Abstract
Summary
Epsilon, a fidelity subunit of Escherichia coli DNA Polymerase III, is encoded by dnaQ+. dnaQ49 is a recessive allele that confers temperature-sensitive and salt-suppressible phenotypes for both replication fidelity and viability. SOS mutagenesis in E. coli is regulated by LexA and requires activated RecA (RecA*) and the products of the umuDC operon, dnaQ49 strains with various recA, lexA and umuDC alleles were constructed to determine if activities induced as part of the SOS response influence epsilon activity. We found: (1) both UmuDC and RecA* independently enhance the dnaQ49 mutator phenotype, and (2) expression of RecA* activity in the absence of UmuDC function increases the temperature sensitivity for viability of dnaQ49. These results support the hypothesis that RecA and one or both of the UmuDC proteins interact with the replication complex during SOS mutagenesis.
Keywords: Mutagenesis, Replication, dnaQ, recA, umuDC
Introduction
The RecA protein of Escherichia coli plays at least three roles in mutagenesis. When activated to its proteolytic state (RecA*) genetically or by DNA damage, it enhances the self-cleavage of the LexA protein, the common repressor of the genes of the rec-lex regulon (Little 1984). The induction of these genes, which include the mutagenesis promoting umuDC operon as well as recA and lexA, constitutes the SOS response (reviewed in Walker 1984). RecA* also facilitates the cleavage of UmuD (Burckhardt et al. 1988; Nohmi et al. 1988; Shinagawa et al. 1988), which activates UmuD for its function in mutagenesis (Nohmi et al. 1988). Since a cleavage-independent umuD mutant does not restore mutability to a recA defective mutant, RecA appears to have at least one more role in mutagenesis (Nohmi et al. 1988).
SOS mutagenesis is hypothesized to be mediated by an error-prone polymerase (Radman 1975). That RecA may interact with and modify the replication complex is suggested by the following evidence: (1)in vivo RecA is required after DNA damage for the recovery of DNA synthesis (Khidhir et al. 1985; Witkin et al. 1987) and the initiation and maintenance of stable DNA replication (Witkin and Kogoma 1984), and (2) in vitro purified RecA protein reduces the fidelity of DNA synthesis by DNA polymerase III (Pol III) holoenzyme, apparently by inhibiting the 3′ → 5′ exonuclease activity of the epsilon subunit (Fersht and Knill-Jones 1983; Lu et al. 1986). One or both of the gene products of umuDC may also interact with the replication complex since: (1) overproduction of UmuDC inhibits DNA synthesis (Marsh and Walker 1985), (2) UmuDC suppresses the inability of a mutant RecA, RecA718, to promote the recovery of DNA synthesis after DNA damage (Witkin et al. 1987), and (3) removal of UV induced blocking lesions by delayed photoreversal allows recovery of mutations in umuDC- bacteria, suggesting that UmuDC is involved in the continuation of DNA synthesis from bases mis-inserted opposite DNA lesions (Bridges and Woodgate 1985). Thus, the several direct roles of RecA may be to “target” the lesion, inhibit the proofreading function of Pol III, and interact with and modify the UmuDC proteins which then aid in or promote lesion bypass (Lu et al. 1986; Bridges and Woodgate 1985; Ennis et al. 1985). However, given the complexity of the system many other models can be generated that fit the experimental data.
The dnaQ gene encodes epsilon, a fidelity subunit of Pol III (Scheuermann et al. 1983). dnaQ49 is a recessive allele of dnaQ+ that confers a temperature-inducible and salt-suppressible mutator phenotype; at higher temperatures in salt-free media, dnaQ49 strains are non-viable due to inhibition of DNA synthesis (Horiuchi et al. 1978). Piechocki et al. (1986) reported that dnaQ49-dependent base substitution mutations were dominated by transversions, the same class of mutation produced when the SOS response is induced in the absence of DNA damage (Miller and Low 1984). They further found that the dnaQ49 mutator phenotype was independent of recA and umuDC, but reduced by a lexA (Ind-) (noninducible) allele. They hypothesized that Pol III complexed with the dnaQ49 gene product is functionally the error-prone SOS polymerase, not requiring the participation of RecA or UmuDC, but requiring some other gene repressed by LexA, to express mutations.
Because the phenotypes conferred by dnaQ49 are conditional, we reasoned that they could be modulated by other proteins that may normally interact with epsilon. We used the classical method of fluctuation tests (Luria and Delbruck 1943) to reexamine interactions between SOS genes and the dnaQ49 mutator phenotype. Our results confirmed those of Piechocki et al. (1986) that the mutation rates of dnaQ49 strains are influenced by the lexA genotype. However, we found that: (1) the effect of LexA was largely if not entirely attributable to its regulation of the umuDC and recA genes, (2) UmuDC+ and RecA* independently enhanced the dnaQ49 mutator phenotype, and (3) the temperature sensitivity for viability of dnaQ49 was exacerbated by RecA* activity in the absence of UmuDC. These results support the hypothesis that RecA* and one or both of the umuDC gene products interact with the replication complex and inhibit proofreading. Furthermore, UmuDC may function to stabilize the replication complex during mutagenic translesion DNA synthesis.
