting that the lack of this negative feedback system by AUF1 indeed results in disease development and detrimental effects.
AUF1, the most extensively studied AU-rich element binding protein, is a shuttle protein (42,43) with multiple functions, such as DNA binding (44), RNA turnover (45) and mRNA translation efficiency (46). The AUF1 family contains four isoforms (p37, p40, p42 and p45) resulting from alternative splicing. Different isoforms vary significantly in their cellular distribution and binding affinity to ARE-containing mRNA. It was suggested that relative level, rather than the absolute amount of individual AUF1isoform, determine the net mRNA stability of ARE-containing transcripts (47). In this study, we showed that the p40/p42 proteins are the predominant isoforms in both HEK293 and human primary endometrial stromal cells. In addition, we have shown that p42 isoform specifically suppresses FGF9 mRNA stability compared with the other isoforms. Our results concur with several lines of evidence that demonstrate certain transcripts are under isoform-specific regulation by the AUF1 family. For example, ectopic overexpression of the p37 isoform selectively degrades ARE-containing mRNA (42), the p45 isoform selectively binds to estrogen receptor mRNA to upregulate gene expression (48) and the p40 isoform selectively and positively regulates interleukin (IL) 10 expression in monocytes (49). Although it is known that different AUF1 isoforms have different binding affinity to specific mRNA 3′-UTRs (50,51), the detailed mechanism by which AUF1 isoforms selectively regulate FGF9 mRNA remains unclear. In addition, a time-dependent decline in cytoplasmic AUF1 was observed along with the increase of FGF9 by PGE2 treatment, whereas the control experiments by ethanol kept at relatively steady-state levels over the time courses. The functional significance of changing cellular distribution of AUF1 still requires further investigation. Nevertheless, similar changes in AUF1 decreasing cytoplasmic level have been associated with hydrogen peroxide (H2O2)-induced p16 expression (35) and in thyroid carcinoma progression (52).
The concept of RNA regulation has currently gained much attention and attempts have been made to explain the complex mechanisms of gene regulation at the post-transcription level (53). The fate of transcripts with similar functions may be under the same control mechanism, such as the PUF RNA-binding protein family (53). The FGF gene family members share a high degree of conservation and are involved in diverse physiological events (37). However, factors that regulate the expression of FGFs remain largely elusive. Previous studies show that many growth factors and signal traducers are ARE-containing transcripts in the human genome [review see (54)]. Using computational analysis (55), our data indicated that nine members of the FGF family (FGF2, 4, 5, 7, 9, 14, 18, 19, 21 and 23) are all ARE-containing genes (Supplementary Table S1). Although only FGF9 ARE is proved to be functional and plays a vital role in FGF9 regulation in this study, these data suggest that ARE-mediated decay is a common pathway to control expression of FGFs. Further, it also suggests that the ARE-mediated decay pathway is another example of RNA regulon that coordinates several physiological conditions through post-transcriptional regulation.
In summary, we have identified a novel mechanism to regulate FGF9 steady-state and stimulus-induced expression by AUF1-mediated mRNA decay. Considering that FGF9 is a potent growth and survival factor, it is not surprising that its expression is tightly controlled by many factors such as transcription factors, microsatellite motifs, and, as reported in this study, the RNA destabilizing protein, AUF1. Our findings provide a new point of view, that some diseases induced by overexpression of peptide growth factors (such as FGF9) might be due to loss-of-function of a negative regulator, for example, an RNA destabilizing protein like AUF1. Nevertheless, it is worth noting that we observed three RNA–protein complexes in the UV cross-linking assay using FGF9 ARE probe. It suggests other ARE-binding proteins may also play some roles in regulation of FGF9 mRNA stability. Among all identified ARE-binding proteins, both HuR (56) and KSRP (57) are known to interact with AUF1 and coordinately regulate ARE-containing mRNA stability. It is of interest to study whether HuR or KSRP binds to FGF9 ARE and control its expression in future investigations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Science Council, Taiwan (NSC 98-3112-B-006-002 to H.S.S.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Iain Bruce for the excellent help in English editing and National RNAi Core Facility, Taiwan for various RNAi constructs.
