Abstract
Background
Both T cell activation during early HIV-1 infection and soluble markers of immune activation during chronic infection are predictive of HIV disease progression. Although the acute phase of HIV infection is associated with increased pro-inflammatory cytokine production, the relationship between cytokine concentrations and HIV pathogenesis is unknown.
Objectives
To identify cytokine biomarkers measurable in plasma during acute HIV-1 infection that predict HIV disease progression.
Design
Study including 40 South African women who became infected with HIV-1 and were followed longitudinally from the time of infection.
Methods
The concentrations of 30 cytokines in plasma from women with acute HIV-1 infection were measured and associations between cytokine levels and both viral load set-point 12 months post-infection and time taken for CD4 counts to fall below 350 cells/μl were determined using multivariate and Cox proportional-hazards regression.
Results
We found that the concentrations of 5 plasma cytokines, IL-12p40, IL-12p70, IFN-γ, IL-7 and IL-15, in women with acute infection predicted 66% of the variation in viral load set-point 12 months post infection. IL-12p40, IL-12p70 and IFN-γ were significantly associated with lower viral load whereas IL-7 and IL-15 were associated with higher viral load. Plasma concentrations of IL-12p40 and GM-CSF during acute infection were associated with maintenance of CD4 counts above 350 cells/μl while IL-1α, eotaxin and IL-7 were associated with more rapid CD4 loss.
Conclusions
A small panel of plasma cytokines during acute HIV-1 infection was predictive of long-term HIV disease prognosis in this group of South African women.
Keywords: HIV-1, cytokines, acute infection, viral load, disease progression
INTRODUCTION
Early immune events during HIV infection are associated with the rate of subsequent disease progression [1]. Peak viremia is accompanied by immune activation and CD4+ T cell depletion, particularly from the gastrointestinal tract [2-4]. Although viral load subsequently declines, immune activation persists, viral replication continues and CD4+ T cells are progressively lost [1, 5]. Immune activation during HIV infection involves activation and proliferation of most immune cells, including T cells, B cells, natural killer (NK) cells and macrophages [6-9]. It also includes increased production of pro-inflammatory cytokines [4, 10, 11]. In the blood of acutely-infected individuals, an intense pro-inflammatory cytokine “storm” is followed by immunoregulatory cytokine production [4]. Pro-inflammatory cytokines enhance HIV replication and CD4+ T cell loss by directly promoting proviral transcription, by recruiting and activating CD4+ T cell targets for HIV infection, and by activation-induced apoptosis of bystander T cells [12-14].
While T cell activation during early HIV infection is known to influence subsequent disease progression [1], the relationship between plasma cytokine production during acute infection and disease prognosis has not been investigated. Biomarkers that can be used to predict the rate of HIV disease progression could be useful in the clinical management of infected individuals, and for the evaluation of candidate HIV vaccines or microbicides aimed at reducing the rate of disease progression, rather than preventing infection [15, 16]. Blood CD4 counts and viral load measurements during primary, chronic and advanced HIV infection are strongly predictive of subsequent disease progression [16-19]. However, it has been argued that coupling CD4 counts and viral load measurements with estimates of T cell proliferation and activation during acute and/or chronic HIV infection could provide significantly increased predictive power [1, 5, 20, 21]. Soluble markers of immune activation, such as tumour necrosis factor-α receptor II p75 (TNF-RII), neopterin and β2-microglobulin, are more easily measurable in plasma samples than cellular activation, and have been found to predict HIV disease progression with comparable efficiency to CD4 counts and viral load measurements [17, 22, 23]. Although the benefits of being able to predict, and possibly modify, disease course during early HIV infection would be substantial, the predictive value of immune activation biomarkers has largely been investigated during chronic HIV infection.
Here we have investigated the association between plasma cytokine concentrations during acute infection and established markers of long-term HIV disease progression such as CD4 counts and viral load measurements. We describe two models based on the easily measurable concentrations of a small number of plasma cytokines that can be used during acute infection to predict HIV disease progression.
