Abstract
Summary
In ubiquitin-like protein (UBL) cascades, a thioester-linked E2~UBL complex typically interacts with an E3 enzyme for UBL transfer to the target. Here we demonstrate a variant mechanism, whereby the E2 Ubc12 functions with two E3s, Hrt1 and Dcn1, for ligation of the UBL Rub1 to Cdc53’s WHB subdomain. Hrt1 functions like a conventional RING E3, with its N-terminus recruiting Cdc53 and C-terminal RING activating Ubc12~Rub1. Dcn1’s “potentiating neddylation” domain (Dcn1P) acts as an additional E3, reducing nonspecific Hrt1-mediated Ubc12~Rub1 discharge and directing Ubc12’s active site to Cdc53. Crystal structures of Dcn1P-Cdc53WHB and Ubc12 allow modeling of a catalytic complex, supported by mutational data. We propose that Dcn1’s interactions with both Cdc53 and Ubc12 would restrict the otherwise flexible Hrt1 RING-bound Ubc12~Rub1 to a catalytically competent orientation. Our data reveal mechanisms by which two E3s function synergistically to promote UBL transfer from one E2 to a target.
Keywords: Ubiquitin, Rub1, NEDD8, E3, RING, ubiquitin ligase, Ubc12, SCF, Cdc53
Introduction
Ubiquitin-like proteins (UBLs) are directed to targets via activation by an E1 enzyme, conjugation to an E2 enzyme, and E3-facilitated ligation to the target (Capili and Lima, 2007; Dye and Schulman, 2007; Kerscher et al., 2006; Knipscheer and Sixma, 2007; Nagy and Dikic, 2010). Because E3s link a UBL with a specific target, a major question is how E3s function. The largest E3 class comprises the RING/U-box/SPRING cohort (Deshaies and Joazeiro, 2009). Here, (1) either a domain or associated adaptor recruits the target, (2) the RING, U-box, or SPRING domain binds a specific E2~UBL (“~” refers to covalent complex), and (3) the UBL is transferred from the E2 Cys to a nucleophile on the target, generally a Lys side-chain. Notably, RING domains can enhance the inherent susceptibility of an associated E2~ubiquitin complex to nonspecific nucleophilic attack (Ozkan et al., 2005; Petroski and Deshaies, 2005b).
Despite many advances in our knowledge of UBL ligation machineries, it remains unclear how some UBLs are directed to targets. A particularly intriguing UBL is budding yeast Rub1 (NEDD8 in mammals) (Hochstrasser, 1998). Rub1’s best characterized target, Cdc53, is a member of the cullin family (Lammer et al., 1998; Liakopoulos et al., 1998). Cdc53 nucleates prototypic SCF E3 ubiquitin ligases: Cdc53’s N-terminal region binds Skp1-F box protein complexes, which recruit substrates for ubiquitination (Feldman et al., 1997; Skowyra et al., 1997); Cdc53’s C-terminal region binds the RING protein Hrt1 (Rbx1 in mammals) (Kamura et al., 1999b; Ohta et al., 1999; Seol et al., 1999; Skowyra et al., 1999; Tan et al., 1999). The Hrt1 RING domain binds the core domain of the ubiquitin E2, Cdc34, and activates Cdc34~ubiquitin for nucleophilic attack, even by a nonspecific nucleophile such as hydroxylamine (Petroski and Deshaies, 2005b). When bound to Skp1-Fbox protein-substrate complexes, Cdc53-Hrt1 is the catalytic module for processive Cdc34-mediated substrate polyubiquitination (Petroski and Deshaies, 2005b). Modification of Cdc53’s Lys760 by Rub1 counteracts SCF inhibition by Lag2 (CAND1 in mammals) (Liu et al., 2009; Siergiejuk et al., 2009).
How is Rub1 ligated to Cdc53? Rub1 is initially activated by its E1, Ula1-Uba3, and transferred to the catalytic Cys of the E2, Ubc12 (Lammer et al., 1998; Liakopoulos et al., 1998). However, a detailed mechanistic view is lacking, in part because two different proteins have been reported as being the E3 for Rub1 ligation to Cdc53. One candidate E3 is Hrt1, which binds Cdc53 and has a RING domain. Mutations in the zinc-binding ligands of the RING domain from mammalian Rbx1 hinder Rub1 ligation to Cdc53 in vitro, and fail to complement Hrt1 null yeast (Kamura et al., 1999a). In higher eukaryotes, the Rbx1 RING domain was shown to bind weakly to Ubc12, and to be required for NEDD8 modification of cullins (Gray et al., 2002; Morimoto et al., 2003). However, the question arises as to how Hrt1 could be an optimal E3 for both Ubc12 and Cdc34, which have distinct features in their catalytic domains.
Dcn1 was also identified as a Rub1 E3 (Kurz et al., 2005). The Dcn1 crystal structure revealed two domains, a UBA domain, and a “potentiating neddylation” (PONY) domain (Kurz et al., 2008). The PONY domain alone (termed Dcn1P here) was reported to bind Ubc12 and Cdc53, and is sufficient to enhance Cdc53~Rub1 levels in vivo and in lysates. Upon this discovery, it was suggested that Hrt1/Rbx1 may play a passive structural role in cullin modification by Rub1 (Kurz et al., 2008). However, Cdc53 can be modified in vitro without Dcn1, raising questions as to Dcn1’s function as an E3.
Here we dissect the roles of Hrt1 and Dcn1 in Rub1 ligation to Cdc53: each E3 has its own activities, and the two E3s function synergistically. We find that Hrt1 functions as a conventional RING E3, by recruiting Cdc53 and promoting Rub1 transfer from Ubc12’s catalytic Cys. Only in the presence of Hrt1, Dcn1 has unique functions to bring Ubc12’s catalytic Cys into close proximity with the Cdc53 target. Through structure/function analyses, we identify mechanisms by which two E3s can function with a single E2 to promote optimal UBL modification of a target.
Results
Hrt1 functions as a RING-type E3 toward Ubc12~Rub1
Properties of RING E3s include: 1) RING domains activate inherent, substrate-independent E2~UBL “discharge”, which is transfer of a UBL from an E2 catalytic cysteine to a nucleophile such as a primary amine; and 2) RING E3s also have separate domains that bind substrates, thus linking RING-catalyzed transfer of a UBL from the E2 to the substrate (Deshaies and Joazeiro, 2009). In principle, Hrt1 could fulfill both functions, with a C-terminal RING domain and an N-terminal strand binding Cdc53. Thus, we wondered whether Hrt1 is a RING-type E3 for Rub1 ligation to Cdc53.
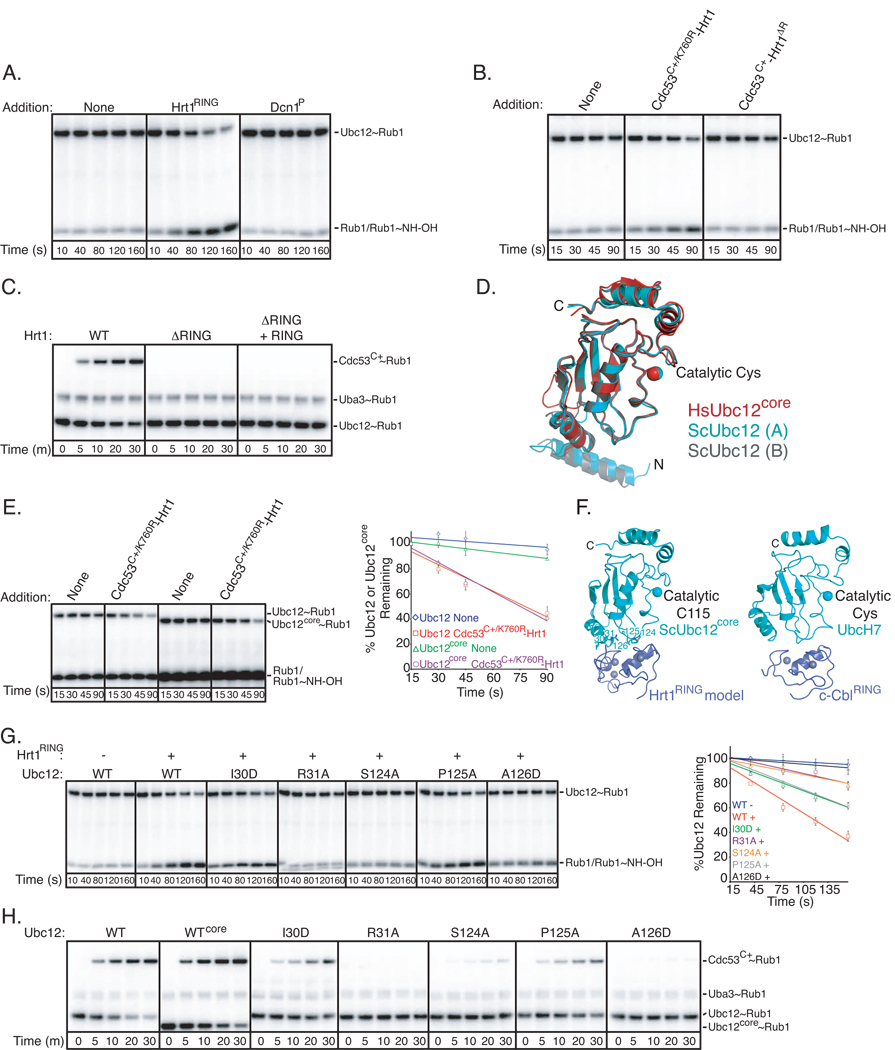
To test whether Hrt1 accelerated substrate-independent catalysis of Rub1 transfer, we adapted an assay for ubiquitin discharge from an E2 Cys to the nucleophile hydroxylamine (Fig. S1A) (Petroski and Deshaies, 2005b). Hydroxylamine serves as a nonspecific amine that can collide with and discharge the thioester-linked E2~UBL conjugate, producing free E2 and isopeptide-linked UBL~NHOH. Briefly, we loaded Ubc12 with [32P]-Rub1 using Ula1-Uba3 and MgATP, and quenched generation of thioester-linked Ubc12~Rub1 with EDTA. Upon addition of hydroxylamine, Ubc12~Rub1 discharge was examined. In the absence of any added factors, Ubc12~Rub1 was relatively stable to attack by hydroxylamine. However, the isolated Hrt1 RING domain enhanced discharge of Ubc12~Rub1 (Fig. 1A, 1G red line).