REFERENCES
- 1.Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat. Rev. Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldfarb M. Functions of fibroblast growth factors in vertebrate development. Cytokine Growth Factor Rev. 1996;7:311–325. doi: 10.1016/s1359-6101(96)00039-1. [DOI] [PubMed] [Google Scholar]
- 3.Gerwins P, Skoldenberg E, Claesson-Welsh L. Function of fibroblast growth factors and vascular endothelial growth factors and their receptors in angiogenesis. Crit. Rev. Oncol. Hematol. 2000;34:185–194. doi: 10.1016/s1040-8428(00)00062-7. [DOI] [PubMed] [Google Scholar]
- 4.Allouche M, Bikfalvi A. The role of fibroblast growth factor-2 (FGF-2) in hematopoiesis. Prog. Growth Factor Res. 1995;6:35–48. doi: 10.1016/0955-2235(95)00041-0. [DOI] [PubMed] [Google Scholar]
- 5.Galzie Z, Kinsella AR, Smith JA. Fibroblast growth factors and their receptors. Biochem. Cell Biol. 1997;75:669–685. [PubMed] [Google Scholar]
- 6.Song J, Slack JM. XFGF-9: a new fibroblast growth factor from Xenopus embryos. Dev. Dyn. 1996;206:427–436. doi: 10.1002/(SICI)1097-0177(199608)206:4<427::AID-AJA8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 7.Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–569. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Kanda T, Iwasaki T, Nakamura S, Kurokawa T, Ikeda K, Mizusawa H. Self-secretion of fibroblast growth factor-9 supports basal forebrain cholinergic neurons in an autocrine/paracrine manner. Brain Res. 2000;876:22–30. doi: 10.1016/s0006-8993(00)02563-4. [DOI] [PubMed] [Google Scholar]
- 9.Naruo K, Seko C, Kuroshima K, Matsutani E, Sasada R, Kondo T, Kurokawa T. Novel secretory heparin-binding factors from human glioma cells (glia-activating factors) involved in glial cell growth. Purification and biological properties. J. Biol. Chem. 1993;268:2857–2864. [PubMed] [Google Scholar]
- 10.Tsai SJ, Wu MH, Chen HM, Chuang PC, Wing LY. Fibroblast growth factor-9 is an endometrial stromal growth factor. Endocrinology. 2002;143:2715–2721. doi: 10.1210/endo.143.7.8900. [DOI] [PubMed] [Google Scholar]
- 11.Miyamoto M, Naruo K, Seko C, Matsumoto S, Kondo T, Kurokawa T. Molecular cloning of a novel cytokine cDNA encoding the ninth member of the fibroblast growth factor family, which has a unique secretion property. Mol. Cell Biol. 1993;13:4251–4259. doi: 10.1128/mcb.13.7.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colvin JS, Feldman B, Nadeau JH, Goldfarb M, Ornitz DM. Genomic organization and embryonic expression of the mouse fibroblast growth factor 9 gene. Dev. Dyn. 1999;216:72–88. doi: 10.1002/(SICI)1097-0177(199909)216:1<72::AID-DVDY9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 13.Hendrix ND, Wu R, Kuick R, Schwartz DR, Fearon ER, Cho KR. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 2006;66:1354–1362. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]
- 14.Kathpalia VP, Mussak EN, Chow SS, Lam PH, Skelley N, Time M, Markelewicz RJ, Jr, Kanduc D, Lomas L, Xiang Z, et al. Genome-wide transcriptional profiling in human squamous cell carcinoma of the skin identifies unique tumor-associated signatures. J. Dermatol. 2006;33:309–318. doi: 10.1111/j.1346-8138.2006.00075.x. [DOI] [PubMed] [Google Scholar]
- 15.Wing LY, Chuang PC, Wu MH, Chen HM, Tsai SJ. Expression and mitogenic effect of fibroblast growth factor-9 in human endometriotic implant is regulated by aberrant production of estrogen. J. Clin. Endocrinol. Metab. 2003;88:5547–5554. doi: 10.1210/jc.2003-030597. [DOI] [PubMed] [Google Scholar]
- 16.Huang JY, Hong YT, Chuang JI. Fibroblast growth factor 9 prevents MPP+-induced death of dopaminergic neurons and is involved in melatonin neuroprotection in vivo and in vitro. J. Neurochem. 2009;109:1400–1412. doi: 10.1111/j.1471-4159.2009.06061.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen TM, Kuo PL, Hsu CH, Tsai SJ, Chen MJ, Lin CW, Sun HS. Microsatellite in the 3′ untranslated region of human fibroblast growth factor 9 (FGF9) gene exhibits pleiotropic effect on modulating FGF9 protein expression. Hum. Mutat. 2007;28:98. doi: 10.1002/humu.9471. [DOI] [PubMed] [Google Scholar]
