Abstract
BioPAX (Biological Pathway Exchange) is a standard language to represent biological pathways at the molecular and cellular level. Its major use is to facilitate the exchange of pathway data (http://www.biopax.org). Pathway data captures our understanding of biological processes, but its rapid growth necessitates development of databases and computational tools to aid interpretation. However, the current fragmentation of pathway information across many databases with incompatible formats presents barriers to its effective use. BioPAX solves this problem by making pathway data substantially easier to collect, index, interpret and share. BioPAX can represent metabolic and signaling pathways, molecular and genetic interactions and gene regulation networks. BioPAX was created through a community process. Through BioPAX, millions of interactions organized into thousands of pathways across many organisms, from a growing number of sources, are available. Thus, large amounts of pathway data are available in a computable form to support visualization, analysis and biological discovery.
Keywords: pathway data integration, pathway database, standard exchange format, ontology, information system
Introduction
Molecular biology research has yielded detailed knowledge of biomolecular components and their interactions. Increasingly powerful technologies, including genome-wide molecular measurements, have accelerated the progress towards a complete map of molecular interaction networks in cells and between cells of key organisms. A single person can no longer memorize these maps, therefore, they must be represented in a form suitable for computer processing and storage and made easily available to scientists via software systems. Accordingly, the BioPAX (Biological Pathway Exchange) project aims to facilitate knowledge representation, systematic collection, integration and wide distribution of pathway data from heterogeneous information sources and thereby, their incorporation into distributed biological information systems that support visualization and analysis.
Goal: toward complete representation of basic cellular processes
Biology has come a long way since the Boehringer-Mannheim wall chart of metabolic pathways1 and the Nicholson Metabolic Map2. Since then, a number of groups have developed methods and databases for organizing pathway information3-16, but only recently collaborated as part of the BioPAX project to develop a generally accepted standard way of representing these pathway maps. Complete molecular process maps must include all interactions, reactions, dependencies, influence and information flow between pools of molecules in cells and between cells. For ease of use and simplicity of presentation, such network maps are often organized in terms of sub-networks or pathways. Pathways are models that biologists have delineated within the entire cellular biochemical network that help us describe and understand specific biological processes. Thus, a useful definition of a pathway is a set of interactions between physical or genetic cell components, often describing a cause-and-effect or time-dependent process, which explain some observable biological function. How do we represent these pathways in a generally accepted and computable form?
Challenge: strong growth of pathway databases
The total volume of pathway data mapped by biologists and stored in databases has entered a rapid growth phase17, similar to the rapid expansion of biological sequence data after the introduction of automated sequencing technology. The number of pathway and molecular interaction related online resources has grown from 190 in 2006 to 325 in 2010, a 70% increase17. In addition, molecular profiling methods, such as RNA profiling using microarrays or protein quantification using mass spectrometry, provide large amounts of information about the dynamics of cellular pathway components and increase the power of pathway analysis techniques18,19. However, this growth poses a formidable challenge for pathway data collection and curation as well as for database, visualization and analysis software, as these data are often fragmented.
Impediment: fragmentation of pathway databases
The principal motivation for building pathway databases and software tools is to facilitate qualitative and quantitative analysis and modeling of large biological systems using a computational approach. Over 300 pathway or molecular interaction related data resources17 and many visualization and analysis software tools3,20-22 have been developed. Unfortunately, most of these databases and tools were originally developed to use their own pathway representation language, resulting in a heterogeneous set of resources that are extremely difficult to combine and use. This has occurred because many different research groups, each with their own system for representing biomolecules and their interactions in a pathway, work independently to collect pathway data recorded in the literature (estimated from text-mining projects23 to be present in at least 10% of the over 20 million articles currently indexed by PubMed). As a result, researchers waste much time collecting information from different sources and converting its representation from one system to another. They may pay substantial opportunity cost as a result of pathway data fragmentation. For instance, visualization and analysis tools developed for one pathway database cannot be reused for others, making software development efforts more expensive. The situation currently resembles biologists assembling a multi-dimensional puzzle, with thousands of pieces, each one created and shared ad hoc. It is, therefore, imperative to develop computational methods to cope with both the magnitude and fragmented nature of this rapidly expanding and exceedingly valuable pathway information. While independent research efforts are needed to find the best ways to represent pathways, community coordination and agreement on one or a few standard sets of semantics is necessary to be able to efficiently integrate pathway data from multiple sources on a large scale.
Requirement: a shared language for pathways
A common, inclusive and computable pathway data language is necessary to share knowledge about pathway maps and to facilitate integration and use for hypothesis testing in biology24. A shared language facilitates communication by reducing the number of translations required to exchange data between multiple sources (Figure 1). Developing such a representation is challenging due to the large variety of pathways in biology and the diverse uses of pathway information. Pathway representations frequently use abstractions for metabolic, signaling, gene regulation, protein interaction and genetic interaction and these serve as a starting point toward a shared language25. Also, several variants of this common language may be required to answer relevant research questions in distinct fields of biology, each covering unique levels of detail addressing different uses, but these should be rooted on common principles and must remain compatible.
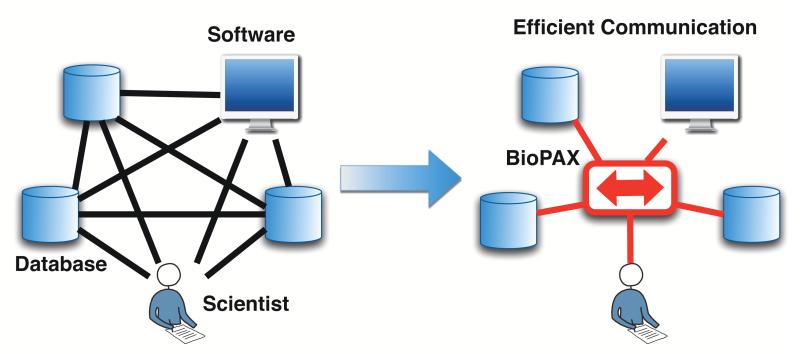
Figure 1.
BioPAX is a shared language for biological pathways. BioPAX reduces the effort required to efficiently communicate between pathway users, databases and software tools. Without a shared language, each system must speak the language of all other systems in the worst case (black lines). With a shared language, each system only needs to speak that language (central red box).
