Abstract
Mouse transgenesis has proven invaluable for analysis of gene function and generation of human disease models. We describe here the development of a pronuclear injection-based targeted transgenesis (PITT) system, involving site-specific integration in fertilized eggs. The system was applied to two different genomic target loci to generate a series of transgenic lines including fluorescent mice, which reproducibly displayed strong, ubiquitous and stable transgene expression. We also demonstrated that knockdown mice could be readily generated by PITT by taking advantage of the reproducible and highly efficient expression system. The PITT system, which circumvents the problem of unpredictable and unstable transgene expression of conventional random-integration transgenic mice, reduces the time, cost and effort needed to generate transgenic mice, and is potentially applicable to both in vivo ‘gain-of-function’ and ‘loss-of-function’ studies.
INTRODUCTION
Transgenic mice have been extensively used for in vivo analysis of gene function and generation of human disease models (1). Pronuclear injection (PI) is the most common method used to generate transgenic mice (2,3), although the wide variability in the level and pattern of transgene expression that can influence the phenotype often varies strongly, due to the random nature of copy number, configuration and integration site of the transgene (4,5). The effect of the insertion site on the transgene expression can be minimized by including insulator sequences into the transgenic construct (6,7), or by using large DNA constructs such as Bacterial Artificial Chromosome (BAC) (8). Even in such strategies, transgene insertion can alter the expression and/or function of endogenous genes at the integrated region. Therefore, researchers must produce at least five lines for screening of lines with adequate transgene expression and for obtaining reproducible and firm results. The targeted integration of a single-copy transgene has become recently possible through homologous recombination or site-specific recombination in embryonic stem (ES) cells. Such targeted transgenesis is advantageous for achieving predictable and reproducible transgene expression (4,9,10). However, the targeted system is more technically demanding and time-consuming than PI-based transgenesis.
The present study established a PI-based targeted transgenesis (PITT) system based on site-specific recombination in fertilized eggs, but not in ES cells. By applying our PITT method, we established a number of transgenic lines, including fluorescent mice of which transgene expression was both predictable and reproducible. We also successfully applied the technique to generate knockdown mice. Because of its simplicity and time- and cost-effectiveness, the PITT system is a potentially useful first choice to achieve ‘gain-of-function’ and ‘loss-of-function’ of gene of interests.
MATERIALS AND METHODS
Plasmid construction
The sequences and maps of the plasmids used in the present study are available upon request. Plasmids containing site-specific recombination sites, drug-resistant genes, reporter genes and other components were generated through a multi-step process of ligation-based cloning. Site-specific recombination sites, FRT, JT15, JTZ17 (11) and lox2272 (12), were generated by using synthesized oligos. Neomycin-resistant gene (neo), hygromycin-resistant gene (hyg), EGFP gene, lacZ gene, internal ribosome entry site (IRES) sequence, tau and splice acceptor (SA) site were derived from pPGK-neo-loxP (Gene Bridges, Heidelberg, Germany), pJR225, pCE-29 (13), pIZA2 (kindly provided by Dr Noguchi, Meiji Milk Co., Tokyo, Japan), pCAGGS-FLPe (14) (Gene Bridges), tauEGFP plasmid (kindly provided by Dr Hitoshi Sakano) and pSAbgeo (15) (kindly provided by Dr Philippe Soriano), respectively. The genes encoding fluorescent proteins used in this study were either kindly provided by Dr Roger Tsien (tdTomato, mCFP, mCitrine) (16–18) or purchased from Clontech (Mountain View, CA) (mOrange, pECFP-Nuc, pEYFP-Nuc) and MBL (Nagoya, Japan) (Kikume Green-Red). The nucleotide sequences of junctional portions after cloning and PCR-amplified regions were confirmed by sequencing.
The plasmids used to check for possible recombination between heterotypic lox sites were generated as follows. The SacB gene (pDNR-Dual, Clontech) was amplified by PCR using the primer sets M730 and M734, M730 and M735, M731 and M736, M732 and M736, and M733 and M736 (Supplementary Table S1). The amplified fragments were cloned into the SmaI site of pUC119 vector, generating pAYT (loxP-loxP), pAYU (loxP-lox511), pAYV (JT15-lox2272), pAYW (JTZ17-lox2272) and pAYX (JT15/JTZ17-lox2272).
The targeting construct for H2-Tw3 gene (19) was generated by modifying the citb585c7 BAC clone (from a CITB library derived from the 129/SV mouse strain, Research Genetics/Invitrogen, Carlsbad, CA, USA) using recombineering method with the DY380 Escherichia coli (20) and the plasmids, pADY and pAEF (21). The protocols for recombineering fragments, of which homology regions (HRs) were PCR-amplified from the BAC clone with the following primer sets: (M393 and M394 for HR1, M395 and M396 for HR2, M397 and M398 for HR3 and M399 and M155 for HR4; Supplementary Table S1), were generated based on the protocols described previously by our group (21). The recombineering protocol was based on the protocol of Liu et al. (20). The resulting targeting vector (pAFP) had the following components in a 5′ to 3′ direction: I-SceI site, long arm, FRT, PGK-Tn5 promoter, neo, JT15, IRES, ECFP-Nuc, polyA site, lox2272, short arm and diphtheria toxin fragment A (DT-A) cassette.
The targeting construct for Rosa26 locus (22) was generated as follows. BAC clones containing this locus were obtained by PCR screening of 129/Ola BAC library (23). The BAC clone 124I18 was used for recombineering. HRs for recombineering fragments were PCR-amplified from the BAC clone with the following primer sets: M058 and M057 for HR1, M056 and M055 for HR2, M060 and M059 for HR3 and M054 and M053 for HR4 (Supplementary Table S1). Recombineering fragments were prepared according to the method described previously by our group (21), and used for targeting vector construction. The resulting targeting vector (pAIW) had the following components in a 5′ to 3′ direction: DT-A cassette, short arm, SA, FRT sequence-fusion neo, JT15, IRES, ECFP-Nuc, polyA site, lox2272, long arm and I-SceI site.
The donor vectors had the following components in a 5′ to 3′ direction: hygromycin-resistance cassette (CAG promoter, hyg, polyA site), FRT, DNA of interest (DOI), lox2272, JTZ17, IRES, reporter gene, polyA site and a plasmid backbone. As for the promoterless reporter gene, lacZ gene was used for pAJK, pAMF, pAKB, pAMG, pAMJ, pANQ, pAOB, pAOF, pAOK, pAOL, pAOM, pAOT and pAOU, EGFP gene was used for pA748 and EYFP-Nuc gene was used for pA617. In the DOI region, the following cassettes were included in each donor vector: ‘dual SacB genes with GFPuv’ (21) in pAJK and pA748, ‘CMV-HcRed1-polyA’ in pA617, ‘CAG-EGFP-polyA’ in pAMF, ‘FRT-fusion EGFP-polyA’ in pAKB, ‘CAG-EGFP-miR(Hr-1)-polyA’ in pAMG, ‘CAG-EGFP-miR(Hr-2)-polyA’ in pAMJ, ‘CAG-tau/kikume-polyA’ in pANQ, ‘CAG-mCherry-polyA’ in pAOB, ‘CAG-mCitrine-polyA’ in pAOF, ‘CAG-mCFP-polyA’ in pAOK, ‘CAG-mOrange-polyA’ in pAOL, ‘CAG-tdTomato-polyA’ in pAOM, ‘CAG-EGFP-miR(Tyr-1/2)-polyA’ in pAOT and ‘CAG-EGFP-miR(Tyr-3/4)-polyA’ in pAOU.
