Abstract
MicroRNAs are short, approximately 22 nucleotide non-coding RNAs binding to partially complementary sites in the 3'UTR of target mRNAs. This process generally results in repression of multiple targets by a particular microRNA. There is substantial interest in methods designed to predict the microRNA targets and effect of single nucleotide polymorphisms (SNPs) on microRNA binding, given the impact of microRNA on posttranscriptional regulation and its potential relation to complex diseases. We developed a web-based application, MicroSNiPer, which predicts the impact of a SNP on putative microRNA targets. This application interrogates the 3'-untranslated region and predicts if a SNP within the target site will disrupt/eliminate or enhance/create a microRNA binding site. MicroSNiPer computes these sites and examines the effects of SNPs in real time. MicroSNiPer is a user-friendly web-based tool. Its advantages include ease of use, flexibility and straightforward graphical representation of the results. It is freely accessible at http://cbdb.nimh.nih.gov/microsniper.
Keywords: microRNA, SNP, FASTA program, 3'UTR, gene expression, computational prediction
Introduction
Short regulatory microRNAs (miRNAs) were originally discovered in plants and thought to be a unique phenomenon for this biological kingdom. However, miRNAs were soon discovered in animals, including mammals. Currently, the number of confirmed miRNAs in human exceeds six hundred [Griffiths-Jones et al., 2008].
Initially, miRNAs are processed from transcript and form hairpin-like loops [Winter et al., 2009]. Then, mature miRNAs, approximately 22 nucleotides long, together with the protein silencing complex (RISC), bind to target sites on the messenger RNA (mRNA). This event induces posttranscriptional repression via mRNA destabilization and/or translational repression.
The elucidation of mRNA targeting mechanism by miRNAs has become an important aim for computational and experimental biologists [Bartel, 2009; Mendes et al., 2009]. Various methods of computational predictions have been developed after it was demonstrated that 3'-untranslated regions (3'UTR) contain miRNA binding sites for those miRNAs which possess a certain degree of complementarity. By including the criterion of evolutionary conservation for these miRNA target sites (miRTSs), a sign of purifying selection, it has been possible to filter out many false positive targets.
It has been demonstrated that perfect complementarity in region 2 through 7 nucleotides starting from the 5'-end of the miRNA, the so-called `seed', significantly enriches true positive predictions of miRTS. Experiments have shown that base pairing disruption in this seed region significantly impairs target mRNA down-regulation. However, the requirement of a conserved, contiguous, stretch of 7 nucleotides in the 3'UTR, complementary to the seed region, would capture most of the mammalian miRNA binding sites. By increasing this length from 7 to 8 nucleotides improves the specificity, yet increases the number of false negatives, since most mRNA binding sites require only 7 nucleotides [Brennecke et al., 2005; Lai, 2002; Lewis et al., 2003]. Using a region of 7 conserved nucleotides, matched to the miRNA seed, Lewis et al. estimated the ratio of correctly predicted targets to false positive as 3.5:1 using a five species alignment of 3'UTRs [Lewis et al., 2005]. It has been estimated that the preservation of 3'-UTR pairing to miRNAs under purifying selection, is a characteristic of the majority of the human protein-coding genes [Chen and Rajewsky, 2006; Saunders et al., 2007].
Currently, approximately 1300 experimentally supported miRNA target sites (miRTS) have been reported [Papadopoulos et al., 2009]. Computational predictions based purely on matching seeds, regardless of other evolutionary and biological considerations such as conservation, spatial, temporal distribution of targets and miRNAs, would yield many false positive or false-negative binding sites. To add to this complexity, a single gene can be targeted by several miRNAs in multiple sites [Brennecke et al., 2005; Grimson et al., 2007; Hon and Zhang, 2007; Krek et al., 2005].
The duality of mechanism of down-regulating gene expression adds an additional layer of complexity to the experimental validation of computational predictions since high throughput expression analysis measures degradation of target mRNA, ignoring the translational repression mechanism. Eventual confirmation requires the analysis of the expression of a reporter gene, e.g. luciferase, with a cloned putative miRNA targets and co-expression of miRNA and target mRNA [Cheng and Li, 2008; Liu and Kohane, 2009; Sethupathy and Collins, 2008].
Target-prediction computational tools mostly fall into two categories. The first category includes tools that use the conservation of binding site as the main criteria. Some well known programs in this category are TargetScan [Friedman et al., 2009], which considers stringent seed pairing, number of pairs, and the secondary structure accessibility of sites, and PicTar [Lall et al., 2006], which also takes into account the stability of predicted pairs. The target prediction algorithm used by DIANA-microT [Maragkakis et al., 2009] depends on the principles of binding and conservation, i.e. on conserved and non-conserved number of binding sites. This tool also integrates biological pathways and the analysis of predicted interactions of target genes.
RNAhybrid [Rehmsmeier et al., 2004] identifies multiple miRNA target sites with the most favorable energy of binding. It is able to determine the optimal and the suboptimal binding energies where the user can define the degree of complementarity. Miranda [Betel et al., 2008] and miRBase [Griffiths-Jones et al., 2008] are not as strict about seed pairing though these tools require additional pairing beyond the seed region. Recent programs such as Pita Top [Kertesz et al., 2007] and mirWIP [Hammell et al., 2008] require reasonable seed pairing while considering the site context.
