Abstract
Genome-wide analysis of single nucleotide polymorphism (SNP) markers is an extremely efficient means for genetic mapping of mutations or traits in mice. However, this approach often defines a relatively large recombinant interval. To facilitate the refinement of this interval, we developed the program SNP2RFLP. This program can be used to identify region-specific SNPs in which the polymorphic nucleotide creates a restriction fragment length polymorphism (RFLP) that can be readily assayed at the benchtop using restriction enzyme digestion of SNP-containing PCR products. The program permits user-defined queries that maximize the informative markers for a particular application. This facilitates fine-mapping in a region containing a mutation of interest, which should prove valuable to the mouse genetics community. SNP2RFLP and further details are publicly available at http://genetics.bwh.harvard.edu/snp2rflp/
Introduction
The positional cloning and characterization of mutations in the mouse is a powerful means for functional annotation of the mammalian genome. Many mouse gene mutations cause phenotypes that serve as models of human genetic disorders. Mapping and positional cloning of these potentially accelerate our understanding of the mouse gene, its human ortholog, and the underlying etiology of the disorder. The utilization of single nucleotide polymorphism (SNP) markers has markedly facilitated genetic mapping because they are abundant throughout the genome and can be analyzed in a high-throughput manner using automated technology (Wang et al. 1998). However, mutation mapping analysis using a genome-wide SNP panel does not generally yield high-resolution localization (Moran et al. 2006), and “benchtop” technologies for fine-mapping using SNPs and microsatellite markers are often inefficient. We have developed a web-based tool we call SNP2RFLP, which can extract region-specific SNPs from the dbSNP database (Sherry et al. 1999) and identify those SNPs that would create restriction fragment length polymorphisms (RFLPs) when assayed by restriction enzyme digestion of SNP-containing PCR products. The input to SNP2RFLP is the two mouse strains used in the cross, the chromosomal region, and a user-defined set of restriction endonucleases. SNP2RFLP extracts the SNPs from dbSNP that are polymorphic between the two strains in the region in question. The program simulates a restriction digest of the SNP-containing sequences with each enzyme to determine whether the SNP creates an RFLP. Informative markers are then analyzed using Primer3 (Rozen and Skaletsky 2000), which finds suitable PCR primers surrounding the SNP. The output of SNP2RFLP is the informative SNPs that create RFLPs and the forward and reverse PCR primers. This information can then be used to readily perform the RFLP assays and further refine the region containing the mutation of interest.
Methods
A local PostgreSQL database was constructed to hold all mouse SNPs from the NCBI dbSNP (Mouse Build 126) along with their flanking sequences. The database contains 8 million unique mouse SNPs, with 200–400 bp of flanking sequence for each SNP. SNP-containing flanking sequences were analyzed by Primer3, which identifies optimal PCR primers surrounding each SNP that meet standardized criteria for product size, primer melting temperature (Tm) (~60°C), and GC content (~50%) (Rozen and Skaletsky 2000). These forward and reverse primers are stored in the database along with each SNP.
There are 68 million known strain genotypes for the SNPs in the database, which holds genotype data for 99 different mouse strains. Seventeen strains, including A/J, DBA/2 J, 129S1/SvlmJ, C3H/HeJ, BALB/cByJ, AKR/J, NZW/LacJ, CAST/EiJ, BTBR T + tf/J, WSB/EiJ, FVB/NJ, NOD/LTJ, KK/HIJ, PWD/PhJ, MOLF/EiJ, C57BL/6 J, and 129X1/SvJ, were interrogated using a high-density array and each has approximately 2–6 million SNP genotypes (Sherry et al. 1999). The other 82 strains have only on the order of hundreds or thousands of SNP genotypes.
Restriction digest simulation is done by scanning each SNP-containing sequence for the recognition sites of select restriction enzymes. A SNP is considered to result in an informative RFLP assay if an enzyme site is found in the sequence of one strain but not in the other strain due to the alteration of the restriction site by the polymorphism. The default enzymes are AluI, AflII, ClaI, DdeI, EcoRV, Fnu4HI, HaeIII, HhaI, HinfI, KpnI, MboI, MseI, MspI, PstI, PvuI, PvuII, RsaI, SacII, SalI, ScaI, ScrFI, and Sau96I. This list comprises efficient, frequently cutting restriction enzymes that have a high probability of providing a robust RFLP assay for any given SNP. In addition, the user can select an option that includes all the enzymes in the simulated restriction digest. Analysis of the number of restriction enzyme sites within a given amplicon is performed to avoid assays with very high complexity or very small size differences of restriction fragments. All the restriction enzymes and recognition sequences used by SNP2RFLP were obtained from the restriction enzyme database (REBASE) (Roberts et al. 2003).