Implementation: the BioPAX biological pathway exchange language
BioPAX was developed to address these challenges. We have developed BioPAX as a shared language to facilitate communication between diverse software systems and to establish standard knowledge representation of pathway information. BioPAX supports representation of metabolic and signaling pathways, molecular and genetic interactions and gene regulation. Relationships between genes, small molecules, complexes and their states (e.g. post-translational protein modifications, mRNA splice variants, cellular location) are described, including the results of events. Details about the BioPAX language are available in online documentation at http://www.biopax.org. The BioPAX language provides a set of terms, with associated descriptions, to represent many aspects of biological pathways and their annotation. It is implemented as an ontology, a formal system of describing knowledge (Box 1) that helps structure pathway data so that it is more easily processed by computer software (Figure 2). It provides a standard syntax used for data exchange that is based on OWL (Web Ontology Language) (Box 1). Finally, it provides a validator that uses a set of rules to verify whether a BioPAX document is complete, consistent and free of common errors. BioPAX is the only community standard for biological pathway exchange to and from databases, but coordinates with other standards in related areas (Figure 6).
Box 1. What is an ontology?
An ontology is a formal system for representing knowledge62. Formal representation is required for computer software to make use of information. Formal knowledge systems have been used in science for thousands of years, for example, Aristotle’s representation of the basic elements of all things (the five elements Fire, Earth, Air, Water and Ether). Well known modern examples include organism taxonomies63 or the Gene Ontology64. A formal representation allows for consistent communication of knowledge between individuals or computer systems and helps manage complexity in information processing as knowledge is broken down into clear concepts that can be considered independently. Ontologies also enable integration of knowledge between independent resources linked on the World Wide Web (WWW). Such linked, structured data form the basis of the semantic web, an extension of the WWW that promises improved information management and search capability61. Representing and sharing knowledge using ontologies is simplified by availability of the standard web ontology language (OWL) (http://www.w3.org/TR/owl-features/). Tools to edit OWL, such as Protégé65, have been developed by the Semantic Web community and adopted in the life sciences.
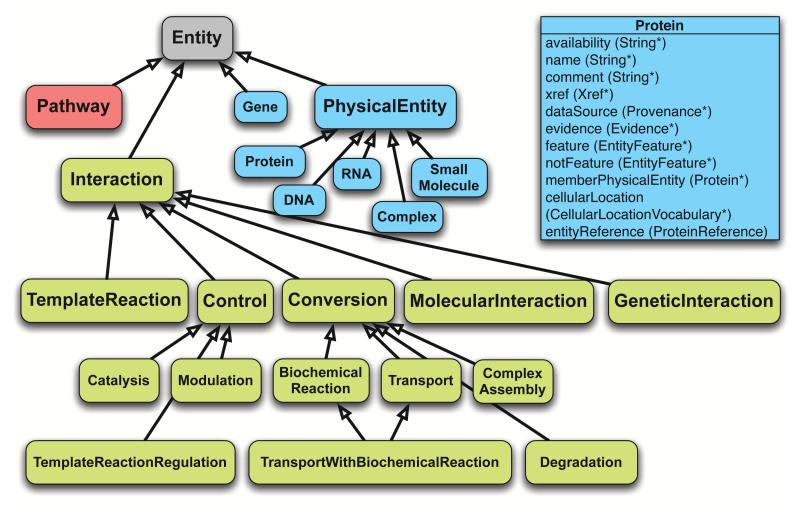
An ontology is composed of classes, properties (representing relations) and restrictions and is used to define individuals (instances of classes, also known as objects) and values for their properties. Classes (also known as concepts, types) are often arranged into a specialization hierarchy (or taxonomy) where child classes are more specific than, and inherit the properties of, parent classes. For example, in BioPAX, the Biochemical Reaction class is a ‘subclass of’ the Conversion class. Classes may have properties (also known as fields, attributes or slots), which express possible relations to other classes (i.e. the may have values of specific types). For example, a Small Molecule is related to the Chemical Structure class by the property structure. Restrictions (also known as constraints) define allowable values and connections within an ontology. For example, Molecular Weight must be a positive number. Individuals are instances of classes where values occupy the properties of those instances. BioPAX defines the classes, properties and restrictions required to represent biological pathways and leaves creation of the individuals to users (data providers and consumers). Advantages of implementing BioPAX using OWL are that both the ontology and the individuals and values can be stored in the same XML-based format, which makes data transmission easier. Also, OWL is a standard ontology language that is supported by useful software tools for editing, transmitting, querying, reasoning and visualizing.
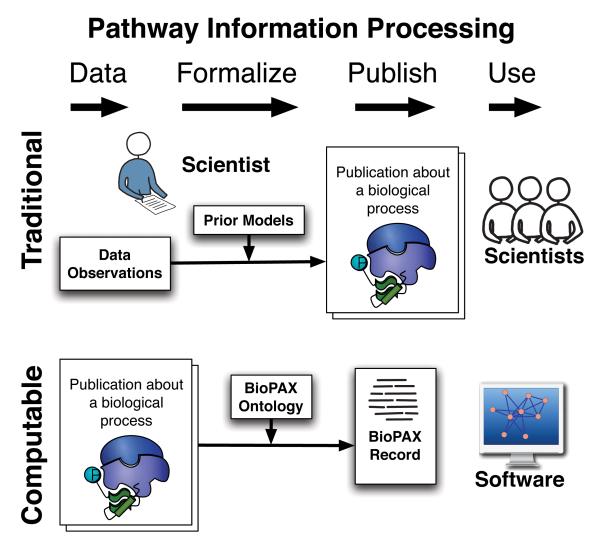
Figure 2.
BioPAX enables computational data gathering, publication and use of information about biological processes. Traditional pathway information processing: Observations considering prior models published as text and figures. Computable pathway information processing: Scientist’s description represented using formal, computable framework (ontology) published in a computer software readable format for analysis by scientists.
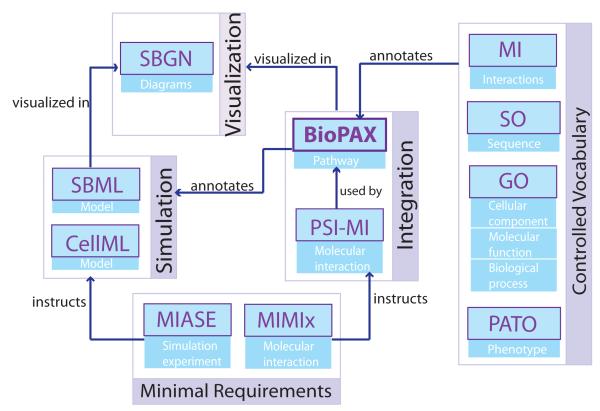
Figure 6.
The relationship among popular standard formats for pathway information. BioPAX and PSI-MI are designed for data exchange to and from databases and pathway and network data integration. SBML and CellML are designed to support mathematical simulations of biological systems and SBGN represents pathway diagrams.