Materials and methods
Bacterial strains and plasmids
All strains used in this study are derivatives of DM1187 [=AB1157 (Bachmann 1987) but argE+ recA441 sulA11 lexA51 (formerly tif-1 sfiA11 spr-51) (Mount 1977)]. The various alleles used and their origins are given in Table 1. Strains were constructed by P1vir transduction (Miller 1972). The presence of the desired allele was confirmed by sensitivity or resistance to UV light for recA and lexA, non-mutability by methyl methanesulfonate for umuC-, or high spontaneous mutation frequency for dnaQ49 and mutL. All strains are Srl+; lexA+ and lexA3 strains are mal::Tn5 or mal+; dnaQ49 strains are zae::Tn10; and umuC36 strains are zcf-286::Tn5.
Table 1.
The alleles used in this study
| Allele | Alternative name | Phenotype | Source | Reference |
|---|---|---|---|---|
| recA alleles | ||||
| recA430 | lexB30 | Deficient in RecA* | GW2010, G. Walker, USA | Elledge and Walker (1983a); Roberts and Roberts (1981) |
| recA + | – | Wild type | AB1157, G. Walker, USA | Bachmann (1987) |
| recA441 | tif-1 | Thermally inducible RecA* | DM1187, E. Eisenstadt, USA | Mount (1977); Castellazzi et al. (1972); Roberts et al. (1978) |
| recA730 | – | Constitutive RecA* | RH4655, G. Maenhaut-Michel, Belgium | Witkin et al. (1982) |
| lexA alleles | ||||
| lexA3 | lexA (Ind–) | Cleavage resistant repressor | RB800, R. Brent, USA | Mount et al. (1972); Little et al. (1980); R. Brent (personal communication) |
| lexA + | – | Wild type | AB1157, G. Walker, USA | Bachmann (1987) |
| lexA51 | lexA (Def), spr51 | Defective repressor | DM1187, E. Eisenstadt, USA | Mount (1977); Castellazzi et al. (1972); Roberts et al. (1978) |
| umuC alleles | ||||
| umuC122::Tn5 | – | Nonmutable | GW2100, G. Walker, USA | Elledge and Walker (1983b) |
| umuC36 | – | Nonmutable | EE134, E. Eisenstadt, USA | Kato and Shinoura (1977); G. Walker (personal communication) |
| Other alleles | ||||
| dnaQ49 | – | Thermally inducible mutator, temperature sensitive viability | KH1366, B. Bachmann, USA | Horiuchi et al. (1978; 1981) |
| mutL::Tn10 | – | Mismatch repair defective | ES1484, E. Eisenstadt, USA | Radman and Wagner (1986); E. Siegel (personal communication) |
Media
LB media was as given in Miller (1972). Salt-free LB was as LB but without NaCl. Drugs were added in the following concentrations: tetracycline, 20 mg/l; kanamycin, 45 mg/l; streptomycin, 100 mg/l; rifampicin (rif), 100 mg/l.
Fluctuation tests
Strains were grown overnight in LB broth plus appropriate drugs at 32° C. Cultures were then plated on LB-rif to test for rifampicin resistance (rifr) and diluted 10–5 or 10–6, depending on viability, into salt-free LB broth to give an initial inoculum of 104–103 cells per ml. Aliquots were then grown at 32° C and 37° C. When saturated, the cultures were plated on LB-rif and titered on LB plates incubated at 32° C. Experiments in which there was a greater than 0.01 probability that the original inoculum contained a rifr cell were discarded. Mutation rates are based on 9–29 cultures and were calculated by the methods given in Lea and Coulson (1949).
Results
Mutation rates of dnaQ+ and dnaQ49 bacteria with various recA and lexA alleles
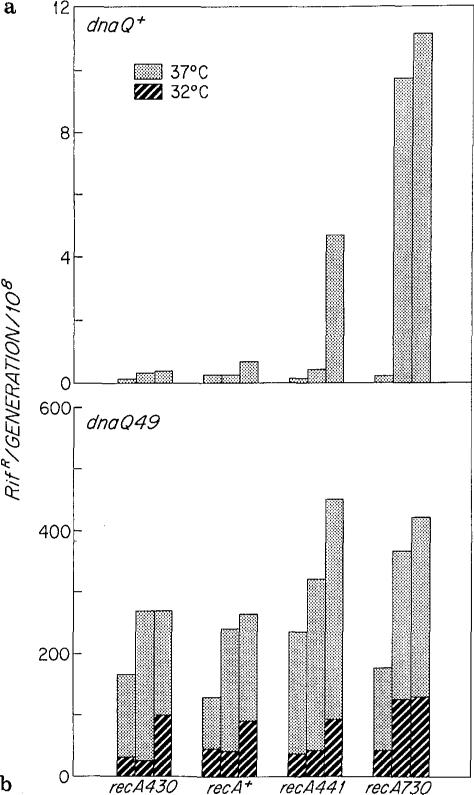
As detailed in Table 1, the strains used in these experiments have the following phenotypes: (1) deficient (recA430), DNA-damage inducible (recA+), temperature inducible (recA441), and constitutive (recA730) for RecA* activity, (2) repressed (lexA3), RecA*-inducible (lexA+), and constitutive (lexA51) for expression of the SOS response, and (3) wild type (dnaQ+) and temperature sensitive (dnaQ49) for epsilon activity. The spontaneous mutation rates to rifr of these strains are given in Fig. 1, which is organized so that the recA alleles from left to right have increasing RecA* activity and for each recA allele SOS derepression increases from left to right.