Figure 1. Hrt1 RING-type E3 activity for Rub1 modification of Cdc53.
Error bars - +/− 1 standard deviation from 3 independent experiments.
A. Time course of Ubc12~Rub1 discharge to hydroxylamine. Ubc12 was loaded with [32P]-Rub1 by Ula1-Uba3 in the presence of MgATP. The reaction was treated with EDTA to prevent subsequent [32P]-Rub1 loading of Ubc12, and hydroxylamine was added in the presence or absence of Hrt1RING or Dcn1P. Aliquots were removed at the indicated times and added to nonreducing SDS sample buffer prior to analysis by SDS-PAGE/Phosphorimager.
B. Time course of Ubc12~[32P]-Rub1 discharge to hydroxylamine in the presence or absence of Cdc53C+/K760R-Hrt1 or Cdc53C+-Hrt1ΔR. Note different time-scale from (A).
C. Phosphorimager data for time-course of pulse-chase assay monitoring [32P]-Rub1 transfer from Ubc12 to Cdc53C+ with the indicated versions of Hrt1, and the isolated Hrt1RING as indicated.
D. Crystal structure of S. cerevisiae Ubc12. The two molecules in the asymmetric unit (Chain A cyan; Chain B grey) are shown superimposed with the prior structure of the catalytic core domain of human Ubc12 (red) (Huang et al., 2005).
E. Same as (B), but comparing the function of the Ubc12 core domain lacking the N-terminal helix (Ubc12core), and with quantification. Standard error from 3 independent experiments is shown.
F. Structural model of the yeast Ubc12 core domain (cyan, residues 25 to the C-terminal 188) associated with the Hrt1 RING (blue, with zinc atoms as grey spheres, left) based on UbcH7 (cyan)-cCbl RING (blue, with zinc atoms as grey spheres, right) (Zheng et al., 2000).
G. Same as (A), but with the indicated versions of Ubc12 mutated at the predicted interface with the Hrt1 RING, and with quantification included for comparison between Ubc12 and mutants. Note different time-scale from (E).
H. Phosphorimager data for time-course of pulse-chase assay monitoring [32P]-Rub1 transfer from Ubc12 to Cdc53C+ with the indicated versions of Hrt1 and Ubc12.
We next assayed Hrt1 activity in the presence of a Cdc53 fragment referred to as Cdc53C+, which encompasses the entire cullin C-terminal domain that mediates interactions with Hrt1 and contains the Rub1 modification site (Lys760), plus 2 1/2 N-terminal cullin repeats. In order to test potential Rub1 E3 activity of Hrt1, we made several variant complexes, including Cdc53C+/K760R, in which the Lys760Arg substitution prevents Rub1 modification of Cdc53. Also, Hrt1ΔR binds Cdc53 but lacks the RING domain. In complex with Cdc53C+/K760R or Cdc53C+ respectively, Hrt1, but not Hrt1ΔR, accelerated Ubc12~Rub1 discharge to hydroxylamine (Fig. S1A, Fig. 1B, 1E red line).
We also examined the role of the Hrt1 RING domain for Rub1 transfer from Ubc12 to Cdc53C+ Lys760, using a pulse-chase protocol (Fig. S1B). Briefly, after “pulse” generation of the Ubc12~[32P]-Rub1 thioester conjugate, Cdc53C+-Hrt1 was added, and Rub1 was “chased” from Ubc12 to Cdc53C+. No Rub1 transfer was observed with the Hrt1 deletion mutant lacking the RING (Fig. 1C). Addition of the isolated RING domain in trans was not sufficient to overcome this defect (Fig. 1C). Thus, Hrt1 must contain both the Cdc53-binding region and the RING to stimulate Rub1 transfer to Cdc53.
To obtain insights into how the Hrt1 RING domain mediated E3 activity toward Ubc12, we wished to model a Hrt1-Ubc12 complex. First, we determined the crystal structure of Ubc12 (Fig. S1). The asymmetric unit had two molecules, each with two domains (Fig. 1D). The C-terminal “core” closely resembles that of human Ubc12 (0.97 Å rmsd), containing a standard E2 catalytic domain fold (Huang et al., 2005). Ubc12’s N-terminal 20 residues formed a helix (Fig. 1D). To ascertain which region of Ubc12 functions with Hrt1, we performed deletion analysis using both our assay for Ubc12~Rub1 discharge and our pulse-chase protocol to exclusively monitor effects of mutations on Rub1 transfer to Cdc53C+ (Fig. S1A, B). These assays exclude any mutagenic effects on the earlier step of forming the Ubc12~Rub1 thioester intermediate. Our data suggested that the Ubc12 core domain alone was largely responsible for Hrt1-mediated Rub1 discharge and transfer to Cdc53 Lys760 (Fig. 1E, 1H).
A structural model for the Hrt1 RING, based on 63% sequence identity to Rbx1, was obtained from swissmodel.expasy.org. Given the similarity of RING domains and among E2-RING complexes (Mace et al., 2008; Yin et al., 2009; Zheng et al., 2000), a model was obtained by docking our Ubc12 and the prior UbcH7-cCbl structures, and the Hrt1 RING domain model onto the cCbl RING (Zheng et al., 2000). The model predicted that Hrt1 interacted with Ubc12 core domain’s 1st α-helix and α2/α3-loop (Fig. 1F). Thus, we tested the effects of Ile30Asp and Arg31Ala mutations from the 1st α-helix in Ubc12’s core domain, and of Ser124Ala, Pro125Ala, and Ala126Asp mutations from the α2/α3-loop. These Ubc12 mutations at the canonical RING-binding surface hinder both Hrt1-mediated Rub1 discharge from Ubc12 (Fig. 1G), and Rub1 transfer to the substrate, Cdc53 Lys760 (Fig. 1H).
Taken together, our data indicate that Hrt1 functions as a RING-type E3 for Rub1 transfer from Ubc12 to Cdc53.
Dcn1 activation of Rub1 transfer is Ubc12- and Cdc53-specific and Hrt1-dependent
Dcn1 was reported to be an E3 for Rub1 with in vitro assays utilizing a 45-fold molar excess of GST-Dcn1 compared to Cdc53 (Kurz et al., 2008). In vivo, the PONY domain alone (termed Dcn1P here) showed similar ability as full-length Dcn1 to enhance Rub1 modification of Cdc53 (Kurz et al., 2008). Thus, we sought to map the regions of Dcn1 required for E3 activity using stoichiometric amounts of Dcn1 and Cdc53. We observe the same levels of Rub1 modification for both full-length Cdc53-Hrt1 produced in insect cells and Cdc53C+-Hrt1 produced in E. coli. In both cases, Rub1 modification was significantly enhanced by a stoichiometric amount of Dcn1 (Fig. S2A). We focus hereafter on Dcn1P (residues 70 to the C-terminus at 269) as its effects were indistinguishable from full-length Dcn1 (Fig. S2A).