Example of a pathway in BioPAX
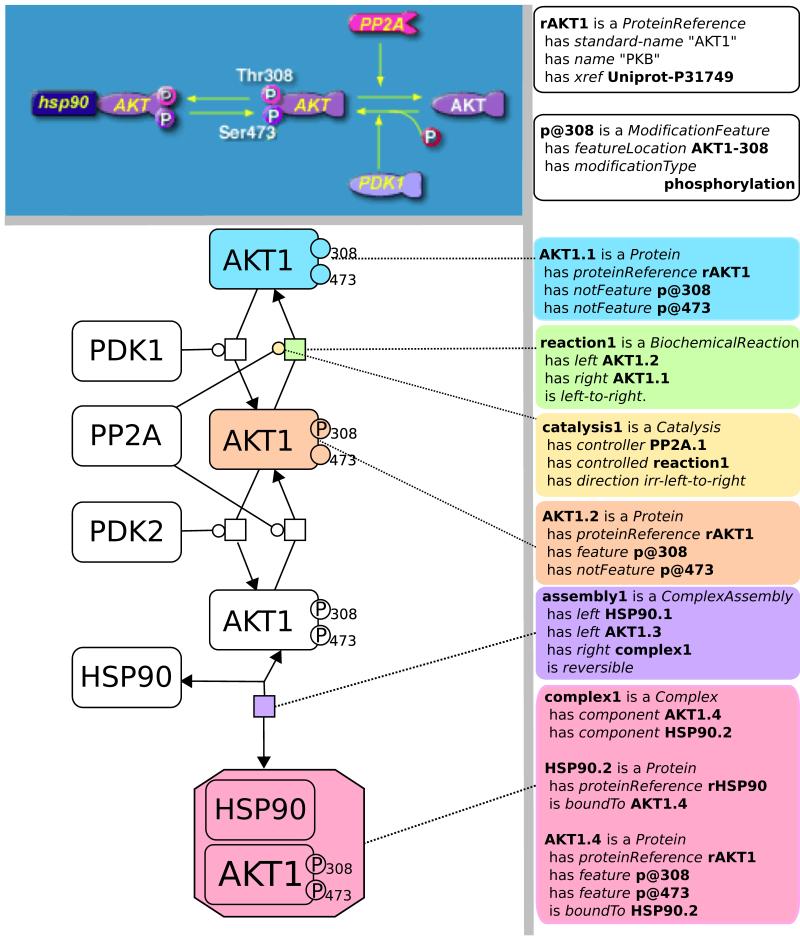
Pathway models described by biologists are generally expressed in scientific language and as network diagrams. An example is the AKT signaling pathway, important in regulating proliferation in many eukaryotic cells and often deregulated in cancer26,27. The AKT pathway is a cell surface receptor activated signaling cascade that transduces signals from the outside to the inside of a cell via a series of molecular binding and protein post-translational regulation events. These include protein-protein interactions and protein kinase mediated phosphorylation events that successively activate downstream kinases to phosphorylate additional proteins and activate or inhibit molecular interactions. The activated pathway eventually results in activation of multiple transcription factors, which turn on sets of genes to promote cell survival. A typical AKT signaling pathway diagram with associated text description can only be interpreted by people, and not computationally. By representing the pathway using the BioPAX language (Figure 3), it can also be interpreted by computer software and made available for numerous uses, such as pathway analysis of gene expression data. Representing a pathway using the BioPAX language sometimes necessitates being more explicit to avoid capturing inconsistent data. For instance, the typical notion of an ‘active protein’ is context dependent, as the same molecule could be active in one cellular context, such a cellular compartment with a set of potential interacting molecules, and inactive in another context. Thus, capturing the specific mechanism of activation, such as phosphorylation modification, is usually required, and the presence of downstream events that include the modified form signifies that the molecule is active. Interactions where the mechanism of action is unknown can also be specified.
Figure 3.
The AKT pathway as represented by a traditional method (top left, from http://www.biocarta.com), a formalized SBGN diagram (http://www.sbgn.org 84) (left), and using the BioPAX language (right). An important advantage of the BioPAX representation is that it can be interpreted by computer software and used in multiple ways, including automatic diagram creation, information retrieval and analysis. Online documentation at http://www.biopax.org contains more details about how to represent diverse types of biological pathways. Actual samples of pathway data in BioPAX OWL XML format are available in Supplementary Tables S2 and S3.
What does BioPAX include?
BioPAX covers all major concepts familiar to biologists studying pathways, including metabolic and signaling pathways, gene regulatory networks and genetic and molecular interactions (Table 3). The BioPAX language is distributed as an ontology definition (Figure 4) with associated documentation, a validator and other software tools (Table 1). Frequently used pathway abstractions in multiple pathway databases and software are supported as follows:
Metabolic pathways are described using the enzyme, substrate, product abstraction28 where substrates and products of a biochemical reaction are often small molecules. An enzyme, often a protein, catalyzes the reaction and inhibitors and activators can modulate the catalysis event.
Signaling pathways involve molecules and complexes participating in biochemical reactions, binding, transportation and catalysis events (Figure 3)5,9,29-31. Molecular states (cellular location, covalent and non-covalent modifications as well as sequence fragments) and generic molecules (such as the homologous family of Wnt proteins) may be described.
Gene regulatory networks involve transcription and translation events and their control12,14. Transcription, translation and other template-directed reactions involving DNA or RNA are captured in a template reaction in BioPAX, which maps a template to its encoded products (e.g. DNA to mRNA). Multiple sequence regions on a single strand of the template, such as promoters, terminators, open reading frames, operons and various reaction machinery binding sites, are active in a template reaction. Transcription factors (generally proteins and complexes), microRNAs and other molecules, participate in a template reaction regulation event.
Molecular interactions, notably protein-protein32-36 and protein-DNA interactions37, involve two or more physical entities. BioPAX follows the standard representation scheme of the Proteomics Standards Initiative Molecular Interaction (PSI-MI) format38.
Genetic interactions occur between two genes when the phenotypic consequence of perturbing both genes is different than expected given the phenotypes of each single gene perturbation39. BioPAX represents this as a pair of genes that participate in a genetic interaction measured using an observed phenotype.
Table 3.
BioPAX covers five main types of biological pathways and coverage has increased over time with new levels of the ontology.
| Type of Biological Pathway |
Main BioPAX Classes Used | Introduced |
|---|---|---|
| Metabolic pathways |
All types of physical entities (most common use of protein, small molecule, complex), All types of conversion events (most common use of BiochemicalReaction, ComplexAssembly and Transport), Catalysis, Modulation and Pathway |
Level 1 |
| Signaling pathways |
All types of physical entities (most common use of protein, complex), All types of conversion events (most common use of BiochemicalReaction, ComplexAssembly, Transport and Degradation), Control, Catalysis, Modulation, MolecularInteraction, Pathway |
Level 2 |
| Molecular interactions |
All types of physical entities (most common use of protein, complex, small molecule), MolecularInteraction, Pathway |
Level 2 |
| Gene regulatory networks |
All types of physical entities, TemplateReaction, TemplateReactionRegulation |
Level 3 |
| Genetic interactions |
Gene, GeneticInteraction | Level 3 |
Figure 4.