Tools in the second category such as versions of TargetScan [Grimson et al., 2007] and Pita [Kertesz et al., 2007] compute miRNA targets regardless of the target conservation among species. These programs require not only strict seed pairing but also site accessibility. RNA22 [Miranda et al., 2006] uses a pattern-based approach where patterns are generated from sets of known miRNAs. This program bypasses conservation analysis and only considers base pairing stability. Another web-tool, MicroInspector [Rusinov et al., 2005], detects miRNA binding sites in 3'UTRs based on complementarity using two sliding windows of six nucleotides with dynamic hybridization, allowing G:U wobble base pairs, and the subsequent folding of the duplex with Vienna RNA secondary structure routines [Hofacker, 2003].
Currently, it is accepted that disease-associated functional SNPs alter gene expression. With this in mind, the interplay between SNPs and miRNAs has become more important [Sethupathy and Collins, 2008]. Recently, there have been a number of findings of SNPs in 3'UTRs that affect expression due to binding of miRNA; the binding of miRNA was affected by the SNP, i.e. one allele reduced or eliminated the binding [Abelson et al., 2005; Clop et al., 2006; Sethupathy and Collins, 2008; Wang et al., 2008]. The predicted miRNA binding sites, in conjunction with SNPs, are catalogued in PolymiRTS [Bao et al., 2007] and in the Patrocles database [Hiard et al., 2010] which integrates SNPs, phenotype and expression data.
We have developed a user-friendly online tool, MicroSNiPer, which allows researchers to estimate the impact of a SNP on a putative miRNA target. It provides information about the creation or disruption of putative miRNA binding sites in a gene due to the presence of alternative SNP alleles. This web-tool provides a high degree of flexibility at the input stage. A user can load 3'UTR from the University of California Santa Cruz (UCSC) collection, automatically with their associated SNPs from dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) and the HapMap project [Frazer et al., 2007], or manually enter 3'UTR sequences and SNPs.
MicroSNiPer is useful for labs carrying out in-house discovery and characterization of novel transcripts and SNPs. This package makes it possible to combine catalogued and novel SNPs, as well as to manually enter or edit 3'UTRs based on newly available experimental data. Computational prediction of SNP impact on miRTS, followed by the experimental validation of the miRNA binding, can determine whether this is a functional SNP affecting gene expression. We consider MicroSNiPer as a useful tool that can complement other SNP-centered tools such as an the open source SNP-Laboratory Information Management System [Barenboim et al., 2003] for those laboratories that routinely discover and analyze novel transcripts and SNPs.
Implementation
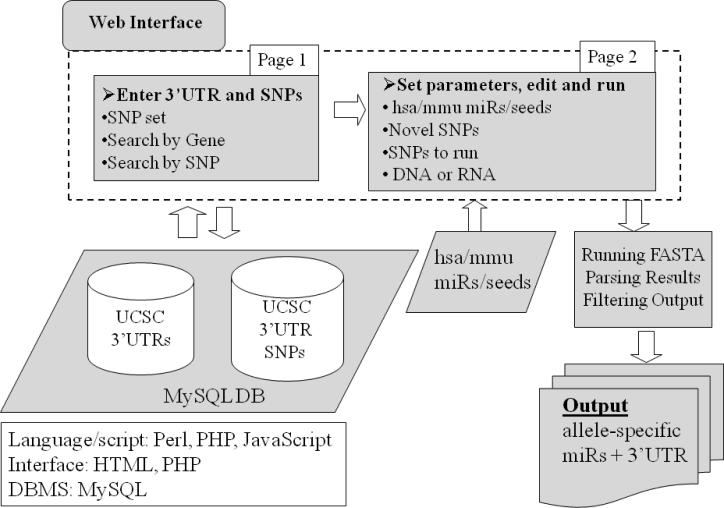
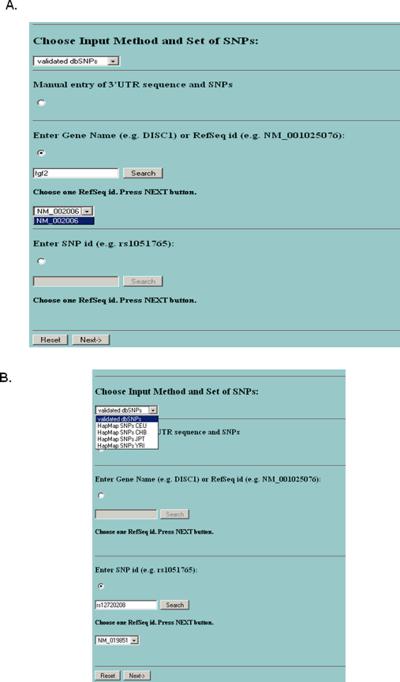
The design of MicroSNiPer is presented in Figure 1. The first step is to choose between two alternative input methods, manual or automatic. The first method is manually entering in a 3'UTR sequence and the corresponding SNPs. The other method is to load a 3'UTR and SNPs automatically from the database. MicroSNiPer allows the user to search 3'UTRs by either specifying the gene name or RefSeq ID (Figure 2A) or alternatively, by entering in the SNP rs ID (Figure 2B). After pressing the Search button, all known RefSeq IDs associated with this gene will be displayed. If a gene consists of several distinct isoforms, multiple RefSeq IDs will be listed. The user must select one RefSeq ID and a SNP collection from an appropriate population (Figure 2). With this choice, a new page will appear with all known SNPs that reside within the 3'UTR (Figure 3).