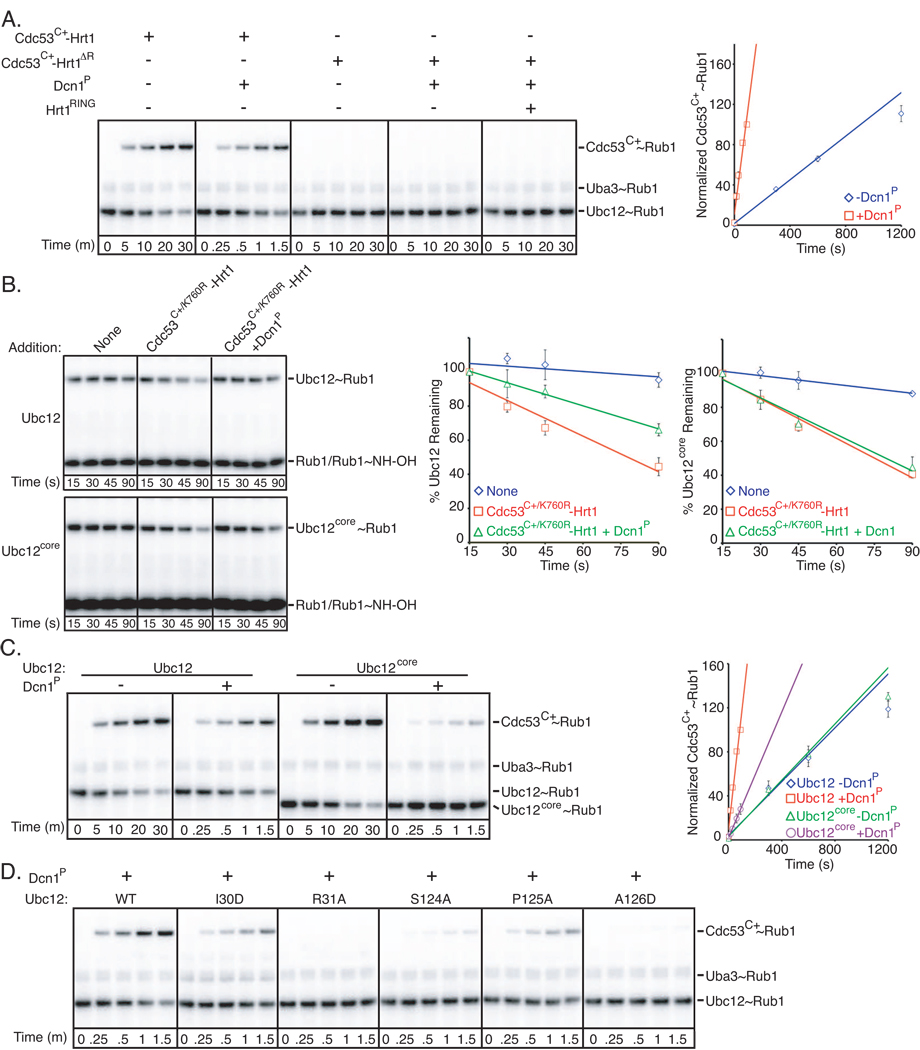
We first tested whether Dcn1P acted like Hrt1 and accelerated inherent Ubc12~Rub1 catalysis, independent of a protein substrate. Unlike Hrt1, Dcn1P did not accelerate Ubc12~Rub1 discharge (Fig. 1A). We also tested whether Dcn1P on its own could promote Rub1 transfer to a Cdc53 substrate. Addition of Dcn1P did not overcome the lack of Cdc53C+ modification observed when the RING domain was deleted from Hrt1, including when the isolated Hrt1 RING domain was provided in trans (Fig. 2A). Thus, Dcn1 itself exhibited E3 activity only in the presence of Hrt1.
Figure 2. Synergistic functions of Dcn1P and Hrt1 as Rub1 E3s.
Error bars - +/−1 standard deviation from 3 independent experiments.
A. Phosphorimager data (left) and quantification (right) for time-course of pulse-chase assay monitoring [32P]-Rub1 transfer from Ubc12 to Cdc53C+ in the presence or absence of Dcn1P and the indicated versions of Hrt1 either in complex with Cdc53C+ or added in trans. To observe comparable levels of Rub1 ligation, a shorter time-course is shown for wild-type Cdc53C+-Hrt1 in the presence of Dcn1P.
B. Time course of Ubc12~Rub1 discharge to hydroxylamine as in Fig. 1E, except in the presence or absence of Dcn1P, for full-length Ubc12 or the isolated core domain (Ubc12core). Raw data – left; quantification – right.
C. Same assay as in (A), but comparing the activity of full-length Ubc12 or the Ubc12 core domain. Raw data – left; quantification – right. Note different times.
D. Same assay as in Fig. 1H, except with earlier time-points and in the presence of Dcn1P.
We wondered whether Dcn1P enhances Hrt1-dependent activities in general. Thus, we considered that a widely-recognized function of Hrt1 is serving as the catalytic component of SCF ubiquitin E3s (Kamura et al., 1999b; Ohta et al., 1999; Seol et al., 1999; Skowyra et al., 1999; Tan et al., 1999). We investigated potential effects of Dcn1P on Hrt1’s ubiquitin E3 activity, using a modified assay for SCFCdc4-mediated polyubiquitination of a high-affinity peptide substrate (Orlicky et al., 2003; Pierce et al., 2009). Unlike effects on Rub1-modification of Cdc53, we did not observe significantly enhanced polyubiquitination activity in the presence of Dcn1P (Fig. S2B).
Two simple models could explain Dcn1 function: Dcn1 could function indirectly, by enhancing Hrt1’s RING E3 activity toward Ubc12. Alternatively, Dcn1 could have distinct E3 activity in the presence of Hrt1. We performed several experiments to test both hypotheses. We ruled out the first model because Dcn1P did not enhance Cdc53C+/K760R-Hrt1-catalyzed Rub1 transfer from Ubc12 to a nonspecific nucleophile (Fig. 2B).
In the presence of Hrt1, Dcn1 acted as a distinct Rub1 E3. First, Dcn1P reduced nonspecific Ubc12~Rub1 discharge in the presence of Cdc53C+/K760R-Hrt1 (Fig. 2B). Second, Dcn1P and Hrt1 E3 activities depended on different regions of Ubc12. For example, Dcn1P had no effect on Rub1 discharge from Ubc12’s isolated core domain; the reduction of nonspecific Ubc12~Rub1 discharge requires Ubc12’s unique N-terminal helical extension (Fig. 2B). In a related vein, although Dcn1P enhances the rate of Rub1 transfer to Cdc53C+ from full-length Ubc12 by 14-fold, the effect of Dcn1P was reduced to only 2.6-fold for Ubc12’s isolated core domain (Fig. 2C, S2C). Third, the extent to which Dcn1P enhanced Cdc53 Lys760 modification was similar for Ubc12-Hrt1 interface mutants and wild-type Ubc12 (Fig. 1H v. 2D). Thus, unlike Hrt1, Dcn1 activity requires Ubc12’s N-terminal helix and core domain, and is specific for Rub1 modification of Cdc53.
Mapping Dcn1-Cdc53-Hrt1 interactions
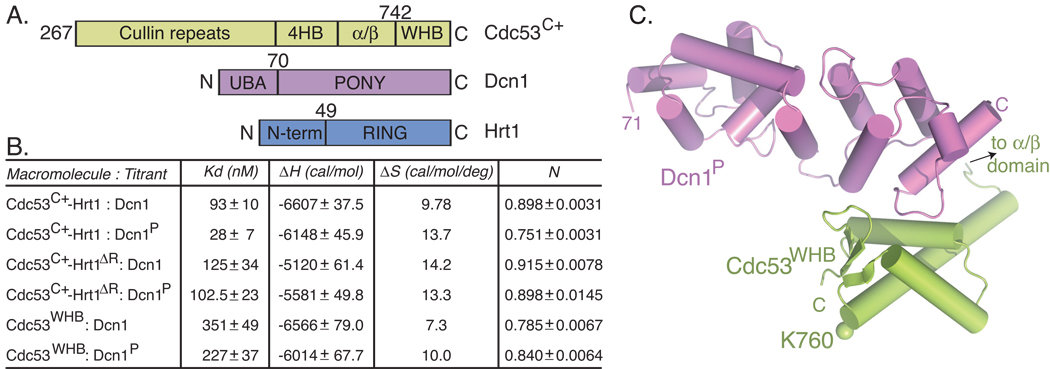
To define the site of interaction between Dcn1 and its substrate, Cdc53, isothermal titration calorimetry (ITC) was used to quantify binding between structure-based deletion mutants. A structural model (Swissmodel) of Cdc53C+ closely resembled other cullins (Angers et al., 2006; Duda et al., 2008; Zheng et al., 2002), with the C-terminal domain assembled from several subdomains: a 4-helix bundle (4HB) that connects to the ntd; an α/β-subdomain that binds Rbx1’s N-terminal strand; and a C-terminal winged-helix subdomain (WHB) that contains the Rub1 acceptor Lys760. Both full-length Dcn1 and Dcn1P were examined for binding to Cdc53C+-Hrt1 and the Hrt1 RING deletion mutant. We also tested binding to an isolated Cdc53 WHB subdomain (Cdc53WHB) because a prior study had identified cullin C-terminal residues as important for Dcn1 binding (Kurz et al., 2008). A range of affinities was observed, potentially arising from structural effects of the deletions. Nonetheless, both full-length Dcn1 and Dcn1P formed tight 1:1 complexes with all of the Cdc53 constructs (Fig. 3A, 3B, S3A). This includes Dcn1P binding to the isolated WHB subdomain with substantially high affinity (Kd value of 227 nM).
Figure 3. A high affinity interaction between Dcn1P and the Cdc53 WHB subdomain.
A. Schematic views of Cdc53C+, Dcn1, and Hrt1 domains.
B. Summary of thermodynamic parameters determined by ITC for binding between the indicated Cdc53-Hrt1 and Dcn1 variants.
C. Overall crystal structure of Dcn1P (violet)-Cdc53WHB (lime). The location of the Rub1 acceptor Lys760 is indicated in sticks, with a sphere for the ε-amino group.
The Dcn1 E3 binds a surface of its Cdc53 substrate distal from the Rub1 acceptor Lys
To understand how Dcn1 binds Cdc53, we performed crystallographic and NMR studies of Dcn1P in complex with Cdc53WHB. Dcn1P- Cdc53WHB complexes from two crystal forms superimposed, so only the structure refined to 2.23 Å resolution is discussed further here (Fig. 3C; Supplemental Information). Dcn1P from the complex with Cdc53WHB superimposed with prior structures (Kurz et al., 2008; Yang et al., 2007) (0.73 and 0.66 Å rmsd, respectively), adopting an elongated domain composed of 2 EF-hand-like folds and 2 C-terminal helices. Likewise, Cdc53WHB superimposed with the WHB subdomain of prior structures of human Cul1 (Zheng et al., 2002) (residues 707–776, 0.84 Å rmsd). The N-terminal helix of Cdc53WHB corresponds to the long H29 helix in Cul1, which protrudes from the WHB and would connect to the cullin α/β subdomain.