PATIENTS AND METHODS
Study participants
Consenting women, recently infected with HIV-1 Subtype C were recruited from HIV negative cohorts which were screened either monthly or 3 monthly for HIV-1 infection as part of the CAPRISA 002 Acute Infection Study [24]. Time of infection was defined as the mid-point between the last HIV antibody negative test and the first HIV antibody positive test, or as 14 days prior to a positive RNA PCR assay on the same day as a negative HIV Enzyme Immunoassay. Plasma samples from 40 women at a median of 6 weeks post-infection (range 1-12), and from 14/40 of these women 25.5 weeks pre-infection (range 2-66) were available for analysis and were included in this study. This study was approved by the University of KwaZulu-Natal and the University of Cape Town Ethics Committees.
Markers of HIV-1 disease progression
Absolute blood CD4+ T cell counts (cells/μl) were measured using a FACSCalibur flow cytometer at regular intervals during HIV-1 infection (weekly for a month following HIV-1 infection, fortnightly for 2 months, monthly for 9 months and quarterly thereafter). Plasma HIV-1 RNA concentrations (copies/ml) were quantified using the COBAS AMPLICOR™ HIV-1 Monitor v1.5 or COBAS Ampliprep/COBAS TaqMan 48 Analyser (Roche Diagnostics, Branchburg, New Jersey, USA). Viral load and CD4 count set-points were defined as the average CD4+ T cell or viral load measurements of 3 consecutive visits between medians of 47 and 55 weeks post-infection (range: 37-69 weeks) overlying the 12 month post-infection time-point.
Measurement of plasma cytokines
Thirty cytokines were measured in plasma from HIV-uninfected and HIV-infected women using High Sensitivity and Human Cytokine LINCOplex kits (LINCO Research, MO, USA): interleukin (IL)-1α, IL-1β, IL-1 receptor agonist (ra), IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12p40, IL-12p70, IL-13, IL-15, IL-17, epidermal growth factor (EGF), eotaxin, fractalkine, granulocyte colony stimulating factor (G-CSF), granulocyte-macrophage (GM)-CSF, interferon (IFN)-γ, IFN-gamma-induced protein (IP)-10, monocyte chemotactic protein (MCP)-1, macrophage inflammatory protein (MIP)-1α, MIP-1β, RANTES, soluble CD40 ligand (sCD40L), transforming growth factor (TGF)-α, TNF-α and vascular endothelial growth factor (VEGF) [25]. The sensitivity of these kits ranged between 0.01 and 27 pg/ml for each of the cytokines measured. All samples were assayed concurrently, on the same plates, in order to avoid intra-assay variability. Each sample was assayed twice using separate High Sensitivity kits, and the average cytokine concentrations of the two assays were used for all analyses. Data was collected using a Bio-Plex™ Suspension Array Reader (Bio-Rad Laboratories Inc®). Cytokine concentrations below the lower limits of detection were reported as the midpoint between the lowest concentration for each cytokine measured and zero [11].
Statistical analyses
Univariate analyses were performed using GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA). Mann-Whitney U and Wilcoxon Signed Rank tests were used for unmatched and matched comparisons, respectively. Spearman Rank tests were used to test for correlations. P-values<0.05 were considered significant. P-values were adjusted using a false discovery rate (FDR) step-down procedure [26] in order to reduce false positive results when multiple comparisons were made.