Tests for possible recombination between heterotypic lox sites
In these experiments, 1/50 volume of the vectors (pAYT, pAYU, pAYV, pAYW and pAYX) isolated from 5 ml of overnight culture, was incubated at 37°C for 60 min with 1 U of Cre recombinase (New England Biolabs, Beverly, MA, USA) in a total volume of 10 µl. After incubation of the reaction mixture at 70°C for 10 min, the presence of Cre-mediated recombination was checked by PCR with primer set M272 and M322 (Supplementary Table S1) using 0.5 µl of reaction mixture as template. From recombined and non-recombined plasmid, 262-bp and 2225-bp fragments were amplified, respectively. Furthermore, 0.5 µl of Cre reaction mixture was also transformed into 5 µl of DH5α (TaKaRa). Following incubation at 37°C in SOC medium, one-fifth of the samples were plated on LB plates containing ampicillin (100 µg/ml), sucrose (7%) and 1/200th of the samples was plated on LB plates containing ampicillin (100 µg/ml). Colonies were counted, and those on ampicillin/sucrose plates were subjected to PCR analysis with M272 or M322 primer set.
Targeting and genotyping of H2-Tw3 and Rosa26 loci
The H2-Tw3 targeting vector (20 µg) linearized with I-SceI was introduced into 1 × 107 E14tg2a ES cells (24) by electroporation. G418-resistant ES cell colonies were picked and subjected to the first screening by PCR with primers #145 and M400 (Supplementary Table S1). The presence of correctly targeted clones was confirmed by Southern blot analysis using a 32P-labeled probe generated by random prime labeling of PCR product amplified from BAC DNA with primers M153 and M154 (Supplementary Table S1). One of the targeted ES clones (3D2) was microinjected into the blastocoels of mouse F1 (C57BL/6J × BDF1) blastocysts to produce chimeric mice. Germ-line transmission was confirmed by crossing male chimeric mice to C57BL/6J female mice. Heterozygous mice were intercrossed to obtain homozygous mice. Genotyping of the targeted allele was performed by PCR with primer set #166 and #142 (Supplementary Table S1). The H2-Tw3 wild-type allele was detected by TaqI digestion of the PCR product amplified with primer set M401 and M402 (Supplementary Table S1).
With regard to the Rosa26 locus, the knock-in vector (20 µg) linearized with I-SceI was introduced into 1 × 107 E14tg2a ES cells by electroporation. G418-resistant ES cell colonies were picked and subjected to the first screening by PCR with primers M195 and M024 (Supplementary Table S1). Two of the obtained targeted ES clones (2A5 and 3D6) were microinjected into mouse F1 (C57BL/6J × BDF1) blastocysts. Germ-line transmission was confirmed by crossing male chimeric mice with C57BL/6J female mice. The heterozygous mice were intercrossed to obtain homozygous mice. The wild-type and targeted allele were genotyped by PCR with primers #145, M273 and M274 (Supplementary Table S1), which gave rise to 429-bp fragment from the wild-type allele and 522-bp fragment from the targeted allele. The presence of successfully targeted mice was confirmed by Southern blot analysis using a DIG-labeled probe amplified by PCR from BAC DNA with primers M372 and M373 for short arm, and M374 and M375 for long arm (Supplementary Table S1).
RMCE in ES cells
Two methods for delivery of plasmid DNA to ES cells were used in this study; electroporation and lipofection. In all experiments, including those of gene targeting, ES cells were cultured in the presence of feeder cells derived from DR-4 strain (25). For electroporation, 5 × 106 of ES cells carrying the targeted locus were electroporated with 10 µg of Cre-expression vector (pCAG/NCre) (26) and 10 µg of donor vector at 200 V and 500 µF. After 24 h, hygromycin was added to the medium to a final concentration of 200 µg/ml to begin selection. Individual clones were picked about 1 week after electroporation, grown in 24-well plates, and finally expanded to generate duplicate plates for freezing, DNA analysis and/or X-gal staining (X-Gal Staining Assay Kit: Genlantis). For lipofection, 1.5–2.6 × 105 of ES cells were seeded onto one well of 6-well plates. After 18 h, 5 µg of the donor vector was Lipofectamin 2000 (Invitrogen)-treated with or without 5 µg of pCAG/NCre, according to the user manual supplied by the manufacturer. The next day, hygromycin selection (200 µg/ml) was initiated. One or 2 days after transfection, ES cells were re-plated onto 10-cm dishes and hygromycin selection continued. Colonies were stained with X-gal and others were picked up for cell propagation used for cell freezing and DNA analysis. Only EGFP-fluorescent ES colonies were picked for transfection with pA748.
PITT
Donor plasmids and pCAG/NCre were prepared using a HiSpeed Plasmid Midi Kit (Qiagen, Hilden, Germany). The optimal concentrations of Cre plasmids for PI were determined by injecting various concentrations (1, 2.5, 5 and 10 ng/µl) of pCAG/NCre into the fertilized eggs obtained from mating ICR female and BDF1 male mice. The injected eggs were cultured up to blastocyst stage, and the concentration of pCAG/NCre (5 ng/µl), which allowed >70% eggs to survive, was employed for further experiments. The concentration of the donor vector used for PITT was determined by injection of various concentrations (5, 10 and 20 ng/µl) of pAOF and 5 ng/µl of pCAG/NCre into the ICR × BDF1 fertilized eggs. These treated eggs were cultured up to the morula stage, and the concentration of pAOF (5–10 ng/µl), which allowed >30% eggs to survive, was employed for PITT experiments.
To perform PITT, the plasmid mixture was diluted to 10–15 ng/µl (5 or 10 ng/µl for donor vector and 5 ng/µl for Cre plasmid) in injection buffer (5 mM Tris/0.1 mM EDTA, pH 7.5). The fertilized eggs were isolated from super-ovulating BDF1 female mice that had been mated with males of the H2-Tw3 or Rosa26 homozygous targeted mice. The plasmid mixture was introduced into eggs by microinjection using a standard protocol (27). The microinjected and surviving embryos were then cultured overnight. The next day, embryos that developed into the two-cell stage were transferred to the oviducts of a foster mother. The resulting offspring was then analyzed for the integration of the transgene into the genome at 3 weeks of age, at the time of weaning.