High-level view of the BioPAX ontology. Classes are shown as boxes and arrows represent inheritance relationships. The three main types of classes in BioPAX are colored, Pathway (red), Interaction (green) and PhysicalEntity and Gene (blue). For brevity, only the properties of the Protein class are shown as an example at the top right. Refer to BioPAX documentation at http://www.biopax.org for full details of all classes and properties.
Table 1.
What is included in BioPAX
| Ontology specification | Web Ontology Language (OWL) XML file, developed using free Protégé ontology editor software65. |
| Language documentation | Explanation of BioPAX entities, example documentation, best practice recommendations, use cases and instructions for carrying out frequently used technical tasks. |
| Example files | Example files for biochemical pathway, protein and genetic interaction, protein phosphorylation, insulin maturation, gene regulation, generic molecules in OWL XML. |
| Graphical representation | Recommendations for graphical representation using Systems Biology Graphical Notation (SBGN) as a guide. |
| Paxtools software | Java programming library supporting import/export, conversion, validation. Can be used to add BioPAX support to software. |
| List of data sources & supporting software | Databases making data available in BioPAX format, software systems for storing, visualizing and analyzing BioPAX pathways. |
The first three pathway abstractions are process-oriented. They imply a temporal order and can be thought of as extensions of the standard chemical reaction pathway notation to accommodate biological information. Molecular and genetic interactions, however, imply a static network of connections among system components instead of the temporally ordered process of reactions that defines a metabolic or signaling pathway. BioPAX supports combining these different types of data into a single model that is useful to gain a more complete view of a cellular process.
BioPAX provides many additional constructs, not shown in Figure 4, that are used to store extra details, such as database cross-references, chemical structure, experimental forms of molecules, sequence feature locations and links to controlled vocabulary terms in other ontologies (Supplementary Figure S1). BioPAX reuses a number of standard controlled vocabularies defined by other groups. For example, Gene Ontology40 is used to describe cellular location, PSI-MI vocabularies38 are used to define evidence codes, experimental forms, interaction types, relationship types and sequence modifications, and Sequence Ontology41 is used to define types of sequence regions, such as a promoter region on DNA involved in transcription of a gene. Other useful controlled vocabularies can be referenced, such as the molecule role ontology42.
BioPAX defines additional semantics that are currently only captured in documentation. For instance, physical entities represent pools of molecules and not individual molecules, corresponding to typical semantics used when describing pathways in textbooks or databases. A molecular pool is a set of molecules in a bounded area of the cell, thus it has a concentration. Pools can be heterogeneous and can overlap, as in the case of a protein existing in multiple phosphorylation states.
BioPAX also defines a range of constructs that are represented as ontology classes. Some of these represent biological entities, such as proteins, and are organized into classes that conceptualize the pathway knowledge domain. Others are used to represent annotations and properties of the database representation of biological entities. For instance, BioPAX provides xref classes to represent different kinds of references to databases that can be useful for data integration. These are represented as subclasses of utility class for convenience. A future version of BioPAX would ideally capture these semantics and structure these concepts more formally.
Uses of pathway information in BioPAX language
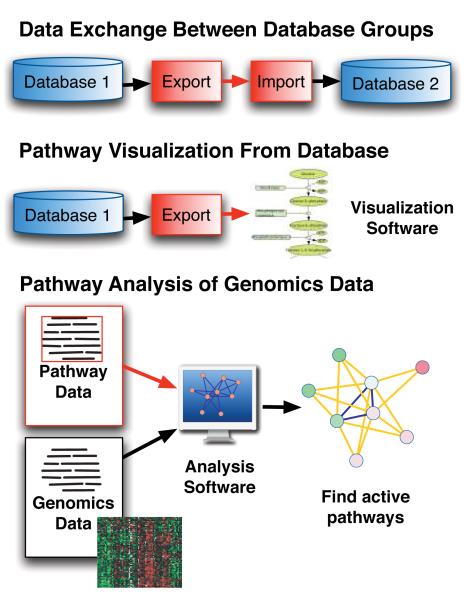
Once pathway data is translated into a standard computable language such as BioPAX, it is easier for software to access it and thereby support browsing, retrieval, visualization and analysis by biologists (Figure 5). This enables efficient re-use of data in different ways avoiding the time-consuming and often frustrating task of translating it between formats (Figure 1). Additionally, it enables uses that would be impractical without a standard format, such as those dependent on combining all available pathway data.
Figure 5.
Example uses of pathway information in BioPAX format. Red colored boxes or lines indicate use of BioPAX.
BioPAX can be used to help aggregate large pathway datasets by reducing the required collection and translation effort, for instance using software such as cPath43. Typical biological queries, such as “What reactions involve my protein of interest?” generate more complete answers when querying these larger pathway datasets. Another frequent use is to find pathways that are active in a particular biological context, such as a cell state, as determined by a genome-scale molecular profile measurement. For instance, pathways with multiple differentially expressed genes, as measured by DNA microarrays, may be transcriptionally active in one biological condition and not in another. Functional genomics and pathway data can be imported into software and combined for visualization and analysis to find interesting network regions. A typical workflow involves overlaying molecular profiling data, such as mRNA transcript profiles, on a network of interacting proteins to identify transcriptionally active network regions, which may represent active pathways44. A number of recent papers have used this pathway analysis workflow to highlight genes and pathways that are active in specific model organisms or diseased tissues, such as breast cancer, using gene and protein expression, copy number variants (CNVs) and SNPs19,44-49. BioPAX has been used in a number of these studies to collect and integrate large amounts of pathway information from multiple databases for analysis. For instance, protein expression data was combined with pathway information to highlight the importance of apoptosis in a mouse model of heart disease50. Multiple groups have found that tumor associated mutations are significantly related by pathway information47,48. And recently, in a study of rare CNVs in 996 autism spectrum disorder affected individuals, a core set of neuronal development related pathways were found to link dozens of rare mutations to autism that were not significantly linked to the disorder on their own by traditional single-gene association statistics49. These studies highlight the importance of pathway information in explaining the functional consequence of mutations in human disease. BioPAX pathway data can also be converted into simulation models, for instance using differential equations51 or rule-based modeling languages52, to predict how a biological system may function after a gene is knocked-out.