Fig 1 a, b.
Mutation rates of the strains in salt-free LB media: a dnaQ+; b dnaQ49. The recA alleles from left to right have increasing RecA* activity. For each recA allele the bars from left to right are the mutation rates of lexA3 [=lexA (Ind-)], lexA+, and lexA51 [=lexA (Def)] derivatives, Thus within each recA background SOS derepression increases from left to right
Figure 1 a shows the mutation rates of dnaQ+ strains at 37° C in salt-free LB media. Derepression of the SOS regulon (lexA51 vs lexA3) increased mutation rates of recA430 and recA+ strains 3-fold but of strains expressing RecA* (recA441 and recA730) 50- to 85-fold. Since even in the lexA51 background the mutation rate conferred by recA730 was still 20-fold greater than that of recA+, the mutator phenotype of this recA allele was largely due to amplified levels of RecA* per se, not to increased expression of other SOS genes due to the cleavage of LexA promoted by RecA*.
Figure 1 b shows the mutation rates of dnaQ49 derivatives of the same strains at 32° C and 37° C in salt-free LB medium. Although the dnaQ49 defect increased mutation rates one to three orders of magnitude, derepression of the SOS regulon (lexA51 vs lexA3) further enhanced mutation rates two- to threefold. In addition, RecA* activity (recA730 and recA441 at 37° C) resulted in a nearly twofold further increase in mutation rates.
The mutator phenotypes of the SOS genes and dnaQ49 are more than additive
The absolute magnitudes of the differences in mutation rates conferred by the various lexA and recA alleles were at least tenfold greater in the dnaQ49 background than in the dnaQ+ background. Thus the mutator phenotypes of dnaQ49 and of the recA/lexA alleles were clearly more than additive, suggesting that the proteins induced as part of the SOS response were influencing epsilon activity. However, an alternative explanation is that the loss of proofreading in dnaQ49 strains could reveal errors induced by SOS expression that are normally corrected in dnaQ+ strains. For example, if the mutation rate of the recA730 lexA51 dnaQ+ strain (Fig. 1 a) reflected only 7% of the potential mutations due to RecA* whereas 93% were normally corrected by proofreading, loss of epsilon activity alone could account for the difference in mutation rates at 37° C between recA+ lexA51 dnaQ49 and recA730 lexA51 dnaQ49 (Fig. 1 b).
If this second hypothesis is correct, the 20-fold difference in mutation rates between recA730 lexA51 dnaQ+ and recA+ lexA51 dnaQ+ (Fig. 1 a) reflects RecA*-induced errors that have escaped both proofreading and a second error correction pathway, mismatch repair (reviewed in Radman and Wagner 1986). Thus if undetected errors were present at tenfold higher levels than observed in recA730 dnaQ+ cells, these errors should also increase the mutation rate of a derivative defective in mismatch repair (mutL-). However, as shown in Table 2, the mutation rate of the recA730 lexA51 mutL::Tn10 strain was only slightly higher than its recA+ counterpart, a difference consistent with an additive effect of mutL- and recA730. Although other models are possible (see Discussion), the simplest interpretation of these results is that the differences among the recA/lexA derivatives of dnaQ49 were not due to failure to correct underlying errors, but due to the influence of the various recA and lexA alleles on the dnaQ49 phenotype itself.
Table 2.