Detection of successful RMCE in ES cells and mice
To detect correct RMCE in recombinants, the primer sets were designed to amplify the junctional region generated by recombination. RMCE-mediated integration of donor vectors into the H2-Tw3 locus was checked by PCR using the primer sets #169 and M396, and #166 and M159 for pAJK integration, #169 and M396, and BbvI digestion of PCR product amplified using primers #166 and M026 for pA748 integration, M025 and M396 for pA617 integration, and #166 and M159, and M396 and M124 for pAMF integration (Supplementary Table S1). X-gal staining of ES clones was also conducted for recombinants with pAJK and pAMF integrations. Donor vector integration into the Rosa26 locus was detected by PCR using the primer sets #169 and M058, and BbvI digestion of PCR product amplified using primers #166 and M026 for pA748 integration, M124 and M058, and #166 and M159 for pAMF integration, #143 and M058, and #166 and M159 for pAKB integration, M124 and M274, and #166 and M159 for pAMG, pAMJ, pANQ, pAOB, pAOF, pAOK, pAOL, pAOM, pAOT and pAOU integration (Supplementary Table S1). X-gal staining of ES clones was also conducted for recombinants with pAJK, pAMF or pAKB integration.
The founder mouse containing targeted transgene integrated by partial RMCE event was crossed with a CAG-Cre transgenic mouse (28) to obtain mice with complete RMCEex allele. The possibility of random integration of the donor vector and the Cre-expression plasmid in founder pups was checked by PCR-based assay using the primer sets M132 and M194, and #142 and M124 for the donor vector, and M515 and M581 for the Cre-expression plasmid (Supplementary Table S1).
Elimination of the extra sequence
FLPe-mediated excision of FRTed sequence including vector sequence and marker genes was performed by introducing FLPe expression plasmid, pCAGGS-FLPe (14), or by crossing RMCEex mouse with FLPe transgenic mouse (29). For the ES cells, pCAGGS-FLPe was transfected into RMCEex cells using Lipofectamin 2000. After selection with puromycin, colonies were picked, grown in 24-well plates, and expanded to generate duplicate plates for cell freezing and DNA analysis. For the mouse with RMCEex allele, pCAGGS-FLPe (5 ng/µl) was introduced into the pronuclei of fertilized eggs by microinjection, and the treated eggs were then transferred to the foster mother. Otherwise, RMCEex male mice were crossed with FLPe transgenic female mice. The resulting offspring was then analyzed for the removal of the FRTed sequence. Excision of FRTed sequence was checked by PCR with the primer sets M393 and #143 for pAMF integration into the H2-Tw3 locus, M022 and M376 for pAMF, pAOF, pAOK, pAOL, pAOM, pAOT integration into the Rosa26 locus (Supplementary Table S1), and some of them were confirmed by Southern blot analysis. The presence or absence of FLPe transgene was confirmed by PCR with primer set M587 and M580 (Supplementary Table S1).
Mice
All mice were kept in the Center of Genetic Engineering for Human Diseases (CGEHD) animal facility at Tokai University School of Medicine. ICR, C57BL/6J and BDF1 mice were obtained from CLEA Japan Inc. (Tokyo, Japan). FLPe transgenic mouse strain (RBRC01835) was provided by RIKEN BRC, which is a participant in the National Bio-Resource Project of the MEXT, Japan (29). CAG-Cre transgenic mouse strain was kindly provided by Dr Jun-ichi Miyazaki (28). Green mouse [C57BL/6-Tg(CAG-EGFP)] was obtained from Japan SLC, Inc. (Hamamatsu, Japan) (30). The H2-Tw3- and Rosa26-targeted mice described above were maintained as homozygotes with a hybrid genetic background of C57BL/6J and 129/Ola.
Observation of fluorescence in color mice
The expression of fluorescence in color mice was first examined by the naked eye under a long-wave UV light (365 nm). For figures (Figure 2B and C, and Supplementary Figures S3 and S5), the images were observed using the Olympus SZX12 Stereo Microscope with filter sets for CFP (Ex438 Dm467-600 Em483), GFP (SZX-MGFP; Ex460-490 Dm505 Em510) and HcRed (SZX-MIY; Ex540-580 Dm600 Em610). For Figure 4C and F, the images were obtained using a Keyence BZ-9000 fluorescence microscope with filter sets (XF114-2; Ex440 Dm455 Em480, OP-66836; Ex470/40 Dm495 Em535/50 and OP-66837; Ex540/25 Dm565 Em605/55). For Figure 4A, D and E, the images were obtained using a Keyence fluorescent stereomicroscopy attached to a cooled CCD camera (VB-G05, VB-7010/7000) with filter sets (OP-42316, OP-51576, OP-42314V, OP-42311, OP-51571, OP-42312, OP-51572, OP-42315 and OP-51575). All mice, except homozygote EGFP transgenic mouse (Rosa26CAG::EGFP) shown in Supplementary Figure S6, were hemizygotes for the transgene.
Figure 2.
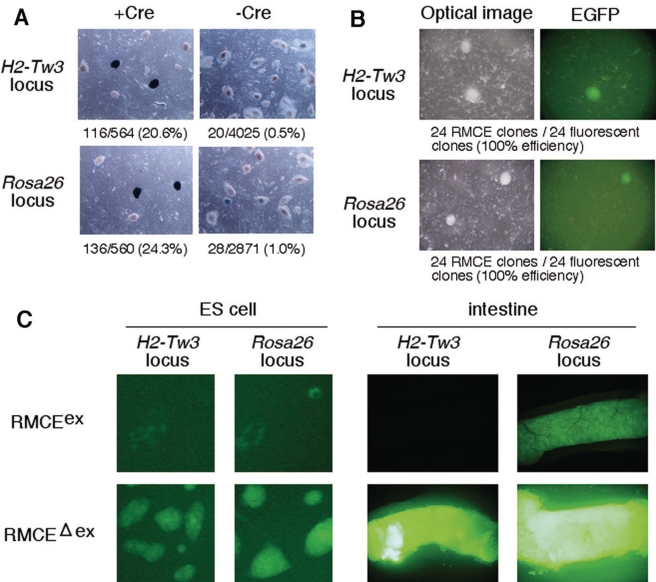
Efficient detection of correct RMCE recombinants in ES cells, and EGFP expression in RMCEex and RMCEΔex alleles. (A) The rate of appearance of the background clones. Donor vector, which contains lacZ gene as promoterless reporter gene (Figure 1A), was introduced into targeted ES cells with or without Cre-expression plasmid. Only 0.5–1.0% of colonies regarded as non-recombined clone stained blue with X-gal after introducing donor vector alone, whereas 20.6–24.3% of colonies showed lacZ expression upon co-introduction of Cre plasmid. (B) Efficient detection of correctly recombined clones. Introduction of a donor vector, in which EGFP gene was included as promoterless reporter gene, into ES cells together with the Cre plasmid resulted in the appearance of hygromycin-resistant EGFP-fluorescent and -non-fluorescent clones. Among them, 48 green fluorescent colonies (24 from H2-Tw3 and 24 from Rosa26) were picked and analyzed by PCR. All the collected clones showed correct recombination (100%; Supplementary Table S2). (C) EGFP expression in ES cells (left) and intestine of the mouse (right) containing a single copy of the ‘CAG-EGFP-polyA’ cassette at the H2-Tw3 and Rosa26 loci. EGFP expression of both ES cells and intestines with RMCEex allele was silenced (weak and mosaic). In the RMCEΔex allele that the vector backbone and the bacterial resistance gene were removed by FLPe administration, ES cells and intestines exhibited robust EGFP fluorescence.