Figure 1.
The schematic pipeline of MicroSNiPer. The web interface consists of the opening screen (Page 1) where manual or automatic selection of the 3'UTR sequence and of a particular set of SNPs is chosen. The 3'UTR and the SNPs within the sequence is loaded from a MySQL database. The sequence and SNPs are passed to the subsequent launching page (Page 2) where the 3'UTR can be edited, novel SNPs can be added, and sets of miRNAs can be chosen. On page 2, the application is launched.
Figure 2.
The initial screen of MicroSNiPer. The user can specify the input method (either manual or from database), a set of SNPs (either validated dbSNPs or HapMap SNP sets). If user use MySQL database he/she can search the database either by gene name or by SNP (rs number). (A) Input by gene name. (B) Input by SNP rs# with a subsequent search of RefSeq IDs.
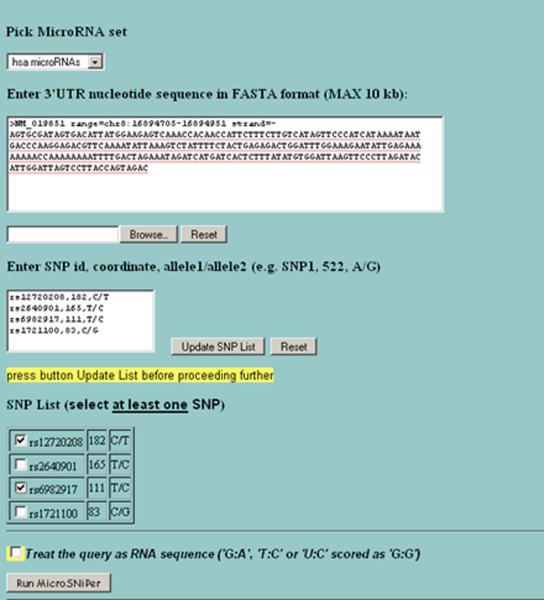
Figure 3.
The second screen of MicroSNiPer. The user chooses a miRNA sets (`hsa' - human, `mmu' – mouse full miRNA or seed sets), edits the 3'UTR. The user can add novel SNPs in the format `SNP id, position, alleles', update the SNP list to include SNPs of interest, and set the wobbling complementary pairs, i.e. the program treats the query as RNA sequences with additional complementarity.
3'UTR sequence and SNP database
To store the 3'UTR sequences and SNPs, a SQL database using the MySQL DBMS has been built. The 3'UTRs and SNPs from dbSNP and HapMap have been downloaded using the UCSC table browser (Table 1). Only SNPs residing within the 3'UTRs have been stored in the database. There are two tables in the database. The first table stores gene names (18,026 genes), and all 3'UTR sequences with RefSeq IDs (26,357 IDs), using the ID as the primary key. The second table stores all RefSeq IDs with their corresponding SNPs. JavaScript code (Ajax) is used to search these tables for the presence of SNPs and 3'UTR sequences without reloading the webpage.
Table 1.
Statistics of the SNPs in the MySQL database
| 3'UTRs | SNPs CEU1 | SNPs JPT1 | SNPs YRI1 | SNPs CHB1 | SNPs dbSNP2 | |
|---|---|---|---|---|---|---|
| Downloaded* | 26,888 | 2,502,319 | 2,354,557 | 2,828,612 | 2,396,521 | 6,620,441 |
| Located in 3'UTRs | N/A | 21,962 | 20,907 | 24,376 | 21,196 | 55,632 |
Only SNPs with MAF ≥ 0.01
Only SNPs validated by any method
from UCSC genome table browser
miRNA sets
Human and mouse miRNAs have been downloaded and stored as a flat file table from miRBase. The user can select either the set of full length miRNAs or the set of miRNAs truncated from 2 to 8 nucleotides (`the seed region') as the library to query (Figure 3).
A user has the option to choose one of four possible sets of SNPs located within the 3'UTR. These sets have been extracted from dbSNP, version 29, (http://www.ncbi.nlm.nih.gov/projects/SNP/) or from the HapMap project [Frazer et al., 2007]. We use only validated SNPs from dbSNP and SNPs from HapMap with MAF ≥ 0.01, i.e. CEU, CHB, JPT and YRI (Table 1, Figure 2).
The next webpage allows the user to edit the 3'UTR and to add or remove the SNPs loaded in from the database (Figure 3). On the same screen, the user can also choose whether to use the full-length miRNAs or only seed regions (2–8 nucleotides from the 5'-end of miRNA).