Comparison of the mutation rates conferred by dnaQ49 and mutL::Tn10 in recA+ and recA730 strains
| Strain | Rifr/generation per 108 cellsa |
|||
|---|---|---|---|---|
| 32° C | n | 37° C | n | |
| dnaQ+ mut+ | ||||
| recA+ lexA51 | 0.56 | 10 | 0.65 | 10 |
| recA730 lexA51 | 10.6 | 12 | 11.1 | 12 |
| dnaQ49 mut + | ||||
| recA+ lexA51 | 90 | 28 | 266 | 22 |
| recA730 lexA51 | 134 | 29 | 417 | 22 |
| dnaQ+ mutL::Tn10 | ||||
| recA+ lexA51 | 115 | 10 | 110 | 9 |
| recA730 lexA51 | 127 | 10 | 135 | 9 |
n, number of determinations
Cultures were grown in salt-free LB at the designated temperatures
The separate effects of UmuDC, RecA* and SOS derepression on the dnaQ49 mutator phenotype
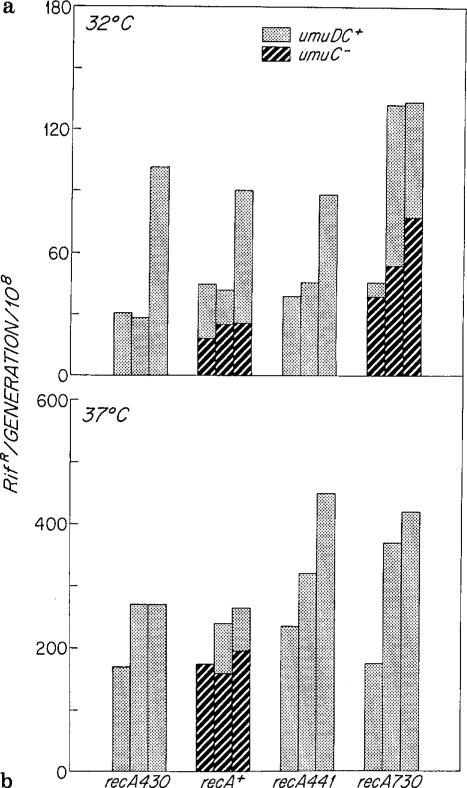
In Fig. 2 the mutation rates at 32° C and 37° C of all the dnaQ49 strains are compared to umuC122::Tn5 derivatives of the recA+ dnaQ49 and recA730 dnaQ49 strains. Because umuDC+ is essential to the mutagenic process but not to the expression of the SOS response (Kato and Shinoura 1977), umuC- strains allow the separate effects of SOS induction, RecA* activity and UmuDC activity to be evaluated. These effects were clearest at 32° C.
Fig. 2a, b.
The mutation rates of dnaQ49 strains compared to umuC::Tn5 derivatives in salt-free LB media: a at 32° C; b at 37° C. The recA alleles from left to right have increasing RecA* activity. For each recA allele the bars from left to right are the mutation rates of lexA3 [=lexA(Ind-)], lexA+, and lexA51 [lexA (Def)] derivatives
As shown in Fig. 2a, in recA+ strains the umuC122::Tn5 defect reduced mutation rates 40%–70%, i.e. about half of the mutations due to dnaQ49 were also dependent on umuC+. Full induction of the umuDC+ operon in the lexA51 strain resulted in a further twofold increase in mutation rate.
RecA* activity enhanced the dnaQ49 mutator pheno-type independently of umuC or lexA (Fig. 2a). The mutation rate of the recA730 umuC122::Tn5 lexA3 strain was twofold greater than its recA+ counterpart; with derepression, this difference increased to threefold. The effect of umuDC+ appeared to be additive to that of RecA* since when SOS was fully induced (lexA51), the magnitude of the difference in mutation rates between umuC122::Tn5 and umuC+ derivatives was almost exactly the same in recA730 as in recA+ strains. The independence of RecA* and UmuDC+ activities in the dnaQ49 background contrasted with their mutator phenotype in dnaQ+ strains, which required both the recA730 and umuDC+ genotypes (data not shown).
There appeared to be little effect of lexA alleles independent of the regulation of recA and umuDC by LexA. Only the small (1.4-fold) difference in mutation rates between lexA3 and lexA51 derivatives of recA+ umuC122::Tn5 dnaQ49 could be attributed to the effect of SOS induction per se (Fig. 2 a). The constitutive RecA* activity of recA730 should result in LexA+ cleavage; in confirmation, there was little difference in overall mutation rates between recA730 lexA+ and recA730 lexA51.
The mutation rates of the recA430 and recA441 strains confirmed the results obtained with recA+ and recA730 (Fig. 2a). At 32° C the RecA441 enzyme is not induced for RecA* activity (Mount 1977), and thus at that temperature recA441 and recA+ strains were essentially the same. However, some RecA* activity appeared to contribute to the mutation rates of both recA+ and recA441 strains since the mutation rates of the RecA*-deficient recA430 strains were 50% lower in lexA3 and lexA+ backgrounds. This difference was entirely overcome by derepression of SOS, suggesting either: (1) a constitutive level of LexA cleavage in recA+ and recA441 strains was allowing some expression of the umuDC operon, or (2) amplified RecA430 was equivalent to RecA+, a result consistent with the residual level of RecA* activity retained by RecA430 (Devoret et al. 1983).
As shown in Fig. 2b, most of the differences among the strains observed at 32° C were also evident at 37° C. Since the mutation rates were higher the effects of the SOS genes at 37° C were relatively smaller, although of equal or greater absolute magnitude to their effects at 32° C. There was again little impact of SOS derepression independent of umuDC+ or RecA*. Although data from the umuC- derivatives of recA730 were not obtainable (see below), it appeared that the magnitude of the increase in mutation rates due to umuDC+ was equivalent at 37° C and 32° C, but the magnitude of the increase due to RecA* was relatively greater at the higher temperature. The RecA* activity of RecA441 was clearly activated at 37° C since mutation rates in this background were similar to those of recA730 strains.
recA730 and umuC- increase the temperature sensitivity for viability of dnaQ49
None of the recA730 umuC122::Tn5 dnaQ49 strains were viable at 37° C in salt-free LB. Survival was somewhat greater on solid media but, as shown in Table 3, the plating efficiency of the recA730 umuC122::Tn5 lexA51 dnaQ49 strain at 37° C was still three orders of magnitude less than its isogenic controls. This temperature sensitivity was not due to overexpression of the umuC::Tn5 fusion since umuC36 strains were likewise non-viable (data not shown).