Figure 4.
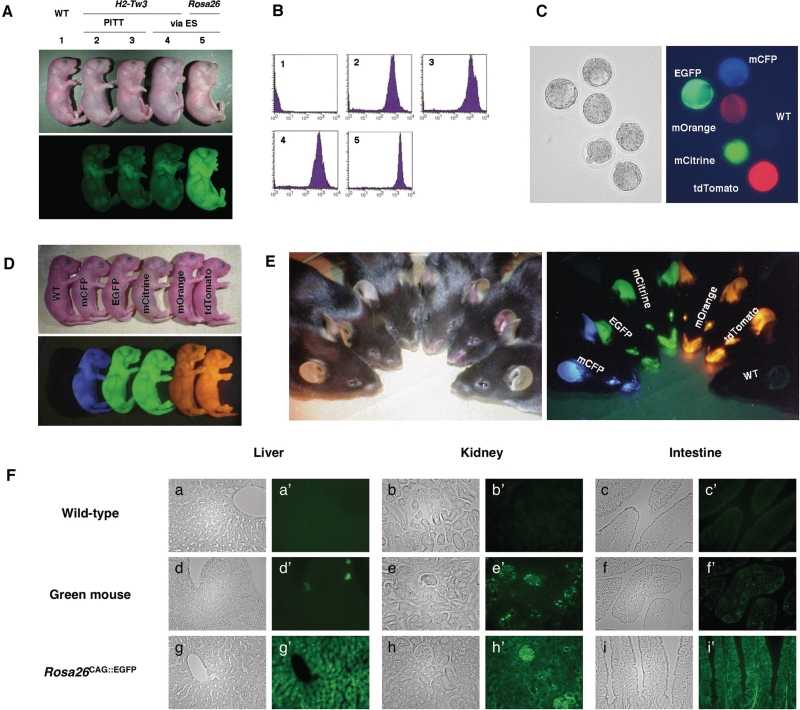
Fluorescent expression in targeted transgenic mice. (A) EGFP fluorescence in H2-Tw3CAG::EGFP and Rosa26CAG::EGFP mice, containing a single copy of the ‘CAG-EGFP-polyA’ cassette at the H2-Tw3 and Rosa26 loci, respectively. Three independent transgenic lines were obtained for H2-Tw3CAG::EGFP: two lines, labeled 2 and 3, were obtained by PITT, while the other, labeled 4, was obtained via RMCE in ES cells. The Rosa26CAG::EGFP line, labeled 5, was obtained via RMCE in ES cells. (B) Analysis of EGFP fluorescence in spleen cells prepared from the targeted transgenic mice. The numbers indicate mouse lines that correspond to those in (A). (C–E) Expression of fluorescent genes in colored mice. All lines containing a single copy of the ‘CAG-fluorescent gene-polyA’ cassette at the Rosa26 locus exhibited ubiquitous and very bright fluorescence in blastocysts (C), newborn mice (D) and adult mice (E). (F) Ubiquitous and uniform fluorescence expression in tissues of Rosa26CAG::EGFP mice. The left panels (a–i) are bright-field images, and the right panels (a′–i′) are dark-field fluorescent images taken with green filters. Rosa26CAG::EGFP (g,g′–i,i′) mice showed generalized pancellular FP expression, while variegated EGFP expression was observed in tissues of the green mouse (d,d′–f,f′). All transgenic mice were hemizygotes for the transgene.
FACS analysis
Spleen cells were prepared and cleared of red blood cells by osmotic lysis. The cells were then applied to flow cytometric analyses performed on a FACSCalibur (BD Biosciences, San Diego, CA, USA). Data were analyzed on a CellQuest program (BD Biosciences).
Histology
Adult tissue samples were dissected and fixed overnight in 4% PFA, then equilibrated in 20% sucrose before freezing. Frozen sections (8 µm) were imaged directly for EGFP fluorescence.
Analyses of Tyrosinase knockdown mouse
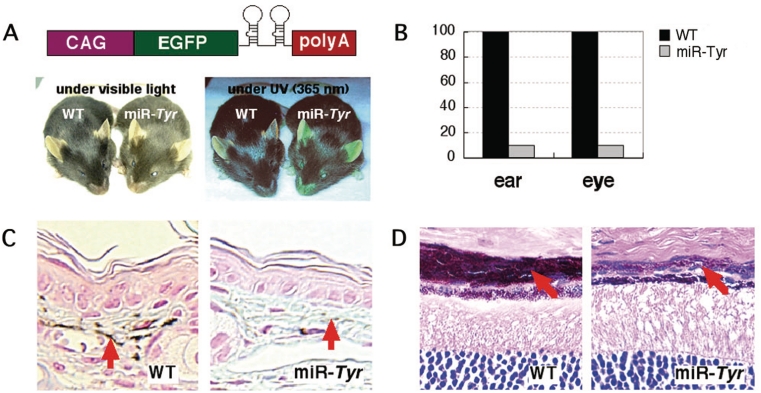
The Tyrosinase (Tyr) knockdown mouse (Rosa26CAG::miR-Tyr) was generated by PITT (pAOT). It contains a single copy of ‘CAG-EGFP-miR(Tyr-1/2)-polyA’ cassette in Rosa26 locus. Artificial microRNA (miRNA) region in pAOT is derived from ‘BLOCK-iT Pol II miR RNAi Expression Vector Kit with EmGFP’ (K4936-00, Invitrogen) with two tandemly arranged oligos for Tyr knockdown (5′-GTAAAGCTGAAATTGGCAGTTCGTTTTGGCCACTGACTGACGAACTGCCTTTCAGCTTTA-3′ and 5′-GTGGACTGGCAAATCCTTCCAGGTTTTGGCCACTGACTGACCTGGAAGGTTGCCAGTCCA-3′). EGFP fluorescence of Rosa26CAG::miR-Tyr mouse was observed under long-wave UV light (365 nm). Paraffin sections were prepared from fixed ears and eyes of Rosa26CAG::miR-Tyr mouse and wild-type littermate, and stained with Alcian Blue-Kernechtrot (for ear) and hematoxylin-eosin (for eye). The mRNA levels of Tyr in the ears and eyes of Rosa26CAG::miR-Tyr mouse and wild-type littermate were assessed by SYBR-green-based real-time reverse transcription (RT)–PCR, with ACTB (β-actin) used as an internal control.