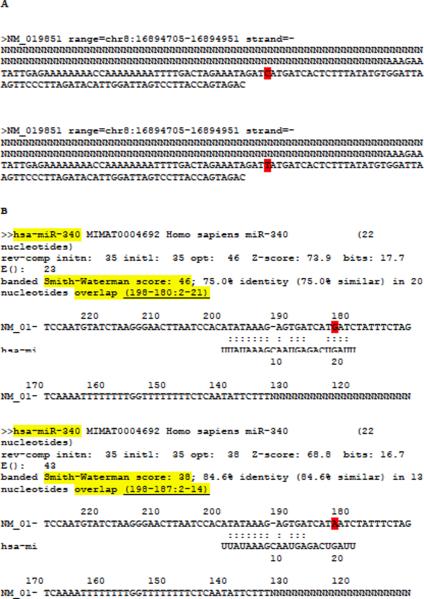
By clicking on the Run button, the MicroSNiPer algorithm and FASTA program is invoked on the datasets. The results are graphically presented within a HTML page (Figure 4). This provides unambiguous visualization of the results. The web-page includes links from the miRNAs to miRBase (http://miRNA.sanger.ac.uk/).
Figure 4.
The output display showing the microRNA-binding sites. The SNPs in the overlapping region are presented on actual page in red. Information about the SNP, rs numbers, position on the 3'UTR and alleles is shown. The 3'UTR sequence and the allele in the sequence are presented in reverse complement.
Specifications
This application is platform independent. MicroSNiPer was developed using Perl, PHP, and JavaScript. The script is embedded within a PHP-based web-interface and it uses the MySQL relational database as the backend database. MicroSNiPer is publicly available from the internet (http://cbdb.nimh.nih.gov/microsniper/).
MicroSNiPer has been developed using the Perl language, a MySQL database system for data storage. The web interface was created using standard development tools (HTML, PHP, Ajax, and JavaScript), and a CGI Perl module running on a web-server Apache 2 (http://www.apache.org) using Red Hat Enterprise Linux 5.3, handled the exchange of information between the user and the database. FASTA (version 35) package was downloaded from the University of Virginia (http://fasta.bioch.virginia.edu).
The current release of mature human and mouse the miRNAs (release 13.0) has been downloaded from miRBase (http://miRNA.sanger.ac.uk/) [Griffiths-Jones et al., 2008]. Sets of mature human and mouse miRNAs and sets of seeds miRNAs (heptamers: from 2 to 8 nucleotides from the 5'-end) were assembled and stored as flat files. The current version of MicroSNiPer allows the user to choose between sets of mature human or mouse miRNAs or just the seed region.
Results
Description of the algorithm
Genomic features used to identify miRNA Target Sites (miRTS)
We used the seed region binding in the miRNA as the major criterion in our algorithm. We applied the FASTA alignment program to determine if a change in an allele would shift the miRNA along the 3'UTR sequence, indicating a creation/disruption event in the miRNA target site. The alignment between the 3'UTR and the miRNA requires an uninterrupted match of at least 6 nucleotides from the 5'-end of miRNA which would mimic what is observed experimentally.
Processing of the 3'UTR sequence and the SNPs
On the initial screen, a user is able to select the SNPs of particular interests, e.g., specific SNPs associated with a given disease or SNPs within a region known for its association with a disease. MicroSNiPer constructs 3'UTR sequence variants with all possible combinations of alleles (haplotypes) in separate files. These 2k sets (haplotypes) of 3'UTR sequence variants are used as input to FASTA, where k is the number of SNPs included in analysis. Running haplotypes through FASTA is particularly important when two SNPs are positioned within a range of 22 nucleotides so their combination might improve or weaken the binding of the miRNAs. Also a user can conveniently run several SNPs in one run. Currently, the limit is set to 6 SNPs running simultaneously.
Modifications of the FASTA algorithm in MicroSNiPer
FASTA is a heuristic algorithm for finding significant matches between a query string (e.g. 3'UTR) and a database string (e.g. miRNAs) [Pearson and Lipman, 1988]. It finds the most significant diagonals in the dot-plot or dynamic programming matrix. A word-size parameter, k (ktup), is set to 6 by default for DNA which is also the minimum size of a miRNA seed. FASTA combines high scoring sub-alignments into a single larger alignment, allowing gaps into the alignment. The miRNA length is ~22 nucleotides leading to high E-values. A banded Smith-Waterman dynamic program is used along the best matched regions. If the banded Smith-Waterman score is equal to zero, the dynamic programming algorithm did not yield any alignment and no alignment will appear in the output.
The FASTA algorithm imposes a penalty for opening a gap and for extending the gap. FASTA outputs the single best alignment and this might occur before the SNP region in the 3'UTR. To force the FASTA algorithm to choose the best match in the SNP region, MicroSNiPer masks all the nucleotides more than 50 nucleotides downstream from the SNP with a character `N's usually reserved for repetitive elements (Figure 5A). FASTA ignores this stretch of `N's.
Figure 5.
MicroSNiPer pre-processing of FGF20 input sequence before running with FASTA program and its FASTA output. (A) Two FGF20 input sequences in separate files generated with SNP rs12720208 in position 182[C/T]. (B) Two FASTA program output files corresponding to each input file, respectively. The 3'UTR sequence and the alternative alleles are complementary to the input 3'UTR sequence. The FGF20 3'UTR sequence is masked upstream with `N'. The alternative alleles are highlighted in red. MicroSNiPer builds a unique lookup key (highlighted with light yellow background) composed of the miRNA name, the banded Smith-Waterman score, and the coordinates of the overlap.