BioPAX is useful for exchanging information among and between data providers and analysis software. Pathway database groups can share the effort of pathway curation by making their pathways available in BioPAX format and exchanging them with others. For example, Reactome8 BioPAX formatted pathways are imported by the NCI/Nature Pathway Information Database (PID)9. Data providers can use existing BioPAX enabled software to add useful new features to their systems. For example, the Cytoscape network visualization software20 can read and display BioPAX formatted data as a network. The Reactome group used this feature to create a pathway visualization tool for their website. Because Reactome data were available in BioPAX format, and Cytoscape could already read BioPAX format, this new feature was easy to implement.
The Paxtools Java programming library for BioPAX has been developed to help software developers readily support the import, export and validation of BioPAX formatted data for various uses in their software (http://www.biopax.org/paxtools/). Using Paxtools and other tools, a range of BioPAX-aware software has been developed, including browsers, visualizers, querying engines, editors and converters (Table 2). For instance, the ChiBE and VisANT pathway visualization tools read BioPAX format22 and the WikiPathways website53, a community wiki for pathways, is working on using BioPAX to help import pathways from numerous sources, including manually edited pathways from biologists. The Pathway Tools software21 and CellDesigner pathway editor54 are developing support for BioPAX-based data exchange. In addition, tools for the storage and querying of Resource Description Framework (RDF - http://www.w3.org/RDF/) datasets, generated within the Semantic Web community, can be used to effectively process BioPAX data.
Table 2.
Databases and software supporting BioPAX. Note, PSI-MI data sources can be converted to BioPAX Level 2 using the PSI-MI to BioPAX converter.
| Database | Type | URL | Format | License | Statistics |
|---|---|---|---|---|---|
| BIND 32 | Protein interactions |
http://tap.med.utoronto.ca/~bind/ | PSI-MI Level 1 |
Free to all |
>85,000 interactions |
| BioCyc databases 66,67 |
Metabolic and signaling |
http://biocyc.org | BioPAX Level 3 |
Free to all |
~500 mostly computationally predicted pathway databases |
| BioGRID 36,68 |
Protein- protein and genetic interactions |
http://www.thebiogrid.org/ | PSI-MI Level 1 and 2.5 |
Free to all |
>265,000 interactions |
| BioModels 69 |
Metabolic and signaling |
http://biomodels.net/ | SBML, BioPAX Level 2 |
Free to all |
>450 pathways, >240 curated pathways, >40,000 interactions |
| Cancer Cell Map |
Signaling Pathways |
http://cancer.cellmap.org | BioPAX Level 2 |
Free to all |
Pathways: 10 Interactions: 2,104 Physical Entities: 899 |
| DIP 33,70 | Protein- protein interactions |
http://dip.doe-mbi.ucla.edu/ | PSI-MI Level 1 |
Free for Academi cs |
>57,000 interactions |
| Ecocyc 14 | Metabolic and Signaling Pathways |
http://ecocyc.org/ | BioPAX, Level 3 |
Free to all |
Pathways: 246 Regulatory interactions: 5,000 Metabolic reactions: 1400 Physical Entities: 3,606 |
| HPRD 71 | Protein- protein interactions |
http://hprd.org/ | PSI-MI Level 2.5 |
Free for Academi cs |
>38,000 interactions |
| IMID | Signaling | http://www.sbcny.org/data.htm | BioPAX Level 2 |
Free to all |
>2000 interactions |
| INOH | Signaling | http://www.inoh.org/ | BioPAX Level 2 |
Free to all |
>60 pathways |
| IntAct 72 | Protein- protein interactions |
http://www.ebi.ac.uk/intact | PSI-MI Level 1 and 2.5 |
Free to all |
>200,000 interactions |
| KEGG Pathway 16 |
Metabolic | http://www.genome.jp/kegg/ | BioPAX Level 1 |
Free for Academi cs |
>330 reference pathways |
| MetaCyc 15 | Metabolic and signaling |
http://metacyc.org/ | BioPAX Level 3 |
Free to all |
1399 curated pathways, >8,100 reactions |
| MINT 73 | Protein- protein interactions |
http://mint.bio.uniroma2.it/mint | PSI-MI Level 1 and 2.5 |
Free to all |
>80,000 interactions |
| MIPS MPact 74 |
Protein- protein interactions |
http://mips.gsf.de/genre/proj/mpact/ | PSI-MI Level 1 and 2.5 |
Free to all |
>12,000 interactions |
| NCI/Nature Pathway Interaction Database 9 |
Signaling | http://pid.nci.nih.gov/ | BioPAX Level 2 |
Free to all |
>400 curated pathways >12800 interactions |
| NetPath 75 | Signaling | http://netpath.org/ | BioPAX Level 2 |
Free to all |
20 large curated pathways |
| OPHID 76 | Protein- protein interaction |
http://ophid.utoronto.ca | PSI-MI Level 1 |
Free for Academi cs |
>424,000 interactions |
| Pathway Commons |
Pathways and interactions |
http://www.pathwaycommons.org | BioPAX Level 2 |
Free to all |
>1,400 collected pathways >421,000 interactions |
| Reactome 77 |
Metabolic and Signaling Pathways |
http://reactome.org/ | BioPAX, Level 2 |
Free to all |
>50 curated pathways |
| RegulonDB 12 |
Regulatory Network |
http://regulondb.ccg.unam.mx | BioPAX Level 3 |
Free to all |
Regulatory interactions: 2,594 Physical Entities: 18,371 Pathways: 2,660 |
| Rhea | Metabolic Reactions |
http://www.ebi.ac.uk/rhea | BioPAX, Level 2 |
Free to all |
>11,000 reactions |
| Software | Type | URL | Format | License | Language |
|---|---|---|---|---|---|
| BiNoM 78 | Editor/Convert er |
http://bioinfo-out.curie.fr/projects/binom/ | BioPAX Level 1 and 2 |
Free to all (open source) |
Java |
| BioPAX validator |
Validator | http://www.ohsucancer.com/biopaxvalidator/index.html | BioPAX Level 1 and 2 |
Free to all (open source) |
Java |
| BioPAX validator |
Validator | http://www.biopax.org/biopax-validator/ | BioPAX Level 3 |
Free to all (open source) |
Java |
| BioUML | Editor/Simulat or |
http://www.biouml.org/ | BioPAX Level 2 |
Free to all (open source) |
Java |
| Biowarehou se |
Biological data warehouse software |
http://biowarehouse.ai.sri.com/ | BioPAX Level 1 and 2 |
Free to all (open source) |
C and Java |
| ChiBE 79 | Visualization and analysis |
http://www.bilkent.edu.tr/~bcbi/chibe.html | BioPAX Level 1 and 2 |
Free for Academi cs |
Java |
| cPath 43 | Pathway database software |
http://cbio.mskcc.org/dev_site/cpath/ | BioPAX Level 1 and 2 |
Free to all (open source) |
Java |
| Cytoscape 20 |
Visualization and analysis |
http://cytoscape.org | BioPAX Level 1, 2, 3 |
Free to all (open source) |
Java |
| ExPlain Analysis System |
Pathway analysis |
http://www.biobase-international.com/pages/index.php?id=286 | BioPAX Level 1 and 2 |
Commerc ial |
|
| GeneSpring GX |
Pathway analysis |
http://www.agilent.com/chem/genespring | BioPAX Level 1 and 2 |
Commerc ial |
Java |
| Navigator 80 | Visualization and analysis |
http://ophid.utoronto.ca/navigator/ | BioPAX Level 1 and 2 |
Free for Academi cs |
Java |