RESULTS
Basic strategy for targeted transgenesis system
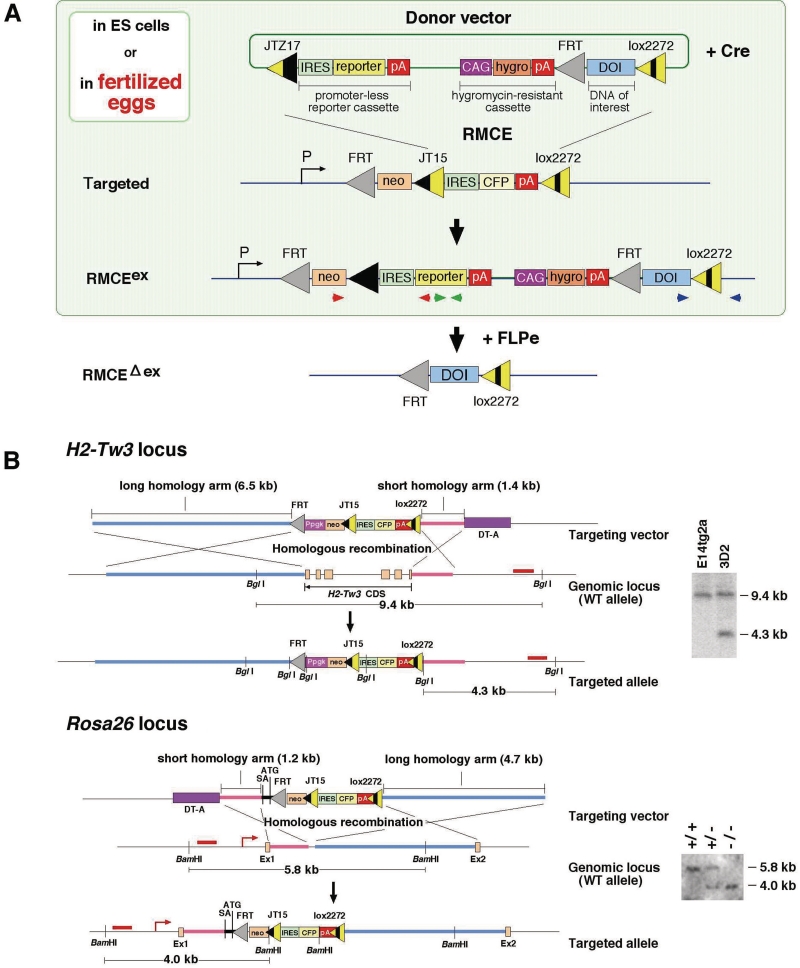
The overall scheme of this system is shown in Figure 1A. ES cells and mice with targeted alleles were initially generated through ES cell targeting to contain recombination-target sequences (loxP-site derivatives, JT15 and lox2272 and FRT) at a predetermined locus. A transgene, flanked by the JTZ17 and lox2272 sites of donor vector, was then integrated into target loci by recombinase-mediated cassette exchange (RMCE) in ES cells or fertilized eggs to generate RMCEex allele. The donor vector also contained hygromycin-resistant cassette (for the detection of both random and targeted integration) and promoterless reporter cassette containing lacZ or fluorescent gene (for the detection of only targeted integration) for selection in ES cells. Finally, the unnecessary sequence flanked by FRT sequences was eliminated by FLPe, resulting in a clean integrated DOI flanked only by FRT and lox2272 (RMCEΔex allele). Among of the multitude of mutant loxP sites reported previously, we chose the ‘JT15/JTZ17 (inverted-repeat variants) and lox2272 (spacer variant)’ combination of loxPs, based on previous studies describing (i) the combination of ‘LE/RE inverted-repeat mutant and lox2272′ showed high recombination efficiency in ES cells (31) and (ii) the JT15/JTZ17 mutant pair promotes the most efficient integration without eliciting intramolecular recombination in E. coli (11).
Figure 1.
(A) Schematic diagram of the integration scheme. The donor vector containing a DOI integrates into a genomically tagged site (Rosa26 in this figure) in the targeted allele by Cre-based RMCE via mutant loxP sites. Extra sequence in the RMCEex allele is removed by FLPe, resulting in an RMCEΔex allele. Red and blue, and the green arrowheads are the PCR primer sets used for detection of correct recombinant (Figure 3B). (B) Targeted insertion of mutant loxP-tagged sequences into the H2-Tw3 and Rosa26 loci. H2-Tw3 and Rosa26 targeting vectors were introduced into E14tg2a ES cells. G418-resistant clones were picked, and the targeted alleles were screened by PCR and certified by hybridization. Southern blot analyses of genomic DNA from ES cells (H2-Tw3) and mice (Rosa26) are shown on the right side of the figure. Homologous recombinations at the H2-Tw3 and Rosa26 loci are detectable using BglI or BamHI-digested genomic DNAs by the probes marked by the red bars, respectively.
Verification of JT15/JTZ17 and lox2272 Combination for the targeted transgenesis in vitro and in ES cells
The particular combination of loxP derivatives, JT15/JTZ17 and lox2272, was chosen to theoretically achieve maximally efficient RMCE with minimal undesired intramolecular recombination, although this particular combination has not been tested before. This study first tested in vitro whether intramolecular recombination occurs in this combination of loxP derivatives. For this purpose, we prepared five plasmids containing SacB gene, which provides a counter selection in E. coli, flanked by the five combinations of loxP derivatives (Supplementary Figure S1A). After in vitro Cre treatment of these plasmids, recombinations between loxP derivatives were checked by two methods: direct PCR of Cre reaction sample, and selection in E. coli grown on the LB sucrose plate. In both tests, Cre-mediated recombination was detected not only in loxP/loxP but also in loxP/lox511 as described previously (32,33), but no intramolecular recombination was observed in the combinations of loxP derivatives used in this study (Supplementary Figure S1B and C).
To establish JT15-lox2272-tagged seed mice, we modified the Rosa26 locus (conferring active and ubiquitous gene expression) (22) and H2-Tw3 locus (conferring tissue-specific expression) (19) using standard ES cell targeting (Figure 1B). The targeting frequencies were 25% (for Rosa26) and 3% (for H2-Tw3).
We next examined RMCE efficiency in mammalian ES cells by transfection of a donor plasmid carrying a hygromycin-resistant cassette and promoterless reporter gene flanked by JTZ17 and lox2272 together with a Cre-expression plasmid (Figure 1A). We used lacZ or EGFP genes as promoterless reporter gene in a donor plasmid, and its expression was initiated by PGK (for H2-Tw3) or endogenous Rosa26 promoter (for Rosa26) after integration into a tagged locus by correct recombination (Figure 1A). Among the hygromycin-resistant colonies, 20–40% of the resistance was attributed to correct RMCE (Supplementary Table S2). As expected, correct RMCE, which can be readily assessed by reporter gene expression, was dependent on the Cre recombinase (Figure 2A and B). No significant differences in the integration efficiency were seen between the two loci and between the two transfection methods (electroporation or lipofection; Supplementary Table S2). These results indicate the availability of JT15/JTZ17 and lox2272 combination for the targeted transgenesis.