Multivariate analyses were performed using STATA™ (StataCorp, Texas, USA). A multivariate regression model was used to determine the cytokines that best predicted 12 month viral load set-points. Log-transformed viral loads and cytokine concentrations were used, except for IL-12p40 which was not normalized following log transformation and was therefore included as a categorical variable [response (1) versus no response (0)]. Using univariate regression as a starting point, cytokines that were significantly associated with viral load set-point, while controlling for each of the cytokines already included, were added to the model in a stepwise manner. Likelihood ratio tests were used to compare nested models. Due to the relatively small size of the study group, the sample of individuals used to develop the model included all women who were followed for at least 12 months post-infection with cytokine datasets (n=31). Model performance evaluation was conducted by repeatedly and randomly sampling subsets (n=10) consisting of three-quarters of the developmental sample and reapplying the model. The validity of the assumptions underlying the model was evaluated and outliers and influential data points were determined using an analysis of residuals. Predicted viral load set-points were calculated for each study participant using the standardized β-coefficients of each of the cytokines included in the model and the observed concentrations or response of each cytokine in the following regression equation:
λi is the predicted viral load set-point of the ith patient whose log-transformed cytokine concentrations or response were X1i, X2i,…Xρi.
A Cox proportional-hazards model was used to determine the cytokines significantly associated with the time taken for study participant CD4 counts to fall below 350 cells/μl for two or more consecutive visits (survival time). Log-transformed cytokine concentrations were used, except in the case of IL-12p40. Following univariate analysis, cytokines which were significantly associated with survival time, while controlling for each of the cytokines already included, were added to the model in a stepwise manner. The likelihood ratio test was used to compare nested models and non-nested models were compared using Aikaike's Information Criterion (AIC), with the lowest AIC indicating the best model in terms of fit. The sample for model development included all women for whom complete cytokine datasets were obtained (n=35). Model performance evaluation was conducted by repeatedly sampling three-quarter subsets (n=10) of the developmental sample and reapplying the model. The validity of the assumptions underlying the model was evaluated and outliers and influential data points were determined by an analysis of residuals.
Risk scores were calculated for each participant using the β-coefficients of the Cox proportional-hazards model [27] and observed concentrations or response of cytokines included in the model according to the following equation:
λi is the risk score of the ith patient whose log transformed cytokine concentrations or response are X1i, X2i,…Xρi. The study group was divided into 3 groups based on risk scores as follows: low risk (0-15); medium risk (15-20); high risk (20-25).
RESULTS
Description of study participants
Forty black women from Durban, South Africa, recently infected with HIV-1 were recruited into this study (Table 1). Most of the women were unmarried (97.5%), 20% reported having more than one partner and 35% were using injectable hormonal contraception at the time of HIV infection. Sexually transmitted infections (STI) were common in this cohort with 94.5% of women having been diagnosed with at least one active infection or bacterial vaginosis. The median acute infection CD4 count and viral load in this group of women were 477 cells/μl and 76 200 copies/ml, respectively (Table 2). The median CD4 count and viral load set-points of women who were followed for at least 12 months post-infection were 415 cells/μl and 39 783 copies/ml, respectively.
Table 1.
Demographic characteristics and prevalence of sexually transmitted infections in HIV-infected study participants
| Characteristics | Value |
|---|---|
| Number of subjects | 40 |
| Age in years [median (IQR)] | 25 (21-37) |
| Marital status [N/Total (% of women married)] | 1/40 (2.5%) |
| Number with >1 partner [N/Total (% of women with >1 partner)] | 8/40 (20%) |
| Contraception use [N/Total (% of women using contraception)] | 14/40 (35%) |
| Prevalence of STIs [N/Total (% of women with lab diagnosed STI)] | 37/39 (94.5%) |
| Prevalence of multiple STIs [N/total (% of women with lab diagnosed multiple STIs)] | 31/36 (86.1%) |
| Trichomonas vaginalis [N/total (% of women PCR positive for T. vaginalis)] | 4/36 (11.1%) |
| Chlamydia trachomatis [N/total (% of women PCR positive for C. trachomatis)] | 6/36 (16.7%) |
| Neisseria gonorrhoea [N/total (% of women PCR positive for N. gonorrhoea)] | 6/36 (16.7%) |
| Treponema pallidum [N/total (% of women with detectable T. pallidum antibody)] | 2/40 (5.0%) |
| Shedding HSV-2 [N/total (% of women PCR positive for HSV-2)] | 1/36 (2.8%) |
| HSV-2 IgG [N/total (% of women with detectable HSV-2 antibody)] | 37/40 (92.5%) |
| Bacterial vaginosis [N/total (% of women gram stain positive for BV)] | 27/36 (75.0%) |
Table 2.