Table 3.
The plating efficiencies of umuC– lexA51 dnaQ49 strains at 32° C and 37° C on salt-free LB media
| Strain | Viable cells per ml × 10–8 |
||
|---|---|---|---|
| LB | Salt-Free LB |
||
| 32° C | 32° C | 37° C | |
| umuC+ lexA51 dnaQ49 | |||
| recA + | 9.9 | 6.6 | 6.9 |
| recA730 | 13.2 | 9.5 | 11.2 |
| umuC122::Tn5 lexA51 dnaQ49 | |||
| recA + | 9.3 | 5.9 | 8.5 |
| recA730 | 6.9 | 2.5 | 0.0085 |
Discussion
The results presented here demonstrate that both UmuDC+ and RecA* independently increase the spontaneous mutation rates of strains with a defective epsilon, DnaQ49. In addition, the temperature sensitivity for viability conferred by dnaQ49 is increased by expression of RecA* activity in the absence of UmuDC+. dnaQ49 is recessive to dnaQ+ for both replication fidelity and viability (Maruyama et al. 1983). Both phenotypes of dnaQ49 are due to a mis-sense mutation that can suppress a dominant mutator allele of dnaQ, mutD51, in cis but not in trans (Takano et al. 1986). These observations and the fact that DNA polymerization by the alpha subunit of Pol III in vitro is intrinsically temperature sensitive (Maki and Kornberg 1985) have led Takano et al. (1986) to propose that DnaQ49 is defective in its ability to bind to the alpha subunit of Pol III. The rationale of the experiments presented here was that if SOS-induced proteins interact with Pol III, then they may exacerbate the conditional phenotypes of the mutant DnaQ49. The results obtained support this model.
Comparing recA+ umuC122::Tn5 lexA3 dnaQ49 and recA730 umuC+ lexA51 dnaQ49 strains at 32° C (Fig. 2a), the combined effects of SOS derepression and RecA* and UmuDC+ activity resulted in a 7-fold increase in mutation rate (equal to a 24-fold increase in average mutation frequency). This 7-fold increase was composed of a small effect of SOS derepression per se (1.4 ×) and about equal effects of UmuDC+ (3 ×) and RecA* (3 ×). At 37° C (Fig. 2b) the relative increase in mutation rates between the strains was smaller (3-fold) and was almost entirely due to UmuDC+ (1.4 ×) and RecA* (1.6 ×).
Our results and conclusions differ in a number of respects from those of a previous study by Piechocki et al. (1986). (1) They found a tenfold lower mutation frequency in lexA (Ind-) dnaQ49 vs lexA+ dnaQ49 strains whereas we found in recA+ or recA430 backgrounds little difference between lexA (Ind-) dnaQ49 and lexA+ dnaQ49 but equivalently higher mutation rates in lexA (Def) dnaQ49 strains. (2) They reported (in a note added in proof) no effect of umuC- on the dnaQ49 mutator phenotype whereas we found that full induction of umuDC+ increased the mutation rates of dnaQ49 strains threefold. (3) They found that recA (Def) alleles did not affect the dnaQ49 mutator phenotype, which is consistent with our finding that there was little difference between recA+ dnaQ49 and recA430 dnaQ49 strains. However, we further demonstrated that fully-induced RecA* activity increased the mutation rates of dnaQ49 strains threefold.
In summary, Piechocki et al. (1986) concluded that the dnaQ49 mutator phenotype was not influenced by RecA or UmuDC but was enhanced by a unknown LexA-repressed gene. In contrast, we conclude that both UmuDC+ and RecA* enhance the dnaQ49 mutator phenotype, that the effects of lexA alleles are almost entirely attributable to regulation of recA and umuDC, and find little evidence for a third gene. While further experimentation is necessary to fully resolve these discrepancies, some may be due to different genetic background or to the large variations in mutation frequencies that obtain at high mutation rates. However, one difference in method - our use of salt-free LB media - is not significant as we confirmed our results in normal LB (data not shown).
Although the obvious interpretation of our results is that RecA* and UmuDC influence epsilon activity, there are alternative explanations. One possibility, that loss of epsilon activity is revealing the underlying errors induced via the SOS pathway, is rendered less likely by the failure of a defect in mismatch repair to likewise reveal these errors (Table 2). This experiment is not definitive since the errors induced by SOS expression may be poorly repaired by mismatch repair but efficiently repaired by proofreading (e. g. see Jones et al. 1987). However, also arguing against this interpretation is the fact that RecA*-dependent mutations in dnaQ+ strains are also umuDC+-dependent (Ciesla 1982; Blanco et al. 1982; our observations), whereas RecA* and UmuDC act independently to enhance the mutation rates of dnaQ49 strains (Fig. 2). Thus the mutant epsilon appears to be revealing separate activities of RecA* and UmuDC that are not evident in dnaQ+ cells.