Stable transgene expression achieved by removal of extra sequences
The RMCEex ES clones were isolated with EGFP expression cassette ‘CAG-EGFP-polyA’. However, the strength of EGFP fluorescence in these clones was weak and often mosaic after several passages, probably due to promoter interference and/or epigenetic silencing (Figure 2C, left). Since the presence of extra sequences including vector sequence and selection markers could affect the expected transgene expression, the clones were next subjected to removal of an extra sequence by administration of FLPe (Figure 1A) (14). The resultant RMCEΔex clones began to exhibit strong and uniform EGFP fluorescence after removal of extra sequence (Figure 2C, left). This robust EGFP fluorescence was noted even after five passages.
This phenomenon was also observed in vivo. After generating RMCEex transgenic mice from the RMCEex ES clones carrying the EGFP expression cassette, the FRT-flanked segment was removed by FLPe to generate RMCEΔex alleles (34). FLPe recombinase was provided either by PI at the zygote stage or by crossing with an FLPe deleter mouse (Supplementary Figure S2) (14,29). In the PI experiment, 69% and 58% of newborns with modified allele showed FLPe recombination in H2-Tw3 and Rosa26 locus, respectively (Supplementary Figure S2B). When RMCEex transgenic mice were crossed with FLPe transgenic mice, all double transgenic progeny obtained so far harbored RMCEΔex alleles (Supplementary Figure S2C). These results indicate that both approaches were highly efficient for our purpose of removing the FRT-flanked sequence in vivo. This elimination step was necessary because EGFP expression driven by a CAG promoter in RMCEex alleles was weak, often mosaic, and inconsistent, while mice with the RMCEΔex allele showed strong and ubiquitous EGFP expression as described below (Figure 2C, right and Supplementary Figure S3).
Generation of targeted transgenic mouse lines by Cre-loxP site-specific recombination in fertilized eggs
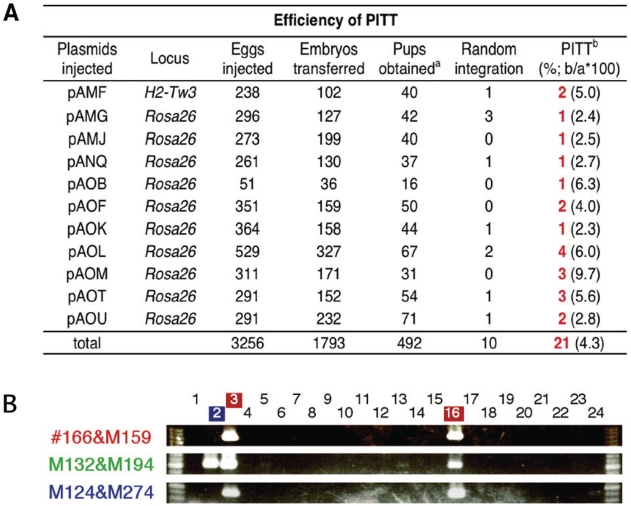
After confirming in ES cells that a transgene can preferentially integrate into the loxP-tagged loci by Cre-mediated site-specific recombination (Figure 2 and Supplementary Table S2), seed mouse lines were established from the Rosa26 and H2-Tw3-targeted ES cells. Both homozygous and heterozygous lines carrying the targeted alleles were viable and developed normally with normal reproductive ability. Fertilized eggs were prepared from homozygous seed male mice and wild-type females to evaluate the success of PITT. We first determined the maximum concentration of Cre-expression plasmid and found that 5 ng/µl, but not 10 ng/µl, could be injected without significant detrimental effects (Supplementary Table S3). Subsequent titration of donor plasmid concentrations showed that up to 5–10 ng/µl was tolerated (Supplementary Table S4). Figure 3B summarizes the results of microinjection of 11 different donor DNAs into 3256 fertilized eggs. Among the 492 newborns obtained, 21 (4.3%) showed targeted integration into the tagged loci, detected by PCR assay (Figure 3B) and some confirmed by Southern blot (Supplementary Figure S4). At least one transgenic founder was obtained for each of the 11 donor DNAs tested. Although the majority of transgenic founders (20/21) showed complete recombination generating correct RMCEex alleles (recombination occurred in both loxP sites: JT15/JTZ17 and lox2272/lox2272), one founder (pANQ in Figure 3A) exhibited partial recombination (recombination occurred only via JT15/JTZ17). In this case, we obtained complete RMCEex allele by crossing partially recombined founder with CAG-Cre transgenic mouse (data not shown) (28). The Rosa26 seed mouse was used in the majority of experiments, with the H2-Tw3 seed mouse used in one trial. Correct recombinants were obtained in all cases (Figure 3A), showing that PITT is not restricted to a particular locus. It is noteworthy that random integration of the donor vector and/or the Cre-expression plasmid was never detected in all targeted transgenic founders based on PCR, indicating that site-specific integration event and random integration event occur independently in mice (data not shown).
Figure 3.
PITT system. (A) PITT efficiency. A total of 11 constructs were microinjected into fertilized eggs containing the targeted allele. The numbers of founder transgenic mice obtained by PITT are shown in red. (B) PCR screening of correct recombinants. Primer sets used to amplify the junction regions and internal sequence of the donor vector, are shown in red and blue, and the green arrowheads in Figure 1A, respectively. Genomic junctions expected for an RMCE and internal regions were amplified in the correct recombinants (Nos. 3 and 16), while only the internal regions were amplified from a pup with random integration (No. 2).
Since the integration mechanism based on site-specific recombination must be different from that on random insertion, and the transgene integration by our system could occur after two-cell stage following the induction of Cre expression, it is important to examine the germ-line transmission efficiency to verify this method for the generation of stable transgenic mouse lines. Therefore, we next evaluated the germ-line transmission of transgenes in 21 founder mice generated. Germ-line transmission was detected with 18 lines from 21 founder mice tested (>85%), indicating an efficiency comparable to that achieved with conventional transgenesis (e.g. 82%) (35). The frequencies of germ-line transmission from individual founders, which was detected by PCR, varied from 0% to 63%, indicating transgene mosaicism (Supplementary Table S5). These results validate the use of PITT for routine transgenesis.
Stable and reproducible transgene expression in the PITT mice
In the current evaluation of PITT, we used a series of fluorescent protein (FP) genes as transgenes. Expression of the EGFP transgene expressed by the RMCEΔex transgenic mice was readily detected under long-wave UV light (365 nm). We compared three H2-Tw3CAG::EGFP mice, two of which were independently generated via PITT while the third was generated via ES cell targeting. All three displayed indistinguishable intensity of EGFP fluorescence (Figure 4A). The expression in the Rosa26CAG::EGFP mouse was higher than the H2-Tw3CAG::EGFP mice probably due to the position effect. EGFP expression examined in spleen cells from the transgenic mice again showed stronger fluorescence in the Rosa26CAG::EGFP mouse compared with the three H2-Tw3CAG::EGFP mice, all of which displayed similar levels of EGFP expression (Figure 4B). The current experiments also generated a series of FP mice, expressing mCFP, EGFP, mCitrine, mOrange or tdTomato with similar specificity (Figure 4C–E and Supplementary Figure S5), and stably for at least five generations. These results indicate that our PITT system provides a reliable method to achieve stable and reproducible transgene expression.