Both Dcn1P- Cdc53WHB crystals utilize similar packing, with only one interface near the C-termini of both proteins consistent with 15N-1H chemical shift perturbations upon titrating unlabeled Dcn1P into uniformly 15N-labeled Cdc53WHB (Table S1, Fig. S3B, S3C). Notably, the interaction occurred on the opposite side of the WHB from the Rub1 acceptor Lys760 (Fig. 3C), and backbone amide chemical shifts for Cdc53 Lys760 and surrounding residues were not significantly altered upon interaction. Thus, Dcn1P may not cause long-range structural changes around Lys760 (Fig. S3B, S3C). Rather, Dcn1P extends away from the WHB and Lys760 (Fig. 3C). One possibility based on the structure is that Dcn1P may function by remotely presenting Cdc53’s Lys760 to Ubc12’s active site, from a distance.
In order to gain insights into which higher-order assemblies might coexist with Dcn1 binding to Cdc53, we superimposed Dcn1P–Cdc53WHB onto the WHB from prior full-length Cul1-Rbx1 complexes with Skp1-F-box, and with CAND1 (Goldenberg et al., 2004; Wu et al., 2003a; Zheng et al., 2002). Consistent with genetic and biochemical studies, Dcn1 binding appeared compatible with cullin complexes with substrate adaptors, and with CAND1 (Fig. S3D, S3E). Indeed, our previous study indicated Dcn1 might not directly compete with the yeast ortholog of CAND1, Lag2, for binding to Cdc53 both in vivo and in vitro (Siergiejuk et al., 2009). We also found that Lag2 inhibits Rub1 transfer to Cdc53 both in the absence or presence of Dcn1P, and that Dcn1P and Lag2 could both exist within a single stoichiometric complex with Cdc53C+-Hrt1 (Fig. S3F–S3I).
Details of Dcn1 E3 recognition of the Cdc53 substrate
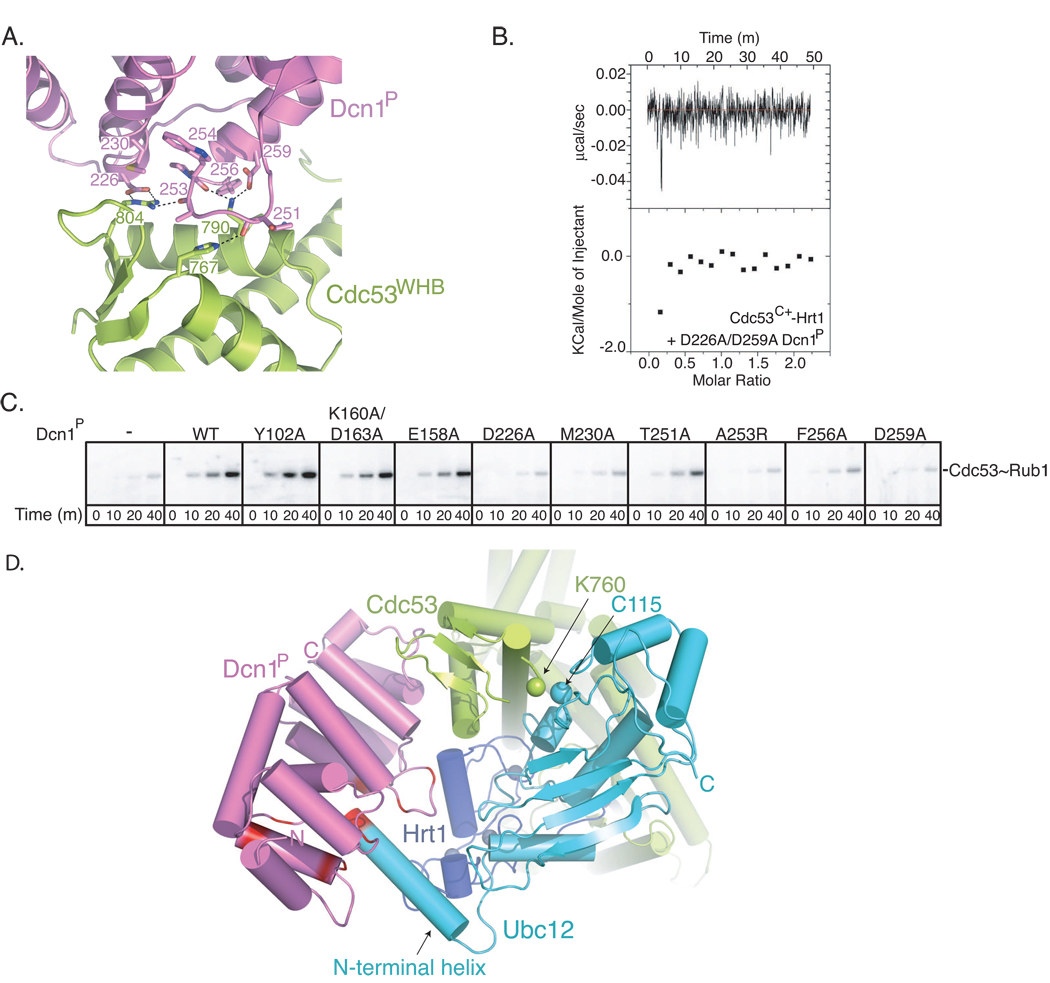
In total, the Dcn1-Cdc53 interactions bury ~985 Å2 of surface. From Dcn1, the extreme C-terminal helix, its preceding loop, and the C-terminal helix of the EF-hand-like-region pack against a surface comprising Cdc53’s most C-terminal helix and the loop between the C-terminal two β-strands. Key interactions are between: Dcn1 Asp226, Met230, and the backbone of Ala253 with Cdc53 Arg804; Dcn1 Ala253 and the Thr251 backbone with Cdc53 His767; Dcn1 Trp254 backbone and Asp259 with Cdc53 Lys790; and Dcn1 Phe256 with Cdc53 Arg791 (Fig. 4A).
Figure 4. E3 interactions for Rub1 ligation to Cdc53.
A. Close-up view crystal structure of Dcn1P (violet)-Cdc53WHB (lime), with contact residues shown in sticks and electrostatic interactions as dashed lines.
B. ITC data for the D226A/D259A double mutant Dcn1P and Cdc53C+-Hrt1.
C. Phosphorimager data from multiple turnover experiments examining time-courses for forming [32P]-Rub1~Cdc53-Hrt1 in the presence of Ula1-Uba3 (Rub1 E1), Ubc12, and the indicated versions of Dcn1P.
D. Cartoon view of structural model of dual Rub1 E3 ligase (Dcn1P, violet / Hrt1, blue with zinc atoms in grey) with Ubc12 (cyan) and Cdc53C+ (lime), generated as shown in Fig. S4. The 4 N-terminal residues of Ubc12 and residues surrounding the Dcn1 groove, which are mutated in Fig. 5, are shown in red. For simplification, Rub1, which would start thioester bound to Ubc12 C115 and end ligated via an isopeptide bond to Cdc53 Lys760 is not shown.
The structure provides a rationale for previous findings of deleterious effects of mutating Dcn1 and Cdc53 residues identical across species. These include key contact residues in Dcn1 (Asp226, Ala253, Asp259) and Cdc53 (Arg804). Simultaneous mutations of contact residues from Dcn1, or the Cdc53 Arg804Ala mutation, led to decreased Rub1 modification of Cdc53 in vivo (Kurz et al., 2008). Indeed, using ITC, we found the interaction significantly impaired upon mutating Dcn1’s Asp226 and Asp259 (Fig. 4B). Also, substitutions of these and other Dcn1P residues at the structurally-observed interface, but not on other surfaces, impaired stimulation of Rub1 transfer to Cdc53C+ (Fig. 4C).
Structural model of dual Rub1 E3 ligase for Cdc53
To gain insights into how Hrt1 and Dcn1 function together as E3s for Cdc53, we built a structural model of a Dcn1-Cdc53C+-Hrt1-Ubc12 complex in a catalytic conformation (Fig. 4D, S4). The model suggests a dual E3 ligase mechanism, as follows. Hrt1’s RING would bind Ubc12 as in other RING E3-E2 complexes. As with other CRLs, flexible tethering of the Hrt1 RING to its N-terminal strand allows rotation of the RING and thus the associated Ubc12~Rub1 to adopt a range of locations relative to Cdc53’s WHB. Amongst the allowed conformations, the Ubc12 active site can approach Cdc53’s Rub1 acceptor Lys760. This would explain our data indicating that Hrt1 alone is sufficient to act as an E3 toward Cdc53.
According to the model, Dcn1 also functions as a standard E3, binding both the substrate and the E2. In addition to specifically interacting with Cdc53, the model predicts that Dcn1 achieves specificity by interacting with Ubc12’s unique N-terminal extension. The model suggests that the first 4 residues of Ubc12’s extreme N-terminal helix would fit into a groove at the junction of the two EF-hand-like folds in Dcn1P (Fig. 4D). Although the minimal interactions would likely be very weak on their own, Dcn1’s binding to Cdc53’s WHB subdomain coupled with Ubc12 binding to the Hrt1 RING would establish a high effective concentration for Dcn1’s groove and Ubc12’s N-terminus. Dcn1P is also poised to make weak contacts at the junction between the Cdc53 WHB domain, the Hrt1 RING domain, and the Ubc12 catalytic core domain. Thus, the model suggests that Dcn1 could restrict the otherwise flexible RING-Ubc12~Rub1 to a catalytically competent orientation.