- 18.Lin YM, Tsai CC, Chung CL, Chen PR, Sunny Sun H, Tsai SJ, Huang BM. Fibroblast growth factor 9 stimulates steroidogenesis in postnatal Leydig cells. Int. J. Androl. 2010;33:545–553. doi: 10.1111/j.1365-2605.2009.00966.x. [DOI] [PubMed] [Google Scholar]
- 19.Chuang PC, Sun HS, Chen TM, Tsai SJ. Prostaglandin E2 induces fibroblast growth factor 9 via EP3-dependent protein kinase Cdelta and Elk-1 signaling. Mol. Cell Biol. 2006;26:8281–8292. doi: 10.1128/MCB.00941-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giri D, Ropiquet F, Ittmann M. FGF9 is an autocrine and paracrine prostatic growth factor expressed by prostatic stromal cells. J. Cell Physiol. 1999;180:53–60. doi: 10.1002/(SICI)1097-4652(199907)180:1<53::AID-JCP6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 21.Seo M, Noguchi K. Retinoic acid induces gene expression of fibroblast growth factor-9 during induction of neuronal differentiation of mouse embryonal carcinoma P19 cells. FEBS Lett. 1995;370:231–235. doi: 10.1016/0014-5793(95)00836-x. [DOI] [PubMed] [Google Scholar]
- 22.Ross J. Control of messenger RNA stability in higher eukaryotes. Trends Genet. 1996;12:171–175. doi: 10.1016/0168-9525(96)10016-0. [DOI] [PubMed] [Google Scholar]
- 23.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 24.Xu N, Chen CY, Shyu AB. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol. Cell Biol. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson T, Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature. 1988;336:396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- 26.Wu MH, Shoji Y, Chuang PC, Tsai SJ. Endometriosis: disease pathophysiology and the role of prostaglandins. Expert Rev. Mol. Med. 2007;9:1–20. doi: 10.1017/S146239940700021X. [DOI] [PubMed] [Google Scholar]
- 27.Chen CY, Xu N, Shyu AB. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bevilacqua A, Ceriani MC, Capaccioli S, Nicolin A. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J. Cell Physiol. 2003;195:356–372. doi: 10.1002/jcp.10272. [DOI] [PubMed] [Google Scholar]
- 29.Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai WS, Kennington EA, Blackshear PJ. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell Biol. 2003;23:3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Ale-Agha N, Galban S, Sobieroy C, Abdelmohsen K, Gorospe M, Sies H, Klotz LO. HuR regulates gap junctional intercellular communication by controlling beta-catenin levels and adherens junction integrity. Hepatology. 2009;50:1567–1576. doi: 10.1002/hep.23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mignone F, Grillo G, Licciulli F, Iacono M, Liuni S, Kersey PJ, Duarte J, Saccone C, Pesole G. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2005;33:D141–D146. doi: 10.1093/nar/gki021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3:REVIEWS0004. doi: 10.1186/gb-2002-3-3-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo GE, Ma LW, Jiang B, Yi J, Tong TJ, Wang WG. Hydrogen peroxide induces p16(INK4a) through an AUF1-dependent manner. J. Cell Biochem. 2010;109:1000–1005. doi: 10.1002/jcb.22474. [DOI] [PubMed] [Google Scholar]
- 36.Wilson GM, Lu J, Sutphen K, Sun Y, Huynh Y, Brewer G. Regulation of A + U-rich element-directed mRNA turnover involving reversible phosphorylation of AUF1. J. Biol. Chem. 2003;278:33029–33038. doi: 10.1074/jbc.M305772200. [DOI] [PubMed] [Google Scholar]
- 37.Itoh N, Ornitz DM. Functional evolutionary history of the mouse Fgf gene family. Dev. Dyn. 2008;237:18–27. doi: 10.1002/dvdy.21388. [DOI] [PubMed] [Google Scholar]