Strong and ubiquitous transgene expression in Rosa26CAG::FP mice
Transgenic mice that express FP in all tissues have been widely utilized as essential tools for transplantation studies and chimera experiments. Although several transgenic lines with ubiquitous expression of FP were developed by random-integration-based transgenesis, highly variegated expression of FP even among similar cell types was noted. This undesirable feature of expression makes it difficult to trace the origin of cells in transplantation and cell-lineage analyses. We also compared EGFP expression in Rosa26CAG::EGFP mouse developed in this study with that in commercially available random-integration EGFP transgenic mouse (green mouse) (30). The intensity of spleen-cell EGFP fluorescence in the Rosa26CAG::EGFP mouse was 2- to 3-fold that in green mouse (Supplementary Figure S6), indicating that our expression system drives very high levels of transgene expression. In other organs, such as liver, kidney and intestine, the EGFP expression in the green mouse was weak and/or sporadic (Figure 4F, d,d′-f,f′), as described previously (36). In contrast, Rosa26CAG::EGFP mice showed stronger and more uniform EGFP expression (Figure 4F, g,g′-i,i′), which may prove critical in some transplantation and chimera experiments. It is noteworthy that all Rosa26CAG::FP mice expressing other FP (e.g. tdTomato) also exhibited similar strong and ubiquitous expression pattern. These results emphasize the potential utility of fluorescent mice successively generated by PITT as new reporter lines for cell lineage analysis.
Application of PITT to in vivo knockdown experiment
Finally, we assessed our strong and ubiquitous expression system for generating miRNA-based knockdown mice. As a test, we generated a mouse expressing artificial miRNAs (Figure 5A) designed to interfere with the expression of Tyrosinase (Tyr), a key enzyme in melanin synthesis. The Rosa26CAG::miR-Tyr mouse had light hair color, reduced ear melanization, and a choroid coat compared to wild-type littermates, consistent with reduced levels of Tyr mRNA expression (Figure 5). The artificial miRNA transgene did not seem to induce apparent adverse phenotype possibly due to not only off-target effect but also oversaturation of the endogenous miRNA pathway (37), suggesting that the Tyr miRNA expression level is high enough to cause pigmentation phenotype but is low to over-saturate the miRNA pathway. This desirable result indicates the potential suitability of our strong and ubiquitous expression system for in vivo loss-of-function experiments.
Figure 5.
Tyrosinase (Tyr) knockdown mouse. (A) The artificial miRNA for Tyr was expressed under the control of the CAG promoter at the Rosa26 locus (Rosa26CAG::miR-Tyr). (B) The levels of Tyr mRNA were quantitated by real-time RT-PCR analysis. Coat color (A), melanin in ears (C) and choroid coat (D) were compared between the wild-type and Rosa26CAG::miR-Tyr mice.
DISCUSSION
The method reported here is the first report that describes the successful generation of a series of targeted transgenic mouse lines via PI. The PITT method described herein showed approximately 0.6% and 4.3% efficiency of correct and stable integration per injected embryo and per newborn, respectively, and a high germ-line transmission rate (>85%). Thus, one germ-line-transmissible targeted transgenic founder is expected to be derived from every 200 embryo-injections on average, representing a similar workload to that needed for generating transgenic lines using the conventional PI-based transgenesis. It is possible to further improve this efficiency by the injection of synthetic Cre mRNA or recombinant Cre protein, or by cell-permeable Cre treatment (38–40).
The PITT strategy has several advantages over previous transgenesis techniques. First, unlike random-integration-based transgenesis, the expression pattern of the transgene is predictable, reproducible and stable. Even when several transgenic lines are obtained, no laborious effort to screen the lines with adequate transgene activity is required and only one targeted transgenic line is sufficient for further analysis. This provides cost saving, space and effort. Second, our PITT method does not require ES cell manipulation once the seed mouse is established. Thus, it saves not only money and effort for ES cell culture, but also substantially reduces the time required to obtain targeted transgenic lines (∼1–2 months). Third, in our PITT method, circular plasmid DNA can be directly injected into the fertilized eggs. Hence, unlike the conventional PI-based transgenesis, it is not necessary to cut out the transgene from the vector backbone and isolate the insert by gel purification.
A previous study investigated the feasibility of targeted transgenesis via PI (33). The study showed successful targeted integration of a transgene at an efficiency comparable to ours. The system, however, suffered from instability of integrated transgene configuration (uncontrollable orientation) due to the combination of loxPs (wildtype loxP and its spacer variant, lox511), leading to unstable transgene expression. In their study, germ-line transmission was not examined. To our knowledge, our study is the first report describing the establishment of a transgenic line generated by targeted transgenesis via PI.
By applying the PITT method, we established a total of 18 transgenic lines, including color FP mice, with stronger and more stable expression than currently available mice. These FP mice will be available to the scientific community. We also demonstrated that knockdown mice can be readily generated by PITT by taking advantage of our reproducible and highly efficient expression system. We also demonstrated that PITT is applicable to at least two different chromosomal loci, and we expect similar success with other loci and many loxP-tagged mice, such as those generated by the International Gene Trap project (http://www.genetrap.org/) (41,42). The PITT method described herein could therefore provide a strong basis for systematic generation of a variety of targeted transgenic lines, without going through ES cells, for reliable transgene expression and for gene knockdown.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-Aid for Young Scientists (B) (20700368) to M.O. from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan; Grant-in-Aid for Exploratory Research (16651102) to M.O. from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan; Tokai University School of Medicine Research Aid to M.O. (by 2005, 2006 and 2007); Research and Study Program of the Tokai University Educational System General Research Organization (2008) to M.O. Funding for open access charge: Research Aid from the Institute of Medical Sciences in Tokai University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Tsien for the mCFP, mCitrine and tdTomato genes, J. Miyazaki for the CAG-Cre transgenic mouse and P. Soriano for the plasmid-containing SA sequence. We also thank S. Yoshimura for the excellent technical assistance.