| Pathway Tools 21 | Pathway prediction, editing, visualization, network analysis, gene expression analysis |
http://bioinformatics.ai.sri.com/ptools/ | BioPAX Level 3 |
Free for Academi cs |
Lisp |
| PATIKA 3 | Visualization | http://web.patika.org | BioPAX Level 1 and 2 |
Free to all |
Java |
| Paxtools | BioPAX input/export library |
http://www.biopax.org/paxtools/ | BioPAX Level 1,2,3 |
Free to all (open source) |
Java |
| PSI-MI to BioPAX converter |
BioPAX translator |
BioPAX Level 2,3 |
Free to all (open source) |
Java | |
| QPACA 81 | Gene expression analysis |
https://cabig.nci.nih.gov/tools/QPACA | BioPAX Level 1 and 2 |
Free to all |
Java |
| SBML to BioPAX converter |
BioPAX translator |
http://www.ebi.ac.uk/compneur-srv/sbml/convertors/SBMLtoBioPax.html | BioPAX Level 2 |
Free to all (open source) |
Java |
| SHARKvie w82 |
Pathway visualizer |
http://www.bioinformatics.leeds.ac.uk/shark/ | BioPAX Level 1 and 2 |
Free to all |
Java |
| The Gateway to Biological Pathways |
Pathway query web service |
http://jlab.calumet.purdue.edu/theGateway/ | BioPAX Level 1 and 2 |
Free to all |
Java |
| VisANT 22,83 | Visualization and analysis |
http://visant.bu.edu/ | BioPAX Level 1 and 2 |
Free to all |
Java |
What is not covered?
The BioPAX language uses a discrete representation of biological pathways frequently used in databases, the literature and textbooks. Dynamic and quantitative aspects of biological processes, including temporal aspects of feedback loops and calcium waves, must also be considered in a complete pathway map. BioPAX does not support this, but coordinates with the SBML and CellML mathematical modeling languages55,56 and a growing software toolset supporting biological process simulation57 which cover these aspects. Detailed information about experimental evidence supporting a pathway map is useful for recognizing the relative levels of support for different pathway aspects. This information is only included in BioPAX for molecular interactions, because that was already defined by the Proteomics Standards Initiative Molecular Interactions (PSI-MI) language58 and it was reused. The BioPAX workgroup makes use of PSI-MI controlled vocabularies and other concepts and coordinates with the PSI-MI workgroup to build these vocabularies in areas of shared interest, such as genetic interactions. Although BioPAX does not aim to standardize how pathways are visualized, work is coordinated with the Systems Biology Graphical Notation (SBGN, http://sbgn.org) community to ensure that SBGN can be used to visualize BioPAX pathways. Currently, most BioPAX concepts can be visualized using SBGN process description (PD) and SBGN activity flow (AF) diagrams and a mapping of BioPAX to SBGN entity relationship (ER) diagrams is under development. BioPAX development is coordinated with the above standardization efforts to ensure complementarity and compatibility. For instance, BioPAX uses controlled vocabularies developed by PSI-MI and can be used to annotate SBML and CellML models (Figure 6). BioPAX aims to be compatible with these and other efforts, so that pathway data can be transformed between alternative representations when needed. For instance, PSI-MI to BioPAX and SBML to BioPAX converters are available (Table 2).
How does the BioPAX community work?
While BioPAX facilitates communication of current knowledge, it is challenging for all knowledge representation efforts to anticipate new forms of information. As new types of pathway data and new knowledge representation languages and tools become available, the BioPAX language must evolve through the efforts of a community of scientists that includes biologists and computer scientists.
BioPAX is developed via community consensus among data providers, tool developers and pathway data users. More than 15 BioPAX workshops have been held since November 2002, attended by a diverse set of participants. Incremental versions (or levels) of the BioPAX language were progressively developed at these workshops to focus the group’s efforts on attainable intermediate goals. Broader input came from mailing lists and a community wiki. Community members participated in developing functionality they were interested in, which was integrated into specific levels (See Supplementary Table S1). Level 1 supports metabolic pathways, Level 2 adds support for molecular interactions and post-translational protein modifications by integrating data structures from the PSI-MI format, and Level 3 adds support for signaling pathways, molecular state, gene regulation and genetic interactions (Table 3). It is anticipated that newer BioPAX levels replace older ones, so use of the most recent BioPAX Level 3 is currently recommended. To ease the burden on users and developers, BioPAX aims to be backwards compatible where practical. Level 2 is backwards compatible with Level 1, however Level 3 involved a major redesign that necessitated breaking backwards compatibility. This said, many core classes have remained compatible with previous levels since Level 1 and software is provided for updating older BioPAX pathways to Level 3 (via Paxtools). All BioPAX material (Table 1) is made freely available under open source licenses via a central website (http://www.biopax.org) in order to encourage broad adoption. The database and tool support (Table 2) of a common language aids the creation, analysis, visualization and interpretation of integrated pathway maps.
In addition to the creation of a shared language for data and software, the process of achieving community consensus spurs innovation in the field of pathway informatics. Community discussion helps resolve technical knowledge representation issues faced by many data providers and users and facilitates the convergence to common terminology and representation. Solutions are discovered in independent research groups and incorporated in new data models and community best practices, which then enable identification of new issues. Thus, community workshops support a positive feedback cycle of knowledge sharing that has led to an accepted BioPAX language and development of better software and databases. We expect this to continue and to support new scientific uses of pathway information, motivated by end user access to valuable integrated pathway information and efficiency gain for database and software development groups. This will especially benefit new pathway databases and software tools that adopt standard representation and software components from the start.
Future community goals
The BioPAX shared language is a starting point on the path to developing complete maps of cellular processes. Additional near and long-term goals remain to be realized to enable effective integration and use of biological pathway information, as described below.