While we are attributing the effects of recA and umuC alleles on the dnaQ49 phenotypes to RecA* and UmuDC, it is possible that other activities are responsible. For example, the impact of umuC- (Fig. 2, Table 3) could be due to loss of UmuC function alone instead of loss of cooperative UmuDC activity. Since RecA* facilitates the cleavage of UmuD (Burckhardt et al. 1988; Nohmi et al. 1988; Shinagawa et al. 1988), the effects of RecA* activity could actually be due to the cleavage product of UmuD. However, UmuD and UmuC probably function together since the phenotypes of umuD- and umuC- are equivalent (Kato and Shinoura 1977; Elledge and Walker 1983 b) and umuD+ and umuC+ fail to complement mutations in their evolutionarily diverged homologues, mucA and mucB respectively (Perry et al. 1985). Nonetheless, other models incorporating more complex interactions can be devised.
With the above caveats, the simplest interpretation of our results is that RecA* and UmuDC directly interact with epsilon or interact with other subunits of the replication complex and thereby affect epsilon activity. The finding that RecA protein can inhibit the 3′ → 5′ exonuclease activity of epsilon in vitro (Fersht and Knill-Jones 1983; Lu et al. 1986) strongly suggests that a direct interaction between RecA* and epsilon occurs in vivo. We further theorize that UmuDC may interact with alpha, the product of dnaE. Our finding that mutations in umuC increase the temperature sensitivity of dnaQ49 when RecA* is active is reminiscent of a similar increase in temperature sensitivity seen in dnaE486 dnaQ49 strains (Horiuchi et al. 1981). In addition, suppressors of lethal dnaQ null mutants have been found in dnaE, i.e. mutations in alpha can relieve its requirement for epsilon (Maurer et al. 1988). Thus an attractive model is that UmuDC may interact with and stabilize alpha when epsilon activity is inhibited.
It was originally proposed by Villani et al. (1978) that exonucleolytic proofreading is inhibited during SOS mutagenesis, thereby allowing replication to continue past nucleotides misincorporated opposite DNA lesions. Bridges and Woodgate (1985) subsequently proposed that misincorporation and lesion bypass are separate processes since only the latter required UmuDC. mutD5, a dominant mutator allele of dnaQ, had no effect on UV mutagenesis, leading Woodgate et al. (1987) to conclude that, as predicted, proofreading by epsilon is not active during SOS mutagenesis. However, the mutD5 defect did not restore UV mutability to umuC- bacteria, suggesting that inhibition of proofreading is not sufficient to allow replication past DNA lesions (Woodgate et al. 1987). The dominance of the mutD5 allele implys that, although defective in proofreading, the MutD5 protein binds alpha at least as tightly as wild-type epsilon (Takano et al. 1986). Thus if SOS mutagenesis involves a modification of the interaction between epsilon and alpha, mutD5 and dnaQ+ bacteria may not differ in that interaction. Similar experiments with the dnaQ49 allele may resolve this issue.
In summary, the results reported here suggest that UmuDC and RecA* interact with the replication complex and inhibit epsilon activity. Thus our data support the hypothesis that SOS mutagenesis involves inhibition of proofreading and a resulting decrease in the replication fidelity of Pol III. However, since most SOS-dependent mutations appear to be targeted to the sites of DNA lesions (Foster et al. 1982; Miller 1982), inhibition of proofreading normally must be transient. The results presented here further suggest that UmuDC may function to stabilize the replication complex during mutagenic translesion DNA synthesis.
Acknowledgements
We thank the people mentioned in Table 1 for bacterial strains. Eric Eisenstadt, John Cairns, Russell Maurer, John Drake and Jeffrey Miller provided many helpful discussions and suggestions. In addition, E. Eisenstadt, R. Maurer and J. Cairns critically read a former version of this manuscript. This work was supported by Public Health Service Grant CA37880 awarded to P.L.F. by the National Cancer Institute, USA
Footnotes
Communicated by R. Devoret
Note added in proof
It has recently been proposed that the UmuDC proteins increase the stability of the replication complex during translesion DNA synthesis by acting as processivity factors [Battista JF, Nohmi T, Donnelly CE, Walker GC (1988) Role of UmuD and UmuC in UV and chemical mutagenesis. In: Friedberg E, Hanawalt P (eds) Mechanism and consequences of DNA damage processing. Liss, New York, in press; Hevroni D, Livneh Z (1988) Bypass and termination at apurinic sites during replication of single-stranded DNA in vitro: a model for apurinic site mutagenesis. Proc Natl Acad Sci USA 85:5046-5050]
References
- Bachmann BJ. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt FC, Ingraham JL, Magasanik B, Low KB, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium cellular and molecular biology. American Society for Microbiology; Washington DC: 1987. pp. 1190–1219. [Google Scholar]