Mutational analysis of Ubc12 N-terminus and Dcn1 groove in Rub1 ligation to Cdc53
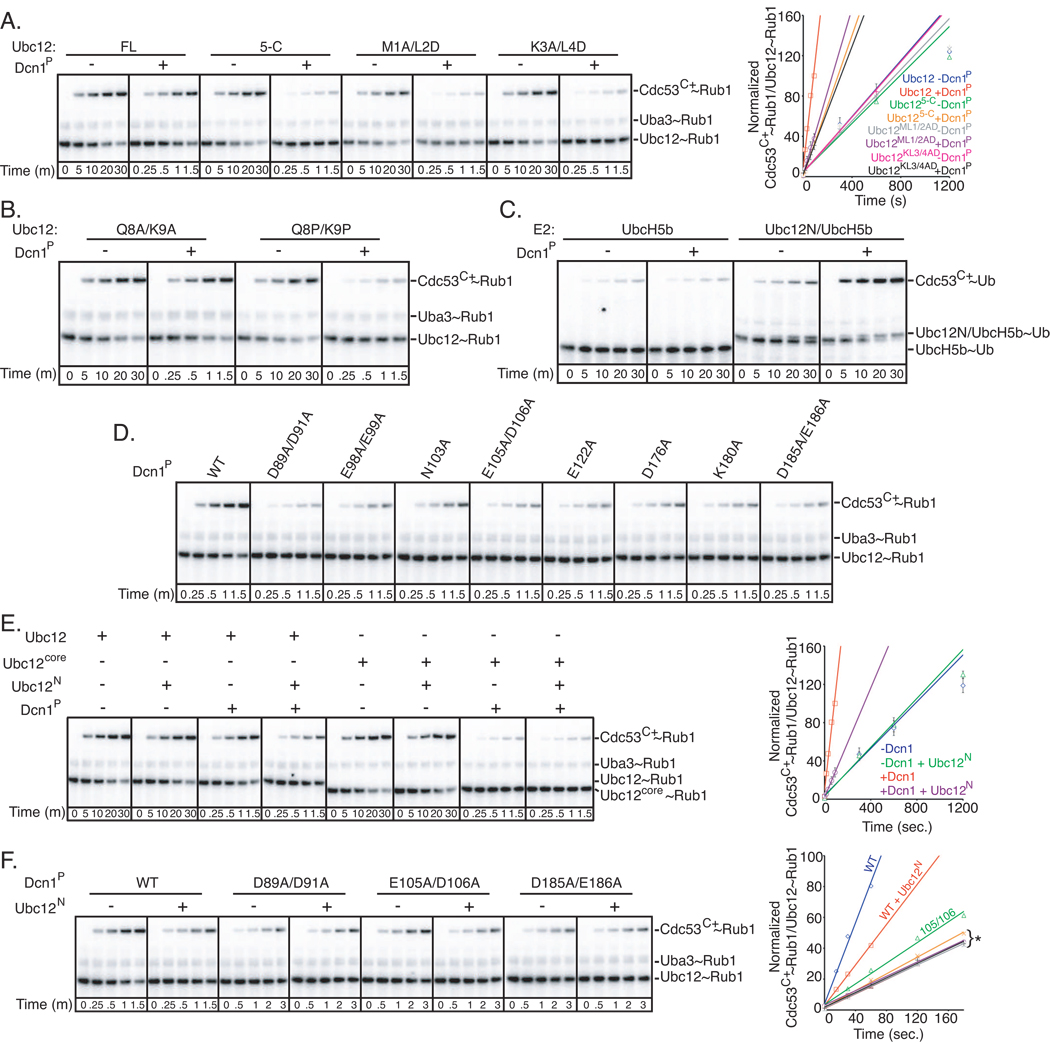
Fundamental features of the structural model are supported by the biochemical studies shown in Figs. 1–4. Thus, we tested predictions based on another key feature of the model, that the 4 most N-terminal residues of Ubc12’s unique helix fit in a groove in Dcn1 (Fig. 4D). The effects of deleting the first four residues of Ubc12 mirrored those observed for deleting the entire N-terminal extension: there was no effect on basal activity, but Dcn1P enhancement of Rub1 transfer to Cdc53C+ was significantly impaired (Fig. 5A). Similar results were obtained with Ala and Asp substitutions in place of Ubc12 residues 1 and 2, or 3 and 4, respectively (Fig. 5A).
Figure 5. Mutational analysis of Ubc12 N-terminal helix and Dcn1 groove in Rub1 ligation to Cdc53.
A. Phosphorimager data (left) and quantification (right) for time-course of pulse-chase assay monitoring [32P]-Rub1 transfer to Cdc53C+ from the indicated versions of Ubc12 in the presence or absence of Dcn1P. To observe comparable levels of Rub1 ligation, shorter time-courses are shown in the presence of Dcn1P. Error bars - +/− 1 standard deviation from 3 independent experiments.
B. Same as (A), but with Ala or Pro mutations in place of Ubc12 residues 8 and 9. Note different time courses.
C. Phosphorimager data for time-course of pulse-chase assay monitoring [32P]-ubiquitin (Ub) transfer from UbcH5b or a chimeric version (Ubc12N/UbcH5b, with Ubc12 residues 1–24 grafted at the N-terminus of UbcH5b) to Cdc53C+ in the absence or presence of Dcn1P.
D. Phosphorimager data for time-course of pulse-chase assay monitoring [32P]-Rub1 transfer from Ubc12 to Cdc53C+ in the presence of the indicated versions of Dcn1P.
E. Phosphorimager data (left) and quantification (right) showing effects of a peptide (Ubc12N), corresponding to Ubc12 residues 1–24 on inhibition of [32P]-Rub1 transfer to Cdc53C+ from full-length or the core domain versions of Ubc12, in the presence or absence of Dcn1P. To observe comparable levels of Rub1 ligation, shorter time-courses are shown in the presence of Dcn1P. Error bars - +/− 1 standard deviation from 2 independent experiments.
F. Assay as in (E), but with wild-type Ubc12 and the indicated versions of Dcn1P. Note that longer time-courses were used for Dcn1P groove mutants to better depict their lack of inhibition by Ubc12N. In quantification (right): *Orange line – effects of adding the Ubc12N peptide in the presence of the E105A/D106A double mutant of Dcn1P. Remainder: data for experiments with the D89A/D91A and D185A/E186A double mutants of Dcn1P, either in the presence or absence of Ubc12N peptide. Error bars - +/− 1 standard deviation from 2 independent experiments.
To test a role for the Ubc12 N-terminal helical structure, effects of Pro substitutions, expected to disrupt the helix, were compared with effects of Alas, which remove side-chains while favoring helical conformation. In the absence of Dcn1P, Rub1 modification of Cdc53C+ was similar by Ubc12 with either Ala or Pro mutations of Gln8 and Lys9 in the middle of the helix. However, the Pro substitutions specifically impaired Dcn1P-mediated Rub1 ligation (Fig. 5B). Thus, Ubc12’s N-terminal helix may contribute to Dcn1P’s Rub1 E3 activity.
Is the Ubc12 N-terminus sufficient to allow an E2 to function with the Dcn1 E3? If so, then transplanting Ubc12’s N-terminal sequence onto a broad specificity E2 that can also bind the Hrt1 RING should impart activation by Dcn1P. To test this, we appended the sequence of Ubc12’s residues 1–24 to the N-terminus of the broadly functioning ubiquitin E2, UbcH5b. Whereas Dcn1P had no effect on the low level of ubiquitin transfer from wild-type UbcH5b to Cdc53’s Lys760, grafting Ubc12’s N-terminal sequence onto UbcH5b allowed enhancement by Dcn1P (Fig. 5C).
Data also support a role for the Dcn1 groove: Ala substitutions in place of individual or pairs of surface residues (Asp89, Asp91, Asn103, Glu105, Asp106, Glu122, Lys180, Asp185, Glu186) around the groove and at the predicted Cdc53/Hrt1/Ubc12 junction diminish Dcn1P-stimulated Rub1 transfer from Ubc12 to Cdc53C+ (Fig. 5D).
The structural model also predicts that blocking Ubc12 access to the Dcn1 groove would inhibit Dcn1P E3 activity. Indeed, very high concentrations of a peptide corresponding to Ubc12’s N-terminus (Ubc12N) inhibited Dcn1P E3 activity by a factor of ~2 (Fig. 5E). The weak inhibitory activity is consistent with the notion that Dcn1 interactions with Ubc12’s N-terminus require a high effective concentration established by other contacts within the catalytic complex. In further agreement with the structural model, the peptide does not display the same inhibitory effect in the absence of Dcn1P, with the Ubc12 core domain, or with the Dcn1P groove mutants (Fig. 5E, 5F). Notably, the Ubc12 N-terminal sequence supplied in trans did not impart Dcn1P E3 activity to the Ubc12 core domain.