Data collection
Data must be collected and translated to a standard format for it to be integrated. This process is underway, as the descriptions of millions of interactions in thousands of pathways across many organisms from multiple databases are now available in BioPAX format. However, vast amounts of pathway data remain difficult to access in the literature and in databases that don’t yet support standard formats. Increasing use of standards requires promoting and supporting data curation teams and automating more of the data collection process using software. Easy to use tools for tasks like pathway editing must also be developed so that biologists can share their data in BioPAX format without substantial investment. Ideally, appropriate software would allow authors to enter data directly in standard formats during the publication process, to facilitate annotation and normalization by curators before incorporation into databases for use by researchers53.
Validation and best practice development
To aid data collection, community best practice guidelines and rules must be developed, led by major data providers, to help diverse groups use BioPAX consistently when multiple ways of encoding the same information exist. This will enable data providers to benefit from automatic syntactic and semantic validation of their data so they can ensure they are sharing data using standard representation and best practices59,60. Data collection and automatic validation will facilitate convergence to generally accepted biological process models.
Semantic integration
Multiple models of the same biological process may usefully co-exist. Ideally, different models could be compared for analysis and hypothesis formulation. However, comparison is difficult because the same concept can be represented in multiple ways due to use of multiple levels of abstraction (such as the hRas protein versus the Ras protein family), use of different controlled vocabularies, data incompleteness or errors. Future research needs to develop semantic integration solutions that recognize and aid resolution of conflicts.
Visualization
Pathway diagrams are highly useful for communicating pathway information, but their automatic construction, in a biologically intuitive way, from pathway data stored in BioPAX is a major challenge. The SBGN pathway diagram standardization effort provides a starting point towards achieving this goal (Figure 3). Intuitive and automatically drawn biological network visualizations may one day replace printed biology textbooks as the primary resource for knowledge about cellular processes.
Language evolution
As uses of pathway information and technology evolve, so must the BioPAX language. For instance, future BioPAX levels should capture cell-cell interactions, be better at describing pathways where sub-processes are not known or need not be represented, more closely integrate third-party controlled vocabularies and ontologies to ease their use and better encode semantics for easier data validation and reasoning.
Many groups within the BioPAX community, including most pathway data providers and tool developers, are working to achieve the above goals. For instance, Pathway Commons (http://www.pathwaycommons.org) aims to be a convenient single point of access for all publicly accessible pathway information and the WikiPathways project (http://www.wikipathways.org/) seeks to enable pathway curation by individuals53. Also, the semantic web community is developing a set of technologies that promise to ease the integration of information dispersed on the World Wide Web (WWW)61. These technologies will aid pathway data integration, since BioPAX is compatible with them through use of the W3C standard Web Ontology Language, OWL. All of the above research and development activities support the vision of data providers sharing computable maps of biological processes in a standard format for convenient use by a community of pathway researchers.
Supplementary Material
Acknowledgements
Funded by the US Department of Energy workshop grant DE-FG02-04ER63931, the caBIG program, the US National Institute of General Medical Sciences workshop grant 1R13GM076939, award number P41HG004118 from the US National Human Genome Research Institute and Genome Canada through the Ontario Genomics Institute (2007-OGI-TD-05). Thanks to many people who contributed to discussions on BioPAX mailing lists, at conferences and at BioPAX workshops, especially Alan Ruttenberg and Jonathan Rees.
Footnotes
Supplementary material Supplementary Table S1. Author contributions.
Supplementary Table S2. An example BioPAX file describing the phosphorylation and activation of CHK2 by ATM in human. Data was originally obtained from the Reactome database8.
Supplementary Table S3. An example BioPAX file describing the two reactions involved in glucose metabolism in Escherichia coli. Data was originally obtained from the EcoCyc database14.
Supplementary Figure S1. Diagram of BioPAX Level 3 utility classes.
References
- 1.Gasteiger E, et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicholson DE. The evolution of the IUBMB-Nicholson maps. IUBMB life. 2000;50:341–344. doi: 10.1080/713803744. [DOI] [PubMed] [Google Scholar]
- 3.Demir E, et al. PATIKA: an integrated visual environment for collaborative construction and analysis of cellular pathways. Bioinformatics. 2002;18:996–1003. doi: 10.1093/bioinformatics/18.7.996. [DOI] [PubMed] [Google Scholar]
- 4.Krull M, et al. TRANSPATH: an information resource for storing and visualizing signaling pathways and their pathological aberrations. Nucleic Acids Res. 2006;34:D546–551. doi: 10.1093/nar/gkj107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukuda K, Takagi T. Knowledge representation of signal transduction pathways. Bioinformatics. 2001;17:829–837. doi: 10.1093/bioinformatics/17.9.829. [DOI] [PubMed] [Google Scholar]
- 6.Davidson EH, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 7.Kohn KW. Molecular interaction map of the mammalian cell cycle control and DNA repair systems. Molecular biology of the cell. 1999;10:2703–2734. doi: 10.1091/mbc.10.8.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matthews L, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaefer CF, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bader GD, Hogue CW. BIND--a data specification for storing and describing biomolecular interactions, molecular complexes and pathways. Bioinformatics. 2000;16:465–477. doi: 10.1093/bioinformatics/16.5.465. [DOI] [PubMed] [Google Scholar]
- 11.Kitano H. A graphical notation for biochemical networks. BIOSILICO. 2003;1:169–176. [Google Scholar]
- 12.Gama-Castro S, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mi H, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keseler IM, et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37:D464–470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caspi R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):D277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34:D504–506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Molecular systems biology. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karp PD, et al. Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief Bioinform. 2010;11:40–79. doi: 10.1093/bib/bbp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Z, et al. VisANT 3.0: new modules for pathway visualization, editing, prediction and construction. Nucleic Acids Res. 2007;35:W625–632. doi: 10.1093/nar/gkm295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann R, et al. Text mining for metabolic pathways, signaling cascades, and protein networks. Sci STKE. 2005;2005:e21. doi: 10.1126/stke.2832005pe21. [DOI] [PubMed] [Google Scholar]
- 24.Racunas SA, Shah NH, Albert I, Fedoroff NV. HyBrow: a prototype system for computer-aided hypothesis evaluation. Bioinformatics. 2004;20(Suppl 1):i257–264. doi: 10.1093/bioinformatics/bth905. [DOI] [PubMed] [Google Scholar]