- Blanco M, Herrera G, Collado P, Rebollo J, Botella LM. Influence of RecA protein on induced mutagenesis. Biochimie. 1982;64:633–636. doi: 10.1016/s0300-9084(82)80102-8. [DOI] [PubMed] [Google Scholar]
- Bridges BA, Woodgate R. Mutagenic repair in Escherichia coli: products of the recA gene and of the umuD and umuC genes act at different steps in UV induced mutagenesis. Proc Natl Acad Sci USA. 1985;82:4193–4197. doi: 10.1073/pnas.82.12.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckhardt SE, Woodgate R, Scheuermann RH, Echols H. UmuD mutagenesis protein of Escherichia coli: overproduction, purification, and cleavage by RecA. Proc Natl Acad Sci USA. 1988;85:1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellazzi M, George J, Buttin B. Prophage induction and cell division in E. coli: further characterization of a thermo-sensitive mutation tif-1 whose expression mimics the effect of UV irradiation. Mol Gen Genet. 1972;119:139–152. doi: 10.1007/BF00269133. [DOI] [PubMed] [Google Scholar]
- Ciesla Z. Plasmid pKM101-mediated mutagenesis in Escherichia coli is inducible. Mol Gen Genet. 1982;186:298–300. doi: 10.1007/BF00331866. [DOI] [PubMed] [Google Scholar]
- Devoret R, Pierre M, Moreau PL. Prophage ϕ80 is induced in Escherichia coli K12 recA430. Mol Gen Genet. 1983;189:199–206. doi: 10.1007/BF00337804. [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Walker GC. The muc genes of pKM101 are induced by DNA damage. J Bacteriol. 1983a;155:1306–1315. doi: 10.1128/jb.155.3.1306-1315.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ, Walker GC. Proteins required for ultraviolet light and chemical mutagenesis: identification of the products of the umuC locus of Escherichia coli. J Mol Biol. 1983b;164:175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- Ennis DG, Fisher B, Edmiston S, Mount DW. Dual role for Escherichia coli RecA protein in SOS mutagenesis. Proc Natl Acad Sci USA. 1985;82:3325–3329. doi: 10.1073/pnas.82.10.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht AR, Knill-Jones JW. Contribution of 3′ → 5′ exonuclease activity of DNA Polymerase III holoenzyme from Escherichia coli to specificity. J Mol Biol. 1983;165:669–682. doi: 10.1016/s0022-2836(83)80273-3. [DOI] [PubMed] [Google Scholar]
- Foster PL, Eisenstadt E, Cairns J. Random components in mutagenesis. Nature. 1982;299:365–367. doi: 10.1038/299365a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T, Maki H, Sekiguchi M. A new conditional lethal mutator (dnaQ49) in Escherichia coli K12. Mol Gen Genet. 1978;163:277–283. doi: 10.1007/BF00271956. [DOI] [PubMed] [Google Scholar]
- Horiuchi T, Maki H, Sekiguchi M. Conditional lethality of Escherichia coil strains carrying dnaE and dnaQ mutations. Mol Gen Genet. 1981;181:24–28. doi: 10.1007/BF00339000. [DOI] [PubMed] [Google Scholar]
- Jones M, Wagner R, Radman M. Repair of a mismatch is influenced by the base composition of the surrounding nucleotide sequence. Genetics. 1987;115:605–610. doi: 10.1093/genetics/115.4.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977;156:121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- Khidhir MS, Casaregola S, Holland IB. Mechanism of transient inhibition of DNA synthesis in ultraviolet irradiated E. coli: inhibition is independent of recA whilst recovery requires RecA protein itself and an additional, inducible SOS function. Mol Gen Genet. 1985;199:133–140. doi: 10.1007/BF00327522. [DOI] [PubMed] [Google Scholar]
- Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–265. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- Little JW. Autodigestion of lexA and phage lambda repressors. Proc Natl Acad Sci USA. 1984;81:1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JW, Edmiston SH, Pacelli LZ, Mount DW. Cleavage of the Escherichia coli lexA protein by the recA protease. Proc Natl Acad Sci USA. 1980;77:3225–3229. doi: 10.1073/pnas.77.6.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Schermann RH, Echols H. Capacity of RecA protein to bind preferentially to UV lesions and inhibit the editing subunit (ε) of DNA polymerase III: a possible mechanism for SOS-induced targeted mutagenesis. Proc Natl Acad Sci USA. 1986;83:619–623. doi: 10.1073/pnas.83.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria SE, Delbruck M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H, Kornberg A. The polymerase subunit of DNA polymerase III of Escherichia coli. J Biol Chem. 1985;260:12987–12992. [PubMed] [Google Scholar]
- Marsh L, Walker GC. Cold sensitivity induced by overproduction of UmuDC in Escherichia coli. J Bacteriol. 1985;162:155–161. doi: 10.1128/jb.162.1.155-161.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama M, Horiuchi T, Maki H, Sekiguchi M. A dominant (mutD) and a recessive (dnaQ49) mutator of Escherichia coli. J Mol Biol. 1983;167:757–771. doi: 10.1016/s0022-2836(83)80109-0. [DOI] [PubMed] [Google Scholar]