Dcn1 E3 helps bring the Ubc12 active site to Cdc53
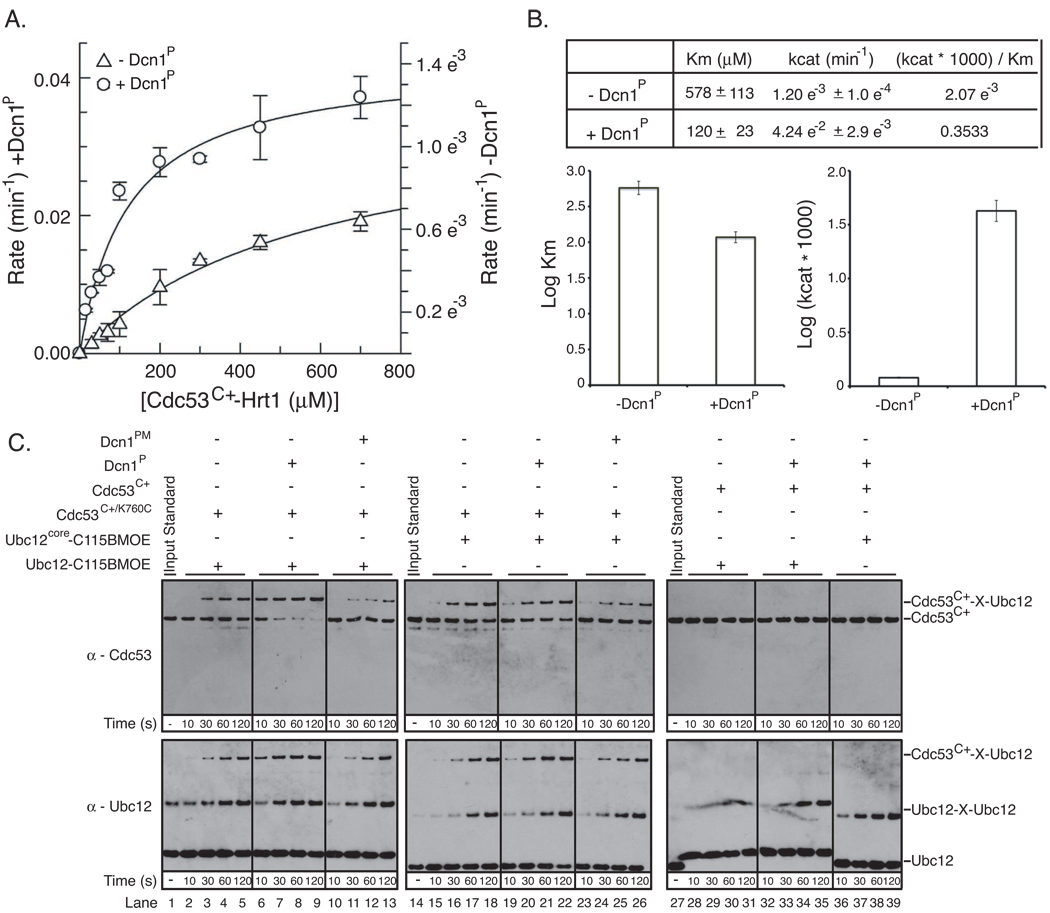
To address the mechanisms by which Hrt1 and Dcn1 cooperate to mediate Rub1 ligase activity, we wished to perform a kinetic analysis. Initial studies indicated that the rapid velocity of Rub1 transfer from Ubc12 to Cdc53 precluded our performing standard kinetic analysis of wild-type proteins. Nonetheless, we did obtain kinetic constants using a mutant version of Ubc12 harboring an Ala mutation in place of a strictly conserved E2 Asn (residue 107) known to be critical for RING E3 stimulation of E2-mediated UBL transfer (Fig. 6A) (Wu et al., 2003b). The kinetic analysis highlights three major points (Fig. 6B). First, under our assay conditions, in comparison to the reaction with only the single Hrt1 E3, the overall kinetic efficiency of the reaction was strikingly ~170-fold greater when the two E3s work in combination. Second, in keeping with the notion that Hrt1 plays the major role of recruiting substrate to the Ubc12, Dcn1P contributed only slightly (~5-fold) to the Km for the Cdc53 substrate. Third, addition of the second E3, Dcn1P, dramatically affected (>35-fold) the kcat of the reaction.
Figure 6. Functions of the Dcn1 Rub1 E3.
Error bars - +/− 1 standard deviation from 3 independent experiments.
A. Michaelis-Menten curves for pulse-chase [32P]-Rub1 transfer to Cdc53C+-Hrt1, from Ubc12 Asn107Ala, in the absence or presence of Dcn1P.
B. Kinetic constants obtained from data in (A).
C. Bis-Maleimidoethane (BMOE) crosslinking (X) of Ubc12C115only (full-length or core domain) to Cdc53C+/K760C, in the absence or presence of Dcn1P or the D226A/D259A double mutant (Dcn1PM). Shown are western blots with antibodies against Cdc53 (top) or Ubc12 (bottom).
How might the Dcn1P E3 make such an enormous contribution to the kcat for the reaction? It is unlikely that Dcn1P dramatically increases the reactivity of Cdc53 Lys760, because Dcn1P binds the opposite side of the Cdc53 WHB subdomain, and our NMR data indicate little effect of Dcn1P on the overall structure around Cdc53 Lys760 (Fig. S3B, S3C). By contrast, the structural data raise the possibility that Dcn1P interacts with Cdc53 and Ubc12 in the context of a complex with Hrt1 to limit the orientations accessible to the Hrt1 RING-Ubc12 complex, and thereby direct Ubc12’s active site to Cdc53.
We tested this notion through a crosslinking approach. First, to be able to attach a crosslinker specifically to the Ubc12 active site, we made a Ubc12 mutant in which the only cysteine is the catalytic Cys115 (Ubc12C115only). Second, we generated Cdc53C+ with a Cys in place of Lys760. After reacting Ubc12C115only with the short-spacer homobifunctional sulfhydryl crosslinker Bis-Maleimidoethane (BMOE) and desalting to remove any free BMOE, Hrt1-Cdc53C+/K760C was added in the presence or absence of Dcn1P. Ubc12 crossslinking to Cdc53 depended on Cys760 (Fig. 6C, lanes 2–5 v. 28–39), and was enhanced by Dcn1P (Fig. 6C, lanes 6–9 v. 2–5). The stimulatory effect is not observed for a Dcn1P mutant (Dcn1PM) that cannot bind Cdc53 (Fig. 4B; 6C lanes 6–9 v. 10–13), and is also impaired by deleting Ubc12’s N-terminal helix (Fig. 6C lanes 6–9 v. 19–22). Thus, the crosslinking results are consistent with our structural model for the Rub1 ligation complex, and support the concept that Dcn1 productively orients Ubc12’s active site toward Cdc53.
Discussion
Hrt1 and Dcn1 function synergistically as a dual Rub1 E3 ligase for Cdc53
We propose that Hrt1 and Dcn1 function together to mediate Rub1 E3 ligase activity toward Cdc53. Hrt1 functions as a conventional RING E3: the N-terminal strand recruits the substrate Cdc53 and the RING binds to and activates Ubc12’s core domain. Based on studies of other cullin-RING complexes (Duda et al., 2008), we expect that the connection between the Hrt1 domains is sufficiently flexible to allow rotation of the RING domain relative to the remainder of the Cdc53 structure. The model of Cdc53-Hrt1, based on homology to human Cul1-Rbx1 structures (Zheng et al., 2002), indicates that the RING is tethered in such a way as to limit the range of orientations that would allow the associated Ubc12 active site to encounter Cdc53’s Lys760.
Our biochemical data are consistent with the notion that the second E3, Dcn1, functions synergistically with Hrt1 to reduce nonspecific Ubc12~Rub1 discharge, and to spatially direct Ubc12’s active site toward Cdc53. Taken together with prior structures of human Cul1-Rbx1 (Zheng et al., 2002) and RING-E2 complexes (Mace et al., 2008; Yin et al., 2009; Zheng et al., 2000), our new structures of Dcn1P-Cdc53WHB and Ubc12 (Table 1) suggest how combinatorial interactions mediate a dual E3 ligase mechanism. Dcn1 binds the Cdc53 WHB subdomain, on the opposite side from the acceptor Lys760, and extends away. The simplest explanation for our mutational data agrees with the structural model that Dcn1’s groove would engage the N-terminal helix of Ubc12 bound to the Hrt1 RING. We intuit that the dual E3 ligase mechanism restricts the Hrt1 RING-Ubc12 orientation for productive encounter with Cdc53’s Lys760 and Rub1 transfer.
Table 1.
Data collection and refinement statistics
| Dcn1P-Cdc53WHB (Form 1) 3O2P.pdb |
Dcn1P-Cdc53WHB (Form 2) 3O6B.pdb |
Ubc12 3O2U.pdb |
|
|---|---|---|---|
| Data Collection | |||
| Beamline | NECAT (ID-24-E APS) |
NECAT (ID-24-C APS) |
SERCAT (BM-22 APS) |
| Space group | P61 | P61 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 57.157, 57.157, 177.725 |
123.914, 123.914, 192.558 |
56.451, 35.938, 87.035 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90.645, 90 |
| Resolution (Å) | 2.23 | 3.1 | 2.0 |
| Rmerge | 6.7 (20.5) | 9.0 (61.9) | 9.2 (49.4) |
| I/σI | 27.4 (11.75) | 22.6 (3.2) | 20.7 (2.5) |
| Completeness (%) | 99.9 (99.9) | 99.1 (99.6) | 98.5 (95.3) |
| Refinement | |||
| Resolution (Å) | 33.0–2.23 | 40.0–3.1 | 25.9–2.0 |
| No. reflections for refinement |
15730 | 28535 | 23515 |
| Rwork/Rfree | 16.9/22.6 | 25.8/30.8 | 18.5 / 23.3 |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.007 | 0.011 | 0.007 |
| Bond angles (°) | 0.95 | 1.7 | 1.16 |
|
Ramachandran plot statistics |
|||
| Most favored regions | 96.8 | 91.8 | 97.0 |
| Additional allowed regions | 2.8 | 6.8 | 2.5 |
| Disallowed regions | 0.4 | 1.4 | 0.5 |
Data for highest resolution shell is shown in parentheses. Rwork = ∑|Fo-Fc|/∑FoRfree is the cross-validation of R-factor, with 5–10% of the total reflections omitted in model refinement.