Using the MicroSNiPer interface, the user has the ability to handle miRNAs or seed sequences as RNA sequences by setting the -U flag. Thus, in addition to the canonical base-pairing, FASTA allows G:U base pairs by scoring “G-A” and “T-C” as “G-G”.
MiRTS-SNP detection and filtering
The FASTA output indicates the coordinates of the base-pair binding interval (overlap) in the 3'UTR sequence. A change in either the overlap, indicating a shift in the 3'UTR target site, or a mismatch arising from the alternative allele, is used to filter the FASTA result file. Six contiguous matches starting from the first to the third nucleotide on the 5'-end of the miRNA is required for each record in the output file (Figure 6). There will be many 6-mers that are fully complementary to a 3'UTR sequence. However, MicroSNiPer selects only those miRNAs which have a SNP located within the target site. This constraint significantly reduces the number of candidate miRNAs in the output. If the alternative allele does not change the base pairing of the miRNA/seed to the 3'UTR, this SNP will have no effect on the alignment (Figure 6A). Using this condition, MicroSNiPer outputs only those miRNAs/seeds where the alignment has been shifted due to a change in the alleles. Another condition MicroSNiPer uses to process the raw FASTA output is to require that at least one SNP occurs between the initial and final nucleotide bond (Figure 6A).
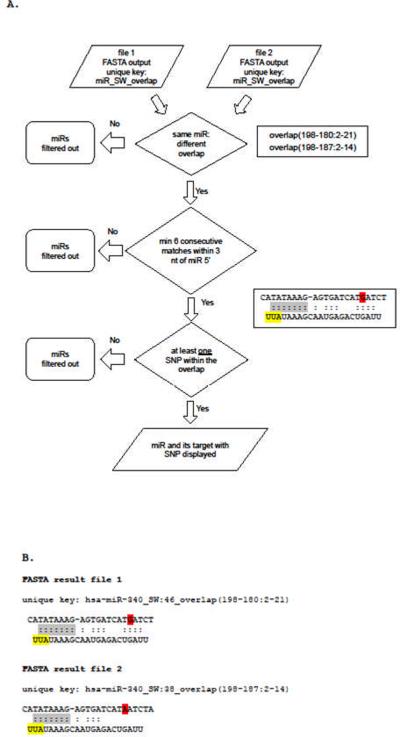
Figure 6.
Process of selection of miRNAs and miRTS for MicroSNiPer output. (A) Flowchart of the selection procedure by MicroSNiPer. The FASTA output indicates the coordinates of the base-pair binding interval (overlap) in the 3'UTR sequence. A change in either the overlap, indicating a shift in the 3'UTR target site, or a mismatch arising from the alternative allele, is used to filter the FASTA result file. Six contiguous matches starting from the first to the third nucleotide on the 5'-end of the miRNA is required. MicroSNiPer selects only those miRNA-target alignments which have a SNP located within the target site. MicroSNiPer outputs only those miRNAs/seeds where the alignment has been shifted due to a change in the alleles. (B) Features used for selection: minimum of 6 consecutive matches (grey) within 3 nucleotides of miRNA 5'-end (yellow); at least one SNP (red) within the overlap.
MicroSNiPer builds a unique lookup key composed of the miRNA name, the banded Smith-Waterman score, and the coordinates of the overlap (key1: hsa-miR-340_SW: 46_overlap (198-180:2-21) and key2: hsa-miR-340_SW:38_overlap (198-187:2-14)).
FASTA `result file 2' is filtered out because the SNP is not in the overlap.
MicroSNiPer builds a unique lookup key composed of the miRNA name, e.g. hsa-miR-340, the banded Smith-Waterman score, e.g. score `46' or `38', and the coordinates of the overlap, e.g. (198-180:2-21) or (198-187:2-14) (Figure 5B, Figure 6B). MicroSNiPer processes the FASTA output files looking for a shift in the overlap region. It will report only those cases where at least one SNP is present within the miRNA overlap.
By default, the output from MicroSNiPer shows matches with a minimum overlap of 6 base pairs. In order to increase the specificity, a user is able to increase the minimum seed length. This action filters out targets with overlaps less than a given threshold.
Limitations of the MicroSNiPer algorithm
The processing of the FASTA output requires a seed region of at least 6 nucleotides (Figure 6A). FASTA reports only one sequence per run. When selecting multiple SNPs for a single run, MicroSNiPer can miss reporting additional target sites if the additional target sites have identical SNPs in them and the FASTA algorithm calculates the identical Smith-Waterman score for a given miRNA. This rare event can be prevented by selecting a single SNP per run.
Validation
Even though there are approximately 1300 known experimentally supported miRTS [Papadopoulos et al., 2009], there are only 7 experimentally confirmed cases where the disease-associated SNPs positioned in the miRTS have an effect on the miRNA binding [Abelson et al., 2005; Adams et al., 2007; Jensen et al., 2009; Kapeller et al., 2008; Mishra et al., 2007; Sethupathy et al., 2007; Tan et al., 2007; Wang et al., 2008]. We have used [Sethupathy and Collins, 2008] which compiled and reviewed most of the known cases where miRTS SNPs affected the miRNA binding (Table 2). From [Sethupathy and Collins, 2008], we choose only in vitro/in vivo validated interactions. From 7 cases, 5 were corroborated using MicroSNiPer while the remaining 2 either had a seed region of less than 6 nucleotides while in the other case the SNP was outside the miRTS.