To avoid nonspecific amplification for a given SNP, the surrounding sequence for each SNP was queried for the presence of known repetitive elements and simple and complex repeats using RepeatMasker, which “masks” these sequences with “N”s (http://www.repeatmasker.org/). The genomic locations of these premasked sequences are stored with each SNP so the user can decide whether to discard SNPs that fall in repeat regions.
The program SNP2RFLP was written in the programming language PERL. PERL was chosen because of its database connections and pattern-matching capabilities. The program was then incorporated into a CGI script that is called from a web interface. This interface was written with the HTML and JavaScript languages.
Results
The input to SNP2RFLP is the two mouse strains used in the cross, the chromosomal region (as defined by base pairs), and a set of restriction endonucleases. A default list of 22 commonly used restriction endonucleases with frequently occurring recognition sites is used by SNP2RFLP to simulate restriction digestion, but additional enzymes can be selected from a list of 1300 endonucleases.
SNP2RFLP extracts the SNPs from dbSNP that are polymorphic between the two strains in the region in question. SNP2RFLP then simulates a restriction digest on the SNP-containing sequences with each enzyme that was selected to determine if the SNP is contained within one or more enzyme recognition sites and creates an RFLP. That is, a SNP-containing sequence is scanned to see if the recognition sequence for any particular enzyme contains the SNP and is found for one strain but not the other due to the alteration of the recognition sequence by the SNP. If this is the case, the SNP is considered informative because the alleles can be distinguished by amplifying the region with PCR, digesting the products with the enzyme, and examining the resulting restriction pattern after agarose gel electrophoresis of the digested product (Fig. 1). Informative SNPs are listed and are accompanied by suggested oligonucleotide primer sequences for PCR amplification of the SNP (extracted from data stored in the database for each SNP), the position of the primers with respect to the SNP, and the number of enzyme recognition sites present in the amplified sequence. The entirety of these data can be visualized as a web-based display (Fig. 2) or can be exported as a spreadsheet document.
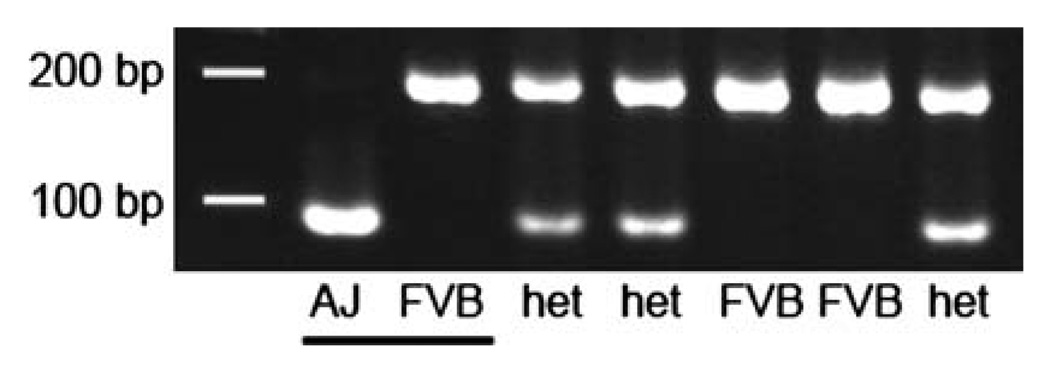
Fig. 1.
A SNP2FRLP-identified RFLP assay used to identify mice carrying a mapped ENU-induced mutation. PCR products of 195 bp encompassing SNP rs37311177 on chromosome 13 were amplified from tail DNA isolated from individual mice and digested with the restriction enzyme MseI. Samples included AJ, FVB strain controls (underlined), and five experimental samples. AJ polymorphism at this SNP creates an RFLP that is not present in the FVB genome
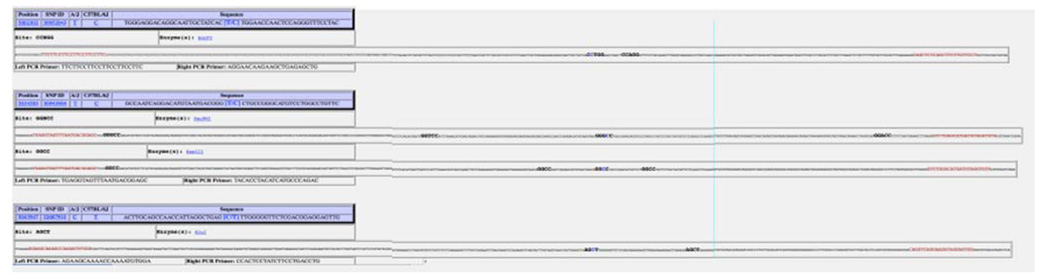
Fig. 2.