REFERENCES
- 1.Gama Sosa MA, De Gasperi R, Elder GA. Animal transgenesis: an overview. Brain Struct. Funct. 2009;214:91–109. doi: 10.1007/s00429-009-0230-8. [DOI] [PubMed] [Google Scholar]
- 2.Gordon JW, Scangos GA, Plotkin DJ, Barbosa JA, Ruddle FH. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc. Natl Acad. Sci. USA. 1980;77:7380–7384. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinster RL, Chen HY, Trumbauer M, Senear AW, Warren R, Palmiter RD. Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell. 1981;27:223–231. doi: 10.1016/0092-8674(81)90376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jasin M, Moynahan ME, Richardson C. Targeted transgenesis. Proc. Natl Acad. Sci. USA. 1996;93:8804–8808. doi: 10.1073/pnas.93.17.8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat. Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 6.Potts W, Tucker D, Wood H, Martin C. Chicken beta-globin 5'HS4 insulators function to reduce variability in transgenic founder mice. Biochem. Biophys Res. Commun. 2000;273:1015–1018. doi: 10.1006/bbrc.2000.3013. [DOI] [PubMed] [Google Scholar]
- 7.Giraldo P, Rival-Gervier S, Houdebine LM, Montoliu L. The potential benefits of insulators on heterologous constructs in transgenic animals. Transgenic Res. 2003;12:751–755. doi: 10.1023/b:trag.0000005089.30408.25. [DOI] [PubMed] [Google Scholar]
- 8.Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 2001;10:83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- 9.Bronson SK, Plaehn EG, Kluckman KD, Hagaman JR, Maeda N, Smithies O. Single-copy transgenic mice with chosen-site integration. Proc. Natl Acad. Sci. USA. 1996;93:9067–9072. doi: 10.1073/pnas.93.17.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorrell DA, Kolb AF. Targeted modification of mammalian genomes. Biotechnol. Adv. 2005;23:431–469. doi: 10.1016/j.biotechadv.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Thomson JG, Rucker EB, 3rd, Piedrahita JA. Mutational analysis of loxP sites for efficient Cre-mediated insertion into genomic DNA. Genesis. 2003;36:162–167. doi: 10.1002/gene.10211. [DOI] [PubMed] [Google Scholar]
- 12.Lee G, Saito I. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene. 1998;216:55–65. doi: 10.1016/s0378-1119(98)00325-4. [DOI] [PubMed] [Google Scholar]
- 13.Sato M, Ishikawa A, Kimura M. Direct injection of foreign DNA into mouse testis as a possible in vivo gene transfer system via epididymal spermatozoa. Mol. Reprod. Dev. 2002;61:49–56. doi: 10.1002/mrd.1130. [DOI] [PubMed] [Google Scholar]
- 14.Schaft J, Ashery-Padan R, van der Hoeven F, Gruss P, Stewart AF. Efficient FLP recombination in mouse ES cells and oocytes. Genesis. 2001;31:6–10. doi: 10.1002/gene.1076. [DOI] [PubMed] [Google Scholar]
- 15.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 16.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 17.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 18.Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 19.Takada T, Kumanovics A, Amadou C, Yoshino M, Jones EP, Athanasiou M, Evans GA, Fischer Lindahl K. Species-specific class I gene expansions formed the telomeric 1 mb of the mouse major histocompatibility complex. Genome Res. 2003;13:589–600. doi: 10.1101/gr.975303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohtsuka M, Mizutani A, Kikuti YY, Kulski JK, Sato M, Kimura M, Tanaka M, Inoko H. One-step generation of recombineering constructs by asymmetric-end ligation and negative selection. Anal. Biochem. 2007;360:306–308. doi: 10.1016/j.ab.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl Acad. Sci. USA. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohtsuka M, Ishii K, Kikuti YY, Warita T, Suzuki D, Sato M, Kimura M, Inoko H. Construction of mouse 129/Ola BAC library for targeting experiments using E14 embryonic stem cells. Genes Genet. Syst. 2006;81:143–146. doi: 10.1266/ggs.81.143. [DOI] [PubMed] [Google Scholar]
- 24.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 25.Tucker KL, Wang Y, Dausman J, Jaenisch R. A transgenic mouse strain expressing four drug-selectable marker genes. Nucleic Acids Res. 1997;25:3745–3746. doi: 10.1093/nar/25.18.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato M, Yasuoka Y, Kodama H, Watanabe T, Miyazaki JI, Kimura M. New approach to cell lineage analysis in mammals using the Cre-loxP system. Mol. Reprod. Dev. 2000;56:34–44. doi: 10.1002/(SICI)1098-2795(200005)56:1<34::AID-MRD5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 27.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. 3rd edn. New York: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- 28.Sakai K, Miyazaki J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem. Biophys. Res. Commun. 1997;237:318–324. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- 29.Kanki H, Suzuki H, Itohara S. High-efficiency CAG-FLPe deleter mice in C57BL/6J background. Exp. Anim. 2006;55:137–141. doi: 10.1538/expanim.55.137. [DOI] [PubMed] [Google Scholar]
- 30.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice' as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 31.Araki K, Araki M, Yamamura K. Site-directed integration of the cre gene mediated by Cre recombinase using a combination of mutant lox sites. Nucleic Acids Res. 2002;30:e103. doi: 10.1093/nar/gnf102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolb AF. Selection-marker-free modification of the murine beta-casein gene using a lox2272 [correction of lox2722] site. Anal. Biochem. 2001;290:260–271. doi: 10.1006/abio.2000.4984. [DOI] [PubMed] [Google Scholar]
- 33.Shmerling D, Danzer CP, Mao X, Boisclair J, Haffner M, Lemaistre M, Schuler V, Kaeslin E, Korn R, Burki K, et al. Strong and ubiquitous expression of transgenes targeted into the beta-actin locus by Cre/lox cassette replacement. Genesis. 2005;42:229–235. doi: 10.1002/gene.20135. [DOI] [PubMed] [Google Scholar]
- 34.Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 1998;16:657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- 35.Nakanishi T, Kuroiwa A, Yamada S, Isotani A, Yamashita A, Tairaka A, Hayashi T, Takagi T, Ikawa M, Matsuda Y, et al. FISH analysis of 142 EGFP transgene integration sites into the mouse genome. Genomics. 2002;80:564–574. doi: 10.1006/geno.2002.7008. [DOI] [PubMed] [Google Scholar]
- 36.Swenson ES, Price JG, Brazelton T, Krause DS. Limitations of green fluorescent protein as a cell lineage marker. Stem. Cells. 2007;25:2593–2600. doi: 10.1634/stemcells.2007-0241. [DOI] [PubMed] [Google Scholar]
- 37.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 38.Kim K, Kim H, Lee D. Site-specific modification of genome with cell-permeable Cre fusion protein in preimplantation mouse embryo. Biochem. Biophys. Res. Commun. 2009;388:122–126. doi: 10.1016/j.bbrc.2009.07.132. [DOI] [PubMed] [Google Scholar]
- 39.Luckow B, Hanggli A, Maier H, Chilla S, Loewe RP, Dehmel S, Schlondorff D, Loetscher P, Zerwes HG, Muller M. Microinjection of Cre recombinase protein into zygotes enables specific deletion of two eukaryotic selection cassettes and enhances the expression of a DsRed2 reporter gene in Ccr2/Ccr5 double-deficient mice. Genesis. 2009;47:545–558. doi: 10.1002/dvg.20531. [DOI] [PubMed] [Google Scholar]
- 40.de Wit T, Drabek D, Grosveld F. Microinjection of cre recombinase RNA induces site-specific recombination of a transgene in mouse oocytes. Nucleic Acids Res. 1998;26:676–678. doi: 10.1093/nar/26.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skarnes WC, von Melchner H, Wurst W, Hicks G, Nord AS, Cox T, Young SG, Ruiz P, Soriano P, Tessier-Lavigne M, et al. A public gene trap resource for mouse functional genomics. Nat. Genet. 2004;36:543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singla V, Hunkapiller J, Santos N, Seol AD, Norman AR, Wakenight P, Skarnes WC, Reiter JF. Floxin, a resource for genetically engineering mouse ESCs. Nat. Methods. 2010;7:50–52. doi: 10.1038/nmeth.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.