Table 2.
MicroSNiPer approach validated with experimental data modified from table in [Sethupathy and Collins, 2008] based on 3'UTR SNPs
| Associated disease | miRNA | Target gene | Putative risk allele (putative effect on miRNA targeting) | MicroSNiPer Validation | Patrocles PT/PF | PolymiRTS DB |
|---|---|---|---|---|---|---|
| Tourette's syndrome | miR-189 NKA miR-24-1* |
SLITRK1 | var321-SLITRK1 [A] (+) (var321,689,G/A) | YES | NO/YES | N/A (No option to apply novel SNP) |
| Breast cancer | miR-206 | ESR1 | rs9341070 [C] (−) | NO (miRNA does not create a seed with a minimum of 6 nucleotides) | NO | YES (but gives miR-122 instead) |
| Hypertension | miR-155 | AGTR1 | rs5186 [C] (−) | YES | YES (displays only target site, not miRNAs) | YES |
| Methotrexate resistance | miR-24 | DHFR | rs34764978 [T] (−) | N/A ( SNP N/V and not in miRTS) | NO | NO |
| Childhood asthma | miR-148a miR-148b miR-152 |
HLA-G | rs1063320 [G and C] (−) for C allele (+) for G allele |
YES | YES | NO |
| Parkinson's disease | miR-433 | FGF20 | rs12720208 [T] (−) | YES | NO/YES | NO |
| Diarrhea | miR-510 | HTR3E | rs62625044 [A] (−) | YES | NO/YES | NO |
| predominant irritable bowel syndrome | nka rs56109847(76,A/G) |
Several cases are not applicable since the SNP is not in the miRTS or in the 3'UTR. If the SNP is not in the MicroSNiPer database (e.g. it was not validated in dbSNP), it was entered manually in the form 'SNP,position,alleles'. N/A - not applicable, N/V - not validated, NKA - “now known as”. MicroSNiPer ignores the effect of a SNP on the binding of miRNA if it is not in the miRTS or a seed with at least 6 uninterrupted matches in the 5'-end. Allele-specific effects on miRNA binding were supported by functional assays in vitro and/or in vivo.
There exits two other databases with similar purpose to MicroSNiPer: PolymiRTS [Bao et al., 2007] and Patrocles [Hiard et al., 2010]. Comparing their performance with MicroSNiPer, in the absence of large sets of experimentally confirmed miRTS-SNPs, is difficult. We ran validated examples from Table 2 on PolymiRTS and Patrocles. From 7 cases, only 2 were corroborated with either PolymiRTS or Patrocles `Polymorphic Targets' precomputed database (Table 2). In the case of gene SLITRK1, there was no option in PolymiRTS to enter a SNP lacking a dbSNP rs number. Patrocles has a `Finder' utility where a user can manually enter polymorphic 3'UTR sequences. Applying this utility, we obtained results similar to MicroSNiPer's output, though additional matches were included that have not been experimentally validated. The `Finder' utility requires the user to manually enter and alter a DNA sequence which makes it difficult to apply.
It is difficult to quantitatively compare the performance of all these tools using only 7 miRTS-SNPs. PolymiRTS and Patrocles are less flexible than MicroSniPer in giving the user the ability to choose sets of miRNAs based on their experimental data. In our opinion, a researcher studying individual genes would prefer more matches, including even some false positive hits, rather than an empty set. MicroSNiPer has an option to improve the specificity by selecting a longer overlap in the seed region. However, this option can result in fewer true positives because the consensus is that most mRNA binding sites requires only 7 base pairs.
Case study - FGF20
Using MicroSNiPer, we analyzed the human FGF20 3'UTR containing four validated SNPs. We identified hsa-miR-433 having a seed entirely complementary to the target site when containing SNP rs12720208 C-allele (Figure 6B) but not with the T-allele. The T-allele disrupted this seed region for has-miR-433. Binding of hsa-miR-433 to this region has been experimentally confirmed and rs12720208 has been shown to be a functional SNP affecting gene expression [Wang et al., 2008].
Surprisingly, neither Patrocles precomputed dataset nor PolymiRTS resulted in any target sites overlapping rs12720208 in the FGF20 3'UTR. This is one of the best characterized and experimentally validated cases of a SNP effect on miRTS where hsa-miR-433 binds to the C-allele but not to the T-allele. Only by manually entering in the C and T allele variants of the FGF20 3'UTR into the Patrocles's `Finder' utility, did the program yield positive results, albeit including false positives. It is true that MicroSNiPer found 9 miRTS for the same SNP. However, increasing the threshold to an 8-mer seed overlap resulted in the correct prediction. In addition, having knowledge about the directionality of expression of FGF20 relative to an allele, gives a researcher the ability to remove extra miRNAs which putative impact do not conform to this directionality. A user can further eliminate the miRNAs by focusing only on miRNAs expressed in tissues under investigation.
Discussion
Mature miRNAs are non-coding RNAs approximately 22 nucleotides long and derived from stem-loop structures. They are delivered to the silencing protein complex, RISC, repressing the expression of target mRNA at the post-transcriptional level.