A screen shot of three informative SNPs returned by SNP2RFLP. The restriction enzyme recognition sites (bold) cut at the SNP position (bold, blue) in the sequence. The genotype of the SNP for each strain is shown. The suggested primers found by Primer3 are highlighted in red along the sequence
We have used the SNP2RFLP service to assist in developing markers in our mapping of mutants in an ongoing ENU mutagenesis screen. In the process of mapping seven different recessive mutations, we have utilized 32 different RFLPs which are summarized in Table 1. We used primarily SNP2RFLPs identified with the default enzyme set. Twenty-three of these yielded easily interpreted results when digested with the prescribed enzyme, although three required additional optimization. Nine assays were not usable for mapping purposes: One did not give a discrete PCR product and eight assays failed to detect the RFLP as predicted by this program. Overall, this program yielded easily implemented assays with 72% reliability.
Table 1.
Analysis of SNP2RFLP-designed RFLP assays for positional cloning
| Primer No. | Position (chr_Mb) |
Strains tested |
SNP | Enzyme | Success |
|---|---|---|---|---|---|
| 217/218 | 13_32.5 | A/J v FVB | rs29904172 | AluI | Yes |
| 241/242 | 13_33.5 | A/J v FVB | rs29239961 | BbsI | Yes |
| 233/234 | 13_34.2 | A/J v FVB | rs37311177 | MseI | Yes |
| 243/244 | 13_37.2 | A/J v FVB | rs6259014 | HinfI | Yes |
| 211/212 | 2_61.9 | A/J v FVB | rs28002307 | RsaI | Yes |
| BB1207/1208 | 7_102.96 | A/J v FVB | rs37343086 | MseI | No: no RFLP by digestion |
| BB1211/1212 | 7_122.5 | A/J v FVB | rs37274506 | Fnu4HI | Yes |
| BB1203/1204 | 7_67.7 | A/J v FVB | rs36590391 | HaeIII | Yes |
| BB1205/1206 | 7_88.1 | A/J v FVB | rs36897851 | DdeI | No: no RFLP by digestion |
| BB1213/1214 | 1_128.56 | A/J v FVB | rs33427936 | PstI | Yes |
| BB1215/1216 | 1_130.6 | A/J v FVB | rs13476106 | RsaI | Yes |
| 249/350 | 1_195 | A/J v FVB | rs13476313 | MseI | Yes |
| 237/238 | 15_5.14 | A/J v FVB | rs32664631 | MseI | Yes |
| 231/232 | 15_66.0 | A/J v FVB | rs36757821 | BglII | Yes |
| 641/642 | 15_63.3 | A/J v FVB | rs37879829 | AluI | Yes |
| 367/368 | 7_22.0 | A/J v FVB | rs36238918 | NcoI | No: no RFLP by digestion |
| 371/372 | 7_28.2 | A/J v FVB | rs37765358 | PvuI | Yes |
| 403/404 | 7_31.2 | A/J v FVB | rs36772588 | RsaI | No: no discrete PCR bands |
| 369/370 | 7_35.3 | A/J v FVB | rs38119160 | NruI | Yes: optimize for robustness |
| BB1137/1138 | 11_82.8 | B6 v FVB | rs28191426 | RsaI | No: no RFLP by digestion |
| BB1227/1228 | 11_82.83 | B6 v FvB | rs28191333 | HaeIII | Yes |
| BB1229/1230 | 11_82.83 | B6 v FvB | rs28191306 | MspI | Yes |
| BB1231/1232 | 11_82.85 | B6 v FvB | rs28191239 | MspI | No: no RFLP by digestion |
| BB1235/1236 | 11_83.01 | B6 v FvB | rs28210166 | AluI | No: no RFLP by digestion |
| BB1139/1140 | 11_83.6 | B6 v FvB | rs28209292 | HaeIII | No: no RFLP by digestion |
| BB1237/1238 | 11_83.60 | B6 v FvB | rs28209292 | HaeIII | No: no RFLP by digestion |
| BB1133/1134 | 11_87.8 | B6 v FvB | rs28241207 | BssHII | Yes |
| BB1143/1144 | 11_88.2 | B6 v FvB | rs26953277 | Fnu4HI | Yes: optimize for robustness |
| BB1141/1142 | 11_88.3 | B6 v FvB | rs29407170 | AluI | Yes |
| BB1145/1146 | 11_88.5 | B6 v FvB | rs27065145 | Sau96I | Yes: optimize for robustness |
| BB1135/1136 | 11_89.1 | B6 v FvB | rs29401408 | HaeIII | Yes |
| BB1131/1132 | 11_89.6 | B6 v FvB | rs27083857 | HincII | Yes |
Discussion
Because the number of characterized SNPs and their distribution across the genome are highly variable, we have incorporated multiple options to control the output returned by SNP2RFLP to produce a useful number of informative markers. First, each SNP in the database has a validation status. NCBI’s dbSNP defines many different ways that a SNP can be validated. For simplicity, if the “display validated SNPs only” option is selected, SNPs that have no validation information are excluded. This reduces the number of informative SNPs in many cases, but gives higher confidence in the utility of those reported.