Clinical characteristics of study participants
| Clinical Characteristics | Median (IQR) | N |
|---|---|---|
| CD4+ T cell counts: | ||
| Acute infection CD4+ T cell count (cells/ìl) | 477 (385-676) | 40 |
| CD4+ T cell count set-point (ave of 3 visits overlying 12 months post-infection; cells/ìl) | 415 (314-607) | 36 |
|
| ||
| Plasma viral load: | ||
| Acute infection plasma viral load (copies/ml) | 76 200 (117 775-339 250) | 40 |
| Plasma viral load set-point (ave of 3 visits overlying 12 months post-infection; copies/ml) | 39 783 (6 613-104 825) | 36 |
Blood CD4+ T cell counts and plasma viral loads were determined for each woman (n=40) during acute HIV-1 infection. CD4+ T cell count and viral load set-points were defined as the average CD4+ T cell or viral load measurements of 3 consecutive visits overlying the 12 month post-infection time-point for each of the 36 women who were followed for at least 12 months post-infection. IQR: Interquartile range.
Plasma inflammatory cytokine concentrations are elevated during acute HIV-1 infection and are associated with peak viremia
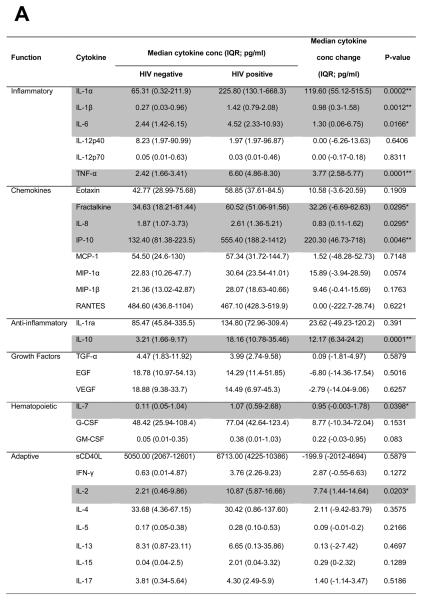
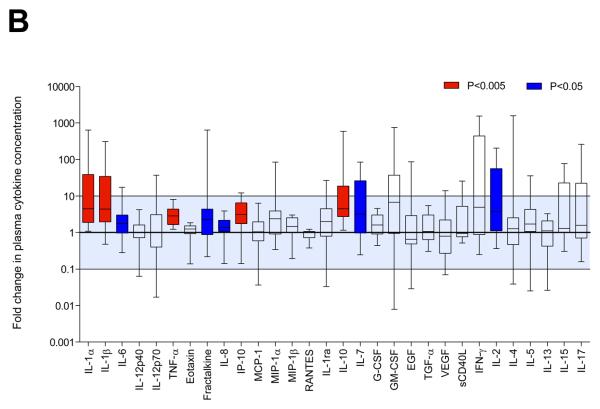
Cytokine concentrations in plasma from HIV-uninfected women (median of 25.5 weeks pre-infection; n=14) were compared with samples from the same women recently infected with HIV-1 (median of 6 weeks post-infection; Fig. 1). Plasma concentrations of several pro-inflammatory cytokines (IL-1α, IL-1β, IL-6, TNF-α, IL-8, fractalkine, IP-10), anti-inflammatory IL-10, and T cell homeostatic cytokines (IL-2, IL-7) were increased during acute HIV-1 infection compared to matched pre-infection samples. After adjusting for multiple comparisons, IL-1α, IL-1β, TNF-α, IP-10 and IL-10 remained significantly elevated (Fig. 1A). Most cytokines (24/30) tended to be elevated during acute infection (Fig. 1B). Acute infection plasma concentrations of each of the 30 cytokines were not associated with any of the demographic characteristics listed in Table 1 (data not shown). No associations between cytokine levels and STIs were found by logistic regression analysis, however, due to the small number of women without an STI (n=2) and the prevalence of multiple infections in this group of participants, a more detailed analysis in a larger cohort of women would be required to confirm these findings.