Combinatorial interactions – an emerging mechanism specifying RING E3-E2 activities
Why would Rub1 ligation to Cdc53 require two E3s? Although we can only speculate, we consider that the dual E3 mechanism is based on four key features of Cdc53-Hrt1-E2 interactions. First, Hrt1 mediates two functions. In addition to its role in Rub1 modification of Cdc53, Hrt1 is the catalytic RING component of the major class of ubiquitin E3s, the cullin-RING E3s that include the large Cdc53-based SCF subfamily (Kamura et al., 1999b; Ohta et al., 1999; Seol et al., 1999; Skowyra et al., 1999; Tan et al., 1999). Thus, Hrt1 must display multifunctional RING E3 activities toward multiple E2s. Second, Hrt1 mediates E3 activity towards different substrates. With Ubc12, Hrt1 mediates Rub1 ligation to Cdc53 (Fig. 1) to counteract the Lag2 inhibitor of SCF assembly (Liu et al., 2009; Siergiejuk et al., 2009). With the ubiquitin E2 Cdc34, Hrt1 mediates polyubiquitination of numerous SCF substrates (Mathias et al., 1996; Verma et al., 1997; Willems et al., 1996). Third, by analogy to human cullin-RING ligases, the Hrt1 RING and N-terminal strand would be flexibly linked so associated E2s access different substrates at a range of relative locations (Duda et al., 2008). Finally, RING E3-E2 interactions are often transient (Christensen and Klevit, 2009; Deshaies and Joazeiro, 2009), likely for cycling between E1s for UBL loading and E3s for UBL ligation (Eletr et al., 2005; Huang et al., 2005).
We propose that combinatorial interactions impart specificity to the multifunctional Cdc53-Hrt1 ligase complex. For Rub1 ligation to Cdc53, this would involve establishing catalytic competence through multiple weak interactions. Notably, the largest effect of Dcn1P is on the kcat for the reaction (Fig. 6). This is consistent with a major role of the dual E3 ligase mechanism to help restrict the orientation of Hrt1 such that the associated Ubc12~Rub1 productively encounters Cdc53’s Lys760.
Combinatorial interactions may be a common feature of RING E3-E2 complexes, with added contacts imparting specificity (Liu and Walters, 2010). Indeed, another Hrt1 partner is the ubiquitin E2 Cdc34, which has distinct active site features for generation of Lys48-linked polyubiquitin chains (Petroski and Deshaies, 2005b). Like Ubc12, Cdc34 also has an extension. Cdc34’s extension interacts with the Hrt1 partner Cdc53 (Kleiger et al., 2009a; Kleiger et al., 2009b). However, here Cdc53 is not the substrate, but rather the cullin component of a multisubunit SCF ubiquitin E3. Electrostatic interactions between the acidic Cdc34 extension and a basic canyon on the cullin enable substrate polyubiquitination. Another example involves the multidomain RING E3 gp78 and the ubiquitin E2 Ube2g2 (Das et al., 2009). In this case, it is the E3 that has a second domain, called “G2BR”, that adds interactions with the “backside” of Ube2g2 distal from the RING binding site. G2BR-binding allosterically alters the conformation of Ube2g2 to modify catalytic activity. Interestingly, providing G2BR to Ube2g2 in trans enhances binding to gp78 and other RINGs. This finding might portend yet to be discovered cases where either the gp78 G2BR, or other sequences like it, could serve in combination with other RING proteins as a component of dual E3 ligase mechanisms for ubiquitin pathways.
Variations on the E2-E3-target theme
We have shown that Rub1 ligation to Cdc53 involves two E3s functioning with one E2. The Rub1 cascade thus joins several other pathways where the model that a single RING E3 bridges a single E2 with targets harboring a common E3-binding motif is too simplistic. For example, some E3s work sequentially with multiple E2s (Garnett et al., 2009; Rodrigo-Brenni and Morgan, 2007; Williamson et al., 2009; Wu et al., 2010a; Wu et al., 2010b), often for the separate steps of ligating a first ubiquitin and then elongating a polyubiquitin chain. Some substrates are modified by different E2-E3 pairs, or by an E2-E3 followed by an E2-E4 complex, each either ligating a different UBL or mediating a different type of ubiquitin modification (Koegl et al., 1999; Parker and Ulrich, 2009; Reindle et al., 2006; Ulrich and Jentsch, 2000; Wertz et al., 2004). Also, one E3 has several distinct substrate binding domains, each recognizing a unique sequence motif (Xia et al., 2008). Variations on E2-E3-target cascades also may be common for E3s outside the RING class. One example is a HECT E3 that utilizes an adaptor protein to recruit both a substrate and particular E2 (Ogunjimi et al., 2005). Thus, while many cascades utilize common E2-E3 modules at their core, additional elements, be they multiple E2s, multiple E3s, E4s, or other factors, seem to regularly be involved in tuning affinities and specificities so that a given target receives a particular UBL modification.
Implications for other cullin-RING ligase pathways
Genetic studies revealed that both Hrt1 and Dcn1 are critical for Rub1 modification of Cdc53; deletion of the Dcn1 gene phenocopies deletion of genes in the Rub1 pathway (Kamura et al., 1999a; Kurz et al., 2008; Kurz et al., 2005). However, Dcn1 has not yet been reported to bind the other yeast cullins Cul3 and Rtt1101, and Rtt101 can also be modified by a UBL other than Rub1 (Laplaza et al., 2004), so it is possible that other E3s function alongside Hrt1 for various modifications of other yeast cullins. The cullin-RING ligase family is even more expanded in higher eukaryotes, with mammals having 2 E2s for the Rub1 ortholog NEDD8 (Huang et al., 2009), 8 cullins (Petroski and Deshaies, 2005a), two Hrt1 orthologs (Kamura et al., 2004), and 5 Dcn1 family members (Kurz et al., 2005).
At least some features we observe for the yeast Cdc53 pathway are conserved in other cullin-RING pathways: mammalian Rbx1 seems to function as a classic RING E3 for NEDD8 modification of Cul1, Cul2, Cul3, and Cul4, with Rbx2 an E3 for Cul5 (Gray et al., 2002; Huang et al., 2009; Morimoto et al., 2003). Also, mammalian Dcn1 homologs bind cullins, consistent with conservation of many residues mediating Dcn1P-Cdc53WHB contacts (Kim et al., 2008; Kurz et al., 2008; Kurz et al., 2005; Meyer-Schaller et al., 2009). Thus, it is likely that mammalian DCNL-cullin complexes will resemble our Dcn1P-Cdc53WHB crystal structure.
There may also be some differences between cullin modification pathways. The Dcn1 groove we find to play a role in Rub1 modification is only partly conserved. Also, to date, human Ubc12’s N-terminal extension has only been observed as an extended structure in complex with UBA3 (Huang et al., 2007; Huang et al., 2004). Although some sequence similarity (Huang et al., 2009) raises the possibility that a helix might form if human Ubc12’s N-terminus were to bind other proteins such as Dcn1P, we do not observe the N-terminus in a crystal structure of free human Ubc12 (D. Huang and BAS, unpublished). Moreover, in the absence of Dcn1, we find NEDD8 modification of human cullins in vitro to be orders-of-magnitude more efficient than of Cdc53 (not shown). Accordingly, knocking out or down mammalian Dcn1s apparently reduces but does not eliminate NEDD8 modification of cullins (Kim et al., 2008; Meyer-Schaller et al., 2009). Also, DCNL1/SCCRO null mice do not share the embryonic lethality of the UBA3 knockout (Kim et al., 2008; Tateishi et al., 2001). Thus, Dcn1 family members may play redundant, less critical, or different roles with mammalian cullins. For example, DCNL3 contains a lipidation site, and can relocalize Cul3 to the plasma membrane for NEDD8 modification (Meyer-Schaller et al., 2009). Notably, despite many similarities, differences in ubiquitination mechanisms have been reported for some paralogous yeast and mammalian cullin-RING ligases (Hao et al., 2007; Orlicky et al., 2003). Irrespective of whether all cullins utilize dual E3 mechanisms for Rub1 or NEDD8 modification, or whether associated Dcn1 family members play other roles in some organisms, it seems likely that there will be many exciting variations on the ways that these and other E3s assemble into active complexes at particular cellular locations to mediate UBL ligation and regulation.
Experimental Procedures
Biochemical assays
Details of reagent generation, reaction conditions, and data analysis are in Supplemental Information. Biochemical assays detecting 32P-labled Rub1 were carried out at 18°C (the ambient room temperature of our room dedicated for radioactive work). Nonradioactive assays were carried out at room temperature. Aliquots were removed at the indicated times and quenched in 2X SDS-sample buffer. Dried radioactive gels were exposed to a Storm (GE) Phosphoimager screen and the amount of Cdc53C+~Rub1 formed quantified as arbitrary units with ImageQuant (GE). For kinetic measurements, four time points were assayed for each reaction to ensure reaction rates were under initial velocity conditions. Kinetic constants were calculated in GraFit v 5.0 (Erathicus) using a standard Michaelis-Menten enzyme kinetics equation. ITC measurements were performed on a MicroCal ITC200 at 22°C.