Second, there are occasions when no informative SNPs are reported between two strains in a specific region. It may be that there are indeed informative SNPs in the region but the genotype may be recorded in only one strain. If the “display SNPs recorded in only one strain” option is selected, then the restriction digest is simulated on the SNPs from the strain where the genotype is known and compared with that for the alternate allele. For those identified as potentially polymorphic, additional methods can be applied by the user to infer the genotype of each SNP in the other strain (such as comparison to haplotypes of well-characterized strains), or they can simply be empirically tested.
Third, as previously noted, SNPs in a repeat region of the genome are often difficult to amplify. The user can select an option for SNP2RFLP to discard SNPs that fall within repeats.
Finally, the desired density of SNP markers returned can be set. SNP2RFLP can be instructed to return all of the informative markers or a subset (e.g., 1 of every 5, 1 of every 10, etc.). This is an extremely valuable option that allows the user to retrieve an adequate and manageable number of markers. As an example, suppose a genome-wide SNP scan, crossing A/J and FVB/NJ, reveals a candidate region on chromosome 13, 14.8–46.7 Mb, in which a particular mutation of interest may be located. Table 2 gives the different numbers of informative SNPs in this region returned by SNP2RFLP by selecting different options in the program. When selecting all of the enzymes and instructing SNP2RFLP to keep all SNPs, SNP2RFLP returns a large and perhaps unmanageable number of SNP markers. By restricting the type and number of enzymes and directing SNP2RFLP to report SNPs at a desired density, a manageable and adequate number of SNP markers can be considered that will facilitate fine-mapping of this candidate region that contains the phenotype-causing mutation of interest.
Table 2.
SNP2RFLP analysis of a recombinant interval from 14.8 to 46.7 Mb on chromosome 13 derived from an A/J × FVB/NJ cross
| No SNPs to output |
No SNPs returned (all enzymes) |
No suitable primers found |
Total SNP2RFLP assays |
No SNPs returned (default enzymes) |
No suitable primers found |
Total SNP2RFLP assays |
|---|---|---|---|---|---|---|
| All | 856 | 29 | 827 | 306 | 10 | 296 |
| 1 every 5 | 172 | 2 | 170 | 62 | 2 | 60 |
| 1 every 10 | 86 | 0 | 86 | 31 | 1 | 30 |
| 1 every 20 | 43 | 0 | 43 | 16 | 1 | 15 |
The SNP2RFLP interface can be accessed through the web at http://genetics.bwh.harvard.edu/snp2rflp. An instruction manual is available on the website or as supplementary data.
Contributor Information
Wesley A. Beckstead, Department of Biology, Brigham Young University, Provo, UT 84602, USA
Bryan C. Bjork, Genetics Division, Brigham and Women’s Hospital, Harvard Medical School, New Research Building, 77 Avenue Louis Pasteur, Boston, MA 02115, USA
Rolf W. Stottmann, Genetics Division, Brigham and Women’s Hospital, Harvard Medical School, New Research Building, 77 Avenue Louis Pasteur, Boston, MA 02115, USA
Shamil Sunyaev, Genetics Division, Brigham and Women’s Hospital, Harvard Medical School, New Research Building, 77 Avenue Louis Pasteur, Boston, MA 02115, USA.
David R. Beier, Email: beier@receptor.med.harvard.edu, Genetics Division, Brigham and Women’s Hospital, Harvard Medical School, New Research Building, 77 Avenue Louis Pasteur, Boston, MA 02115, USA.
References
- Moran JL, Bolton AD, Tran PV, Brown A, Dwyer ND, et al. Utilization of a whole genome SNP panel for efficient genetic mapping in the mouse. Genome Res. 2006;16:436–440. doi: 10.1101/gr.4563306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE: restriction enzymes and methyltransferases. Nucleic Acids Res. 2003;31:418–420. doi: 10.1093/nar/gkg069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward M, Sirotkin K. dbSNP—database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]