Figure 1.
Comparison of plasma cytokine concentrations in women (n=14) before infection (median 25.5 weeks pre-infection) and during acute HIV-1 infection (median 6 weeks post-infection). A) Absolute IL-1α, IL-1β, IL-6, TNF-α, IL-8, fractalkine, IP-10, IL-10, IL-7 and IL-2 concentrations were elevated in women with acute HIV-1 infection relative to concentrations pre-infection. Wilcoxon Signed Ranks test was used for matched comparisons. P-values <0.05 were considered significant and highlighted. *Did not remain significant following false discovery rate adjustment for multiple comparisons. ** Remained significant following adjustment for multiple comparisons. B) Fold upregulation in plasma cytokine concentrations following infection. P-values <0.005 remained significant following adjustment for multiple comparisons (red bars). Blue bars (P<0.05) indicate cytokines that were significantly upregulated before adjustment for multiple comparisons. IQR: Interquartile range.
It was found that IP-10, TNF-α, IL-1α, IL-1β, IFN-γ, and IL-10 correlated with the magnitude of viral load (Fig. 2; a heat map of the association between all 30 cytokines and viral load is shown in Supplementary Fig. 1). After adjustment for multiple comparisons, IP-10 (adjusted p=0.012) and TNF-α (adjusted p=0.0165) concentrations remained significantly associated with viral load.
Figure 2.
Plasma inflammatory cytokine concentrations are associated with concurrent plasma viral loads in women with acute HIV-1 infection. Only cytokines that correlated significantly with viral load before adjustment for multiple comparisons are represented (p-values and Spearman rho values below heat map). Women are ranked according to acute infection viral load. Relative acute infection plasma cytokine concentrations of study participants are shown as a heat map, with each row representing the cytokine concentrations in an individual woman and falling alongside her viral load. For each particular cytokine, the concentrations found in this group of women were ranked and assigned an appropriate colour ranging from white (lowest concentration) to red (highest concentration). Repeated values were assigned the same rank and hence colour. IP-10, TNF-α, IL-1α, IL-1β, IFN-γ and IL-10 correlated with viral load. PID: Patient Identity Number. # IP-10 and TNF-α remained significantly correlated with viral load following adjustment for multiple comparisons. * Not done
Plasma cytokine concentrations during acute HIV-1 infection predict viral load set-point
Univariate regression analysis was used to determine the relationship between plasma cytokine concentrations during acute HIV-1 infection and viral load set-point (Supplementary Table 1). As IL-12p70 was most strongly associated with set-point in the univariate analysis, this cytokine was used as a starting point to develop a multivariate model that included the cytokines that together most strongly predicted viral load set-point. Five cytokines were incorporated into this model, each of which was significantly associated with viral load set-point while controlling for the other cytokines included (p<0.005; Fig. 3A). A positive IL-12p40 response and higher concentrations of IL-12p70 and IFN-γ were associated with lower viral load set-point, while higher concentrations of IL-7 and IL-15 were associated with higher set-point. The model was a good fit (F-statistic p<0.0001) and, together, the concentrations of these 5 cytokines in plasma during acute infection predicted 66% (adjusted R2=0.6577) of the variation in viral set-point at 12 months post-infection.
Figure 3.
Acute infection IL-12p40, IL-12p70, IFN-γ, IL-7 and IL-15 concentrations are predictive of viral load set-point. A) Each cytokine was significantly associated with viral load set-point (p-values <0.005). The model fitted the data significantly (p>F<0.0001), and together the 5 cytokines predicted 65.77% (Adjusted R2 = 0.6577) of the variation in set-point. B) Set-point viral loads (VL) as predicted by the model correlate with observed set-point viral loads.