- Maurer R, Lancy E, Lifsics M, Munson P. Essential function of the editing (ε) subunit of DNA polymerase III in Salmonella typhimurium. In: Moses RE, Summers WC, editors. DNA replication and mutagenesis. American Society for Microbiology; Washington DC: 1988. pp. 247–253. [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1972. [Google Scholar]
- Miller JH. Carcinogens induce targeted mutations. Cell. 1982;31:5–7. doi: 10.1016/0092-8674(82)90398-1. [DOI] [PubMed] [Google Scholar]
- Miller JH, Low KB. Specificity of mutagenesis resulting from the induction of the SOS system in the absence of mutagenic treatment. Cell. 1984;37:675–682. doi: 10.1016/0092-8674(84)90400-8. [DOI] [PubMed] [Google Scholar]
- Mount DW. A mutant of Escherichia coli showing constitutive expression of the lysogenic induction and error-prone DNA repair pathways. Proc Natl Acad Sci USA. 1977;74:300–304. doi: 10.1073/pnas.74.1.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount DW, Low KB, Edmiston SJ. Dominant mutations (lex) in Escherichia coli K-12 which affect radiation sensitivity and frequency of ultraviolet light induced mutations. J Bacteriol. 1972;112:886–893. doi: 10.1128/jb.112.2.886-893.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohmi T, Battista JR, Dodson LA, Walker GC. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and post-translational activation. Proc Natl Acad Sci USA. 1988;85:1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry KL, Elledge SJ, Mitchell BB, Marsh L, Walker GC. The umuDC and mucAB operons whose products are required for UV and chemical mutagenesis: UmuD, MucA and LexA proteins share homology. Proc Natl Acad Sci USA. 1985;82:4331–4335. doi: 10.1073/pnas.82.13.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechocki R, Kupper D, Quinones A, Langhammer R. Mutational specificity of a proof-reading defective Escherichia coli dnaQ49 mutator. Mol Gen Genet. 1986;202:162–168. doi: 10.1007/BF00330533. [DOI] [PubMed] [Google Scholar]
- Radman M. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. In: Hanawalt P, Setlow RB, editors. Molecular mechanisms for repair of DNA, part A. Plenum Publishing Corporation; New York: 1975. pp. 355–367. [DOI] [PubMed] [Google Scholar]
- Radman M, Wagner RE., Jr Mismatch repair in Escherichia coli. Annu Rev Genet. 1986;20:523–538. doi: 10.1146/annurev.ge.20.120186.002515. [DOI] [PubMed] [Google Scholar]
- Roberts JW, Roberts CW. Two mutations that alter the regulatory activity of E. coli recA protein. Nature. 1981;290:422–424. doi: 10.1038/290422a0. [DOI] [PubMed] [Google Scholar]
- Roberts JW, Roberts CW, Craig NL. Escherichia coli recA gene product inactivates phage 2 repressor. Proc Natl Acad Sci USA. 1978;75:4714–4718. doi: 10.1073/pnas.75.10.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann R, Schuman T, Burgers PMJ, Lu C, Echols H. Identification of the ε-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc Natl Acad Sci USA. 1983;80:7085–7089. doi: 10.1073/pnas.80.23.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinagawa H, Iwasaki H, Kato T, Nakata A. RecA protein-dependent cleavage of UmuD protein upon SOS mutagenesis. Proc Natl Acad Sci USA. 1988;85:1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano K, Nakabeppu Y, Maki H, Horiuchi T, Sekiguchi M. Structure and function of dnaQ and mutD mutators of Escherichia coli. Mol Gen Genet. 1986;205:9–13. doi: 10.1007/BF02428026. [DOI] [PubMed] [Google Scholar]
- Villani G, Bouteux S, Radman M. Mechanisms of ultraviolet induced mutagenesis: extent and fidelity of in vitro DNA synthesis on irradiated templates. Proc Natl Acad Sci USA. 1978;75:3037–3041. doi: 10.1073/pnas.75.7.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GC. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984;48:60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin EM, Kogoma T. Involvement of the activated form of RecA protein in SOS mutagenesis and stable DNA replication in Escherichia coli. Proc Natl Acad Sci USA. 1984;81:7539–7543. doi: 10.1073/pnas.81.23.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin EM, McCall JO, Volkert MR, Wermundsen IE. Constitutive expression of SOS functions and modulation of muta-genesis resulting from resolution of genetic instability at or near the recA locus of Escherichia coli. Mol Gen Genet. 1982;185:43–50. doi: 10.1007/BF00333788. [DOI] [PubMed] [Google Scholar]
- Witkin EM, Roegner-Maniscalco V, Sweasy JB, McCall JO. Recovery from ultraviolet light-induced inhibition of DNA synthesis requires umuDC gene products in recA718 mutant strains but not in recA+ strains of Escherichia coli. Proc Natl Acad Sci USA. 1987;84:6805–6809. doi: 10.1073/pnas.84.19.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R, Bridges BA, Herrera G, Blanco M. Mutagenic DNA repair in Escherichia coli XIII: proofreading exonuclease of DNA polymerase III holoenzyme is not operational during UV mutagenesis. Mutat Res. 1987;183:31–37. doi: 10.1016/0167-8817(87)90042-3. [DOI] [PubMed] [Google Scholar]