- 25.Cary MP, Bader GD, Sander C. Pathway information for systems biology. FEBS letters. 2005;579:1815–1820. doi: 10.1016/j.febslet.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 27.Koh G, Teong HF, Clement MV, Hsu D, Thiagarajan PS. A decompositional approach to parameter estimation in pathway modeling: a case study of the Akt and MAPK pathways and their crosstalk. Bioinformatics. 2006;22:e271–280. doi: 10.1093/bioinformatics/btl264. [DOI] [PubMed] [Google Scholar]
- 28.Karp PD. An ontology for biological function based on molecular interactions. Bioinformatics. 2000;16:269–285. doi: 10.1093/bioinformatics/16.3.269. [DOI] [PubMed] [Google Scholar]
- 29.Joshi-Tope G, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33(Database Issue):D428–432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Demir E, et al. An ontology for collaborative construction and analysis of cellular pathways. Bioinformatics. 2004;20:349–356. doi: 10.1093/bioinformatics/btg416. [DOI] [PubMed] [Google Scholar]
- 32.Bader GD, Betel D, Hogue CW. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salwinski L, et al. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chatr-aryamontri A, et al. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerrien S, et al. IntAct--open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stark C, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matys V, et al. TRANSFAC(R) and its module TRANSCompel(R): transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerrien S, et al. Broadening the horizon--level 2.5 of the HUPO-PSI format for molecular interactions. BMC biology. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eilbeck K, et al. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto S, Asanuma T, Takagi T, Fukuda KI. The molecule role ontology: an ontology for annotation of signal transduction pathway molecules in the scientific literature. Comparative and functional genomics. 2004;5:528–536. doi: 10.1002/cfg.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerami EG, Bader GD, Gross BE, Sander C. cPath: open source software for collecting, storing, and querying biological pathways. BMC Bioinformatics. 2006;7:497. doi: 10.1186/1471-2105-7-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nature protocols. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Efroni S, Carmel L, Schaefer CG, Buetow KH. Superposition of transcriptional behaviors determines gene state. PLoS ONE. 2008;3:e2901. doi: 10.1371/journal.pone.0002901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18(Suppl 1):S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 47.Cancer_Genome_Atlas_Research_Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu G, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 2010;11:R53. doi: 10.1186/gb-2010-11-5-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010 doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Isserlin R, et al. Pathway Analysis of Dilated Cardiomyopathy using Global Proteomic Profiling and Enrichment Maps. Proteomics. 2010 doi: 10.1002/pmic.200900412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moraru II, et al. Virtual Cell modelling and simulation software environment. IET systems biology. 2008;2:352–362. doi: 10.1049/iet-syb:20080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hlavacek WS, et al. Rules for modeling signal-transduction systems. Sci STKE. 2006;2006:re6. doi: 10.1126/stke.3442006re6. [DOI] [PubMed] [Google Scholar]
- 53.Pico AR, et al. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kitano H, Funahashi A, Matsuoka Y, Oda K. Using process diagrams for the graphical representation of biological networks. Nat Biotechnol. 2005;23:961–966. doi: 10.1038/nbt1111. [DOI] [PubMed] [Google Scholar]
- 55.Lloyd CM, Halstead MD, Nielsen PF. CellML: its future, present and past. Prog Biophys Mol Biol. 2004;85:433–450. doi: 10.1016/j.pbiomolbio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 56.Hucka M, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 57.Sauro HM, et al. Next generation simulation tools: the Systems Biology Workbench and BioSPICE integration. Omics. 2003;7:355–372. doi: 10.1089/153623103322637670. [DOI] [PubMed] [Google Scholar]
- 58.Hermjakob H, et al. The HUPO PSI’s molecular interaction format--a community standard for the representation of protein interaction data. Nat Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 59.Racunas SA, Shah NH, Fedoroff NV. A case study in pathway knowledgebase verification. BMC Bioinformatics. 2006;7:196. doi: 10.1186/1471-2105-7-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laibe C, Le Novere N. MIRIAM Resources: tools to generate and resolve robust cross-references in Systems Biology. BMC Syst Biol. 2007;1:58. doi: 10.1186/1752-0509-1-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berners-Lee T, Hendler J. Publishing on the semantic web. Nature. 2001;410:1023–1024. doi: 10.1038/35074206. [DOI] [PubMed] [Google Scholar]
- 62.Sowa JF. Knowledge representation : logical, philosophical, and computational foundations. Brooks/Cole; 2000. [Google Scholar]
- 63.Wheeler DL, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.The_Gene_Ontology_Consortium Gene ontology: tool for the unification of biology. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Knublauch H, Fergerson RW, Noy NF, Musen MA. Third International Semantic Web Conference - ISWC; 2004. [Google Scholar]
- 66.Karp PD, et al. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Romero P, et al. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2005;6:R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Breitkreutz BJ, Stark C, Tyers M. The GRID: the General Repository for Interaction Datasets. Genome Biol. 2003;4:R23. doi: 10.1186/gb-2003-4-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Le Novere N, et al. BioModels Database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res. 2006;34:D689–691. doi: 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xenarios I, et al. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peri S, et al. Development of Human Protein Reference Database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hermjakob H, et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zanzoni A, et al. MINT: a Molecular INTeraction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 74.Guldener U, et al. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kandasamy K, et al. NetPath: a public resource of curated signal transduction pathways. Genome Biol. 2010;11:R3. doi: 10.1186/gb-2010-11-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brown KR, Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21:2076–2082. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- 77.Joshi-Tope G, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zinovyev A, Viara E, Calzone L, Barillot E. BiNoM: a Cytoscape plugin for manipulating and analyzing biological networks. Bioinformatics. 2008;24:876–877. doi: 10.1093/bioinformatics/btm553. [DOI] [PubMed] [Google Scholar]
- 79.Babur O, Dogrusoz U, Demir E, Sander C. ChiBE: interactive visualization and manipulation of BioPAX pathway models. Bioinformatics. 2010;26:429–431. doi: 10.1093/bioinformatics/btp665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brown KR, et al. NAViGaTOR: Network Analysis, Visualization and Graphing Toronto. Bioinformatics. 2009;25:3327–3329. doi: 10.1093/bioinformatics/btp595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Novak BA, Jain AN. Pathway recognition and augmentation by computational analysis of microarray expression data. Bioinformatics. 2006;22:233–241. doi: 10.1093/bioinformatics/bti764. [DOI] [PubMed] [Google Scholar]
- 82.Pinney JW, Shirley MW, McConkey GA, Westhead DR. metaSHARK: software for automated metabolic network prediction from DNA sequence and its application to the genomes of Plasmodium falciparum and Eimeria tenella. Nucleic Acids Res. 2005;33:1399–1409. doi: 10.1093/nar/gki285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu Z, Mellor J, Wu J, DeLisi C. VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinformatics. 2004;5:17. doi: 10.1186/1471-2105-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Le Novere N, et al. The Systems Biology Graphical Notation. Nat Biotechnol. 2009;27:735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.