Structural experiments
Crystals were grown by the hanging-drop vapor-diffusion method, and were flash-frozen prior to data collection at the NECAT and SERCAT beamlines of the Advanced Photon Source and the 8.2.1 beamline at the Advanced Light Source. Details of crystallization conditions, data collection and processing, structure determination and refinement, and NMR experiments are in Supplemental Information.
Highlights.
-
-
2 E3s, Dcn1 and Hrt1, synergize for Rub1 ligation to Cdc53
-
-
Hrt1 functions as a conventional RING E3 for Ubc12~Rub1
-
-
Dcn1 imparts specificity and directs Ubc12’s active site to Cdc53
-
-
Structures of Dcn1P-Cdc53WHB and Ubc12 allow modeling of a dual Rub1 E3 ligase
Supplementary Material
Acknowledgments
This was supported by ALSAC, the St. Jude Cancer Center Core grant, NIH R01GM069530 to BAS, R01CA082491 to RWK, ERC Young Investigator Grant to TK, and the Howard Hughes Medical Institute. BAS is an Investigator of the Howard Hughes Medical Institute. We thank the Schulman lab for discussions, DW Miller, S Bozeman, DJ Miller and J Bollinger for admin/computational support, C–G Park and N Pytel for help with NMR sample, P Klebba for advice regarding kinetics, K Rajashankar, I Kurinov, C Ralston and staff at ALS 8.2.1. NECAT is supported by NIH NCRR RR-15301, APS by US DOE W-31-109-Eng-38.
Coordinates/structure factors for Ubc12, and the high and low resolution forms of Cdc53WHB-DCN1P were deposited to the RCSB under accession codes 3O2U, 3O2P and 3O6B, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- Capili AD, Lima CD. Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Curr Opin Struct Biol. 2007;17:726–735. doi: 10.1016/j.sbi.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DE, Klevit RE. Dynamic interactions of proteins in complex networks: identifying the complete set of interacting E2s for functional investigation of E3-dependent protein ubiquitination. FEBS J. 2009;276:5381–5389. doi: 10.1111/j.1742-4658.2009.07249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Mariano J, Tsai YC, Kalathur RC, Kostova Z, Li J, Tarasov SG, McFeeters RL, Altieri AS, Ji X, et al. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol Cell. 2009;34:674–685. doi: 10.1016/j.molcel.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11:1363–1369. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Gray WM, Hellmann H, Dharmasiri S, Estelle M. Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell. 2002;14:2137–2144. doi: 10.1105/tpc.003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. There's the rub: a novel ubiquitin-like modification linked to cell cycle regulation. Genes Dev. 1998;12:901–907. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DT, Hunt HW, Zhuang M, Ohi MD, Holton JM, Schulman BA. Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature. 2007;445:394–398. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DT, Miller DW, Mathew R, Cassell R, Holton JM, Roussel MF, Schulman BA. A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nat Struct Mol Biol. 2004;11:927–935. doi: 10.1038/nsmb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DT, Paydar A, Zhuang M, Waddell MB, Holton JM, Schulman BA. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1. Mol Cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999a;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Jr, Elledge SJ, Conaway RC, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999b;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Kim AY, Bommelje CC, Lee BE, Yonekawa Y, Choi L, Morris LG, Huang G, Kaufman A, Ryan RJ, Hao B, et al. SCCRO (DCUN1D1) is an essential component of the E3 complex for neddylation. J Biol Chem. 2008;283:33211–33220. doi: 10.1074/jbc.M804440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G, Hao B, Mohl DA, Deshaies RJ. The acidic tail of the Cdc34 ubiquitin-conjugating enzyme functions in both binding to and catalysis with ubiquitin ligase SCFCdc4. J Biol Chem. 2009a;284:36012–36023. doi: 10.1074/jbc.M109.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009b;139:957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Sixma TK. Protein-protein interactions regulate Ubl conjugation. Curr Opin Struct Biol. 2007;17:665–673. doi: 10.1016/j.sbi.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Kurz T, Chou YC, Willems AR, Meyer-Schaller N, Hecht ML, Tyers M, Peter M, Sicheri F. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol Cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Kurz T, Ozlu N, Rudolf F, O'Rourke SM, Luke B, Hofmann K, Hyman AA, Bowerman B, Peter M. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature. 2005;435:1257–1261. doi: 10.1038/nature03662. [DOI] [PubMed] [Google Scholar]
- Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaza JM, Bostick M, Scholes DT, Curcio MJ, Callis J. Saccharomyces cerevisiae ubiquitin-like protein Rub1 conjugates to cullin proteins Rtt101 and Cul3 in vivo. Biochem J. 2004;377:459–467. doi: 10.1042/BJ20030755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Walters KJ. Multitasking with ubiquitin through multivalent interactions. Trends Biochem Sci. 2010 doi: 10.1016/j.tibs.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Mimura S, Kishi T, Kamura T. A longevity protein, Lag2, interacts with SCF complex and regulates SCF function. EMBO J. 2009;28:3366–3377. doi: 10.1038/emboj.2009.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace PD, Linke K, Feltham R, Schumacher FR, Smith CA, Vaux DL, Silke J, Day CL. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J Biol Chem. 2008;283:31633–31640. doi: 10.1074/jbc.M804753200. [DOI] [PubMed] [Google Scholar]
- Mathias N, Johnson SL, Winey M, Adams AE, Goetsch L, Pringle JR, Byers B, Goebl MG. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Schaller N, Chou YC, Sumara I, Martin DD, Kurz T, Katheder N, Hofmann K, Berthiaume LG, Sicheri F, Peter M. The human Dcn1-like protein DCNL3 promotes Cul3 neddylation at membranes. Proc Natl Acad Sci U S A. 2009;106:12365–12370. doi: 10.1073/pnas.0812528106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto M, Nishida T, Nagayama Y, Yasuda H. Nedd8-modification of Cul1 is promoted by Roc1 as a Nedd8-E3 ligase and regulates its stability. Biochem Biophys Res Commun. 2003;301:392–398. doi: 10.1016/s0006-291x(02)03051-6. [DOI] [PubMed] [Google Scholar]
- Nagy V, Dikic I. Ubiquitin ligase complexes: from substrate selectivity to conjugational specificity. Biol Chem. 2010;391:163–169. doi: 10.1515/bc.2010.021. [DOI] [PubMed] [Google Scholar]
- Ogunjimi AA, Briant DJ, Pece-Barbara N, Le Roy C, Di Guglielmo GM, Kavsak P, Rasmussen RK, Seet BT, Sicheri F, Wrana JL. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol Cell. 2005;19:297–308. doi: 10.1016/j.molcel.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci U S A. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JL, Ulrich HD. Mechanistic analysis of PCNA poly-ubiquitylation by the ubiquitin protein ligases Rad18 and Rad5. EMBO J. 2009;28:3657–3666. doi: 10.1038/emboj.2009.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005a;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005b;123:1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- Pierce NW, Kleiger G, Shan SO, Deshaies RJ. Detection of sequential polyubiquitylation on a millisecond timescale. Nature. 2009;462:615–619. doi: 10.1038/nature08595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reindle A, Belichenko I, Bylebyl GR, Chen XL, Gandhi N, Johnson ES. Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J Cell Sci. 2006;119:4749–4757. doi: 10.1242/jcs.03243. [DOI] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siergiejuk E, Scott DC, Schulman BA, Hofmann K, Kurz T, Peter M. Cullin neddylation and substrate-adaptors counteract SCF inhibition by the CAND1-like protein Lag2 in Saccharomyces cerevisiae. EMBO J. 2009;28:3845–3856. doi: 10.1038/emboj.2009.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I kappa B alpha. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- Tateishi K, Omata M, Tanaka K, Chiba T. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J Cell Biol. 2001;155:571–579. doi: 10.1083/jcb.200104035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Feldman RM, Deshaies RJ. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol Biol Cell. 1997;8:1427–1437. doi: 10.1091/mbc.8.8.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Willems AR, Lanker S, Patton EE, Craig KL, Nason TF, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci U S A. 2009;106:18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003a;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- Wu K, Kovacev J, Pan ZQ. Priming and extending: a UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol Cell. 2010a;37:784–796. doi: 10.1016/j.molcel.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Hanlon M, Eddins M, Tsui C, Rogers RS, Jensen JP, Matunis MJ, Weissman AM, Wolberger C, Pickart CM. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003b;22:5241–5250. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci U S A. 2010b;107:1355–1360. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Webster A, Du F, Piatkov K, Ghislain M, Varshavsky A. Substrate-binding sites of UBR1, the ubiquitin ligase of the N-end rule pathway. J Biol Chem. 2008;283:24011–24028. doi: 10.1074/jbc.M802583200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhou J, Sun L, Wei Z, Gao J, Gong W, Xu RM, Rao Z, Liu Y. Structural basis for the function of DCN-1 in protein Neddylation. J Biol Chem. 2007;282:24490–24494. doi: 10.1074/jbc.C700038200. [DOI] [PubMed] [Google Scholar]
- Yin Q, Lin SC, Lamothe B, Lu M, Lo YC, Hura G, Zheng L, Rich RL, Campos AD, Myszka DG, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.