The validity of the assumptions underlying the model was evaluated and outliers and influential data points were determined using an analysis of residuals. Exclusion of potential outliers from the dataset and reapplication of the model did not substantially influence the strength of the model. As the time from infection of sampling varied widely (1-12 weeks post-infection), this was included as a variable in the model. However, incorporation of time from infection did not significantly influence the strength of the model (adjusted R2=0.6435), nor was it significantly associated with viral load set-point (p=0.996). Thus the model presented does not include this variable (Fig. 3A). It was furthermore found that the concentrations of the cytokines included in the model did not demonstrate any association with timing in relation to the estimated date of infection. Upon evaluation of model performance by reapplication of the model to 10 randomly-chosen three-quarter subsets of the study group, the influence of each variable on viral load set-point remained statistically significant and the directionality of the relationships between the variables and set-point remained constant, indicating that the model estimates are stable.
Predicted viral load set-points were calculated for each study participant using the standardized β-coefficients of each of the cytokines included in the model (Fig. 3A) and the observed concentrations or response of each cytokine according to the following equation:
As expected, predicted viral load set-points correlated well with observed set-points (Fig. 3B). The model including IL-12p40, IL-12p70, IFN-γ, IL-7 and IL-15 concentrations during acute infection fitted the viral set-point data of this cohort better (R2=0.6577; p<0.0001) than either acute infection viral load (R2=0.0941; p=0.03) or CD4 counts (R2=0.0734; p=0.0577) or the combination of both (R2=0.0800; p=0.0917). The substantially better R2 value of the cytokine model is due in part to this model having been formulated with this cohort. Although the significance of the relationship between these cytokines and viral load set-point was strongly upheld upon reapplication of the model to three-quarter subsets of the study group, assessment of the true predictive power of this model would require application of the model to an entirely different, larger dataset.
Plasma cytokine concentrations during acute HIV-1 infection predict time taken for CD4 counts to fall below 350 cells/μl
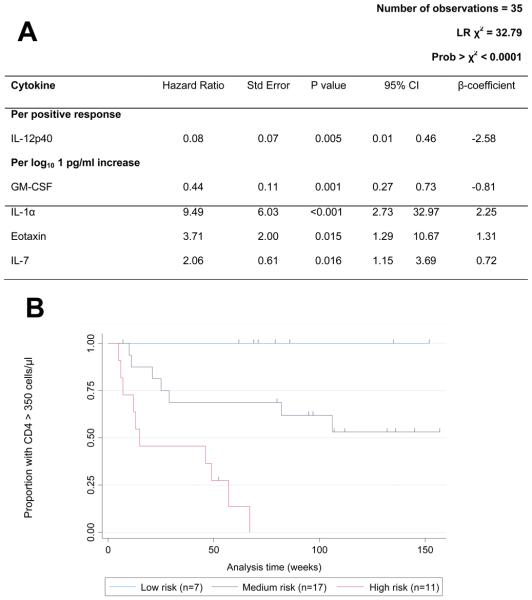
We next determined a subset of cytokines which were most significantly associated with the time taken for participant CD4 counts to fall below 350 cells/μl using a Cox proportional-hazards model. Using univariate survival analysis as a starting point (Supplementary Table 2), cytokines which were significantly associated with survival time were added to the model in a stepwise manner. Plasma concentrations of IL-1α, eotaxin and IL-7 were significantly associated with increased risk of CD4 loss, while GM-CSF and IL-12p40 were associated with reduced risk (p<0.05; Fig. 4A). The model was a good fit (χ2 p<0.0001) and exclusion of potential outliers from the dataset and reapplication of the model did not substantially influence the strength of the model. Time (post-infection) of sampling was included in the model, however this variable was not significantly associated with survival time (p=0.357) while the relationships between each of the cytokines and survival time remained significant. Reapplication of the model