There are two important steps in the computational identification of potential miRNA binding sites: 1) complementarity analysis with a miRNA seed, requiring 6 or more contiguous matches 2) cross-species conservation of a seed [Lewis et al., 2005]. However, if the potential binding site is in a novel transcript and the 3'UTR is not conserved, existing programs might not detect a target site. It is also possible that the allele on the reference sequence does not create a target site while the alternative allele does.
The new studies show that one miRNA could have multiple binding sites on a single 3'UTR. G:U wobbles in the seed region, bulges and 3'-compensatory sites also adds to the complexity of site prediction of miRNA binding [Didiano and Hobert, 2006; Vella et al., 2004; Yekta et al., 2004]. So far, the major criterion increasing the specificity of finding binding-sites is a complementarity between the 5'end of miRNA and its 3'UTR target site [Lewis et al., 2005] and MicroSNiPer is able to utilize this characteristic of miRNA binding.
MicroSNiPer can be applied to evaluate not only 3'UTR as a target sequence but also any RNA/DNA sequence of interest which can be manually entered on the second webpage. This could be useful as new experimental data emerges that miRTSs are present in 5'UTRs or even in the gene ORFs [Kloosterman et al., 2004; Lytle et al., 2007]. This is one of the most important distinctions of MicroSNiPer from the PolymiRTS and Patrocles. These tools has a precomputed output while MicroSNiPer is doing computation on-the-fly allowing to alter sequences and add a novel SNPs in the list of existing dbSNPs. MicroSNiPer is able to compute an output using a haplotypes which are the combinations of all alleles on 3'UTR. It is conceivable that two SNPs positioned within target site range could create a novel miRNA target site with particular combination of their alleles. MicroSNiPer will detect this newly created miRTS automatically.
What to do next with MicroSNiPer results?
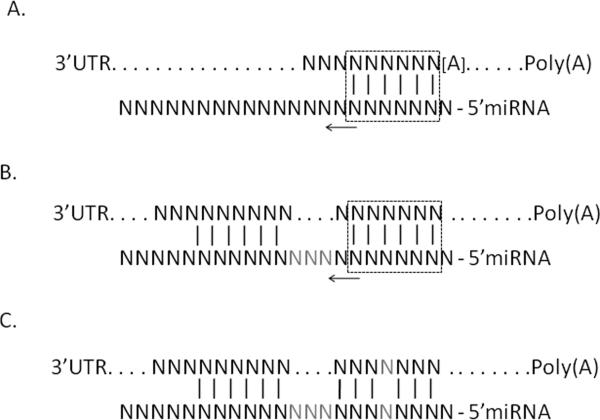
There are several approaches that a scientist can use to apply additional experimental data and databases in order to evaluate the feasibility of the MicroSNiPer prediction to focus on the most plausible miRTS. First of all, it is possible to classify miRTS types according to [Bartel, 2009] as 7mer-A1, 8mer, 6mer sites, etc (Figure 7). The [Bartel, 2009] provides a pie chart with frequencies of each type. Analysis of different modes of miRNA targeting reveals that the 3'-compensatory binding, a pairing on the 3'-end of miRNA lacking a `seed' at 5'-end, comprises less than 1% of in the binding at the conserved sites in mammals [Bartel, 2009].
Figure 7.
Generalized types of miRNA target sites. (A) Canonical and marginal sites together with (B) 3'-supplementary sites comprise approximately 99% of experimentally validated target sites; (C) 3'-compensatory sites lacking 5'-miRNA seed comprise ~1% of known sites. Rectangles mark minimum length of contiguous seed match (6 base pairs); arrows show possible extension of a seed match; grey `N' denotes non-complementary nucleotides between stretches of matching nucleotides; [A] denotes either unbound `A' or any other nucleotide (modified from Figure 1 [Bartel, 2009]).
After using MicroSNiPer, one can estimate the degree of conservation of a predicted miRTS by entering the 3'UTR target sequence into the UCSC genome browser (http://genome.ucsc.edu/) with the conservation track engaged. A high degree of conservation provides additional confidence of the validity of the miRTS. Obviously, the expression of the miRNA and its target has to coincide both spatially and temporally, i.e. be expressed in the same tissue and at the same developmental stage. Several additional databases can facilitate this analysis, namely, miRGator, miRNAmap, smiRNAdb [Hsu et al., 2008; Landgraf et al., 2007; Nam et al., 2008]. Also, it is worthwhile to apply RNA secondary structure programs which estimate the occlusion of miRTS in the secondary structure. Also, alternative alleles could expose miRTS which can facilitate binding of miRNAs and that could be an additional point to study this miRTS further. Commonly used programs for secondary structure predictions are Mfold, RNAfold [Hofacker, 2003; Hofacker and Stadler, 2006; Zuker, 2003]. The program, STarMir, estimates energetic characteristics of hybridization between a target forming secondary structure and a miRNA [Long et al., 2008].
In our opinion, in light of the increasing amount of data regarding the involvement of miRNAs in a wide range of developmental and regulatory processes, miRNA computational tools, including MicroSNiPer, will be more in demand, particular as experiments validate new miRNA- target interactions and elucidate the principles of binding beyond the `seed region' rule.