- 38.Park JY, Pillinger MH, Abramson SB. Prostaglandin E2 synthesis and secretion: the role of PGE2 synthases. Clin. Immunol. 2006;119:229–240. doi: 10.1016/j.clim.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Lapucci A, Donnini M, Papucci L, Witort E, Tempestini A, Bevilacqua A, Nicolin A, Brewer G, Schiavone N, Capaccioli S. AUF1 Is a bcl-2 A + U-rich element-binding protein involved in bcl-2 mRNA destabilization during apoptosis. J. Biol. Chem. 2002;277:16139–16146. doi: 10.1074/jbc.M201377200. [DOI] [PubMed] [Google Scholar]
- 40.Shchors K, Yehiely F, Kular RK, Kotlo KU, Brewer G, Deiss LP. Cell death inhibiting RNA (CDIR) derived from a 3′-untranslated region binds AUF1 and heat shock protein 27. J. Biol. Chem. 2002;277:47061–47072. doi: 10.1074/jbc.M202272200. [DOI] [PubMed] [Google Scholar]
- 41.Lu JY, Sadri N, Schneider RJ. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 2006;20:3174–3184. doi: 10.1101/gad.1467606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarkar B, Lu JY, Schneider RJ. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J. Biol. Chem. 2003;278:20700–20707. doi: 10.1074/jbc.M301176200. [DOI] [PubMed] [Google Scholar]
- 43.Chen CY, Xu N, Zhu W, Shyu AB. Functional dissection of hnRNP D suggests that nuclear import is required before hnRNP D can modulate mRNA turnover in the cytoplasm. RNA. 2004;10:669–680. doi: 10.1261/rna.5269304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Enokizono Y, Konishi Y, Nagata K, Ouhashi K, Uesugi S, Ishikawa F, Katahira M. Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J. Biol. Chem. 2005;280:18862–18870. doi: 10.1074/jbc.M411822200. [DOI] [PubMed] [Google Scholar]
- 45.DeMaria CT, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 46.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 47.Raineri I, Wegmueller D, Gross B, Certa U, Moroni C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ing NH, Massuto DA, Jaeger LA. Estradiol up-regulates AUF1p45 binding to stabilizing regions within the 3′-untranslated region of estrogen receptor alpha mRNA. J. Biol. Chem. 2008;283:1764–1772. doi: 10.1074/jbc.M704745200. [DOI] [PubMed] [Google Scholar]
- 49.Sarkar S, Sinsimer KS, Foster RL, Brewer G, Pestka S. AUF1 isoform-specific regulation of anti-inflammatory IL10 expression in monocytes. J. Interferon. Cytokine Res. 2008;28:679–691. doi: 10.1089/jir.2008.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics. 1998;48:195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- 51.Kajita Y, Nakayama J, Aizawa M, Ishikawa F. The UUAG-specific RNA binding protein, heterogeneous nuclear ribonucleoprotein D0. Common modular structure and binding properties of the 2xRBD-Gly family. J. Biol. Chem. 1995;270:22167–22175. doi: 10.1074/jbc.270.38.22167. [DOI] [PubMed] [Google Scholar]
- 52.Trojanowicz B, Brodauf L, Sekulla C, Lorenz K, Finke R, Dralle H, Hoang-Vu C. The role of AUF1 in thyroid carcinoma progression. Endocr. Relat. Cancer. 2009;16:857–871. doi: 10.1677/ERC-08-0234. [DOI] [PubMed] [Google Scholar]
- 53.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 54.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Halees AS, El-Badrawi R, Khabar KS. ARED Organism: expansion of ARED reveals AU-rich element cluster variations between human and mouse. Nucleic Acids Res. 2008;36:D137–D140. doi: 10.1093/nar/gkm959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan YX, Chen H, Kilberg MS. Interaction of RNA-binding proteins HuR and AUF1 with the human ATF3 mRNA 3′-untranslated region regulates its amino acid limitation-induced stabilization. J. Biol. Chem. 2005;280:34609–34616. doi: 10.1074/jbc.M507802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nechama M, Ben-Dov IZ, Briata P, Gherzi R, Naveh-Many T. The mRNA decay promoting factor K-homology splicing regulator protein post-transcriptionally determines parathyroid hormone mRNA levels. FASEB J. 2008;22:3458–3468. doi: 10.1096/fj.08-107250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.