Conclusion
A web-based application, MicroSNiPer, predicts the impact of a SNP on putative microRNA targets. From linkage and whole genome association studies of disease, the importance of SNPs positioned in the 3'UTR regions is becoming evident. MicroSNiPer straightforward, and adaptable tool, will be useful for a wide range of studies that are characterizing novel transcripts and SNPs linked to disease. This makes MicroSNiPer's output more relevant to the specific research goals. This approach distinguishes MicroSNiPer from precomputed databases predicting impact of SNPs on miRNA targeting. Its benefits also include ease of use, flexibility and simple graphical representation of the results.
Availability and requirements
MicroSNiPer is freely accessible via http://cbdb.nimh.nih.gov/microsniper. It is platform independent and compatible with web-browsers Firefox 3.5, Internet Explorer 7 and higher. MicroSNiPer was developed using Perl, PHP, JavaScript and MySQL DBMS.
Acknowledgments
MB conceived and designed the software. MB, BJZ, YZ implemented the software. DRW developed requirements, planned and directed the project. All authors participated in writing the paper and approved the final version.
The work was supported by the Intramural Research Program of the National Institute of Mental Health, NIH.
Footnotes
Competing interests The authors declare that they have no competing interests.
References
- Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310(5746):317–20. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21(5):1132–47. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- Bao L, Zhou M, Wu L, Lu L, Goldowitz D, Williams RW, Cui Y. PolymiRTS Database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Res. 2007;35(Database issue):D51–4. doi: 10.1093/nar/gkl797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenboim M, Guo Y, Jamison DC. A laboratory information management system (LIMS) for small-scale single nucleotide polymorphism detection. Biophysics. 2003;48(SUPPLEMENT1):S90–S96. [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(Database issue):D149–53. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3(3):e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38(12):1452–6. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- Cheng C, Li LM. Inferring microRNA activities by combining gene expression with microRNA target prediction. PLoS One. 2008;3(4):e1989. doi: 10.1371/journal.pone.0001989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38(7):813–8. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13(9):849–51. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell M, Long D, Zhang L, Lee A, Carmack CS, Han M, Ding Y, Ambros V. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods. 2008;5(9):813–9. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiard S, Charlier C, Coppieters W, Georges M, Baurain D. Patrocles: a database of polymorphic miRNA-mediated gene regulation in vertebrates. Nucleic Acids Res. 2010;38(Database issue):D640–51. doi: 10.1093/nar/gkp926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31(13):3429–31. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, Stadler PF. Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics. 2006;22(10):1172–6. doi: 10.1093/bioinformatics/btl023. [DOI] [PubMed] [Google Scholar]
- Hon LS, Zhang Z. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol. 2007;8(8):R166. doi: 10.1186/gb-2007-8-8-r166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC, Hsu PW, Wong YH, Chen YH, Chen GH, Huang HD. miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res. 2008;36(Database issue):D165–9. doi: 10.1093/nar/gkm1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KP, Covault J, Conner TS, Tennen H, Kranzler HR, Furneaux HM. A common polymorphism in serotonin receptor 1B mRNA moderates regulation by miR-96 and associates with aggressive human behaviors. Mol Psychiatry. 2009;14(4):381–9. doi: 10.1038/mp.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapeller J, Houghton LA, Monnikes H, Walstab J, Moller D, Bonisch H, Burwinkel B, Autschbach F, Funke B, Lasitschka F, et al. First evidence for an association of a functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet. 2008;17(19):2967–77. doi: 10.1093/hmg/ddn195. [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32(21):6284–91. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lai EC. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30(4):363–4. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lall S, Grun D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol. 2006;16(5):460–71. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Liu H, Kohane IS. Tissue and process specific microRNA-mRNA co-expression in mammalian development and malignancy. PLoS One. 2009;4(5):e5436. doi: 10.1371/journal.pone.0005436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long D, Chan CY, Ding Y. Analysis of microRNA-target interactions by a target structure based hybridization model. Pac Symp Biocomput. 2008:64–74. [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc Natl Acad Sci U S A. 2007;104(23):9667–72. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes ND, Freitas AT, Sagot MF. Current tools for the identification of miRNA genes and their targets. Nucleic Acids Res. 2009;37(8):2419–33. doi: 10.1093/nar/gkp145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Mishra PJ, Humeniuk R, Longo-Sorbello GS, Banerjee D, Bertino JR. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci U S A. 2007;104(33):13513–8. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S, Kim B, Shin S, Lee S. miRGator: an integrated system for functional annotation of microRNAs. Nucleic Acids Res. 2008;36(Database issue):D159–64. doi: 10.1093/nar/gkm829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37(Database issue):D155–8. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988;85(8):2444–8. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. Rna. 2004;10(10):1507–17. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinov V, Baev V, Minkov IN, Tabler M. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 2005;33(Web Server issue):W696–700. doi: 10.1093/nar/gki364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A. 2007;104(9):3300–5. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, Hatzigeorgiou AG, Antonarakis SE. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3' untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81(2):405–13. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P, Collins FS. MicroRNA target site polymorphisms and human disease. Trends Genet. 2008;24(10):489–97. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, Solway J, Gern JE, Lemanske RF, Nicolae D, et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81(4):829–34. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3'UTR. Genes Dev. 2004;18(2):132–7. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, van der Walt JM, Mayhew G, Li YJ, Zuchner S, Scott WK, Martin ER, Vance JM. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82(2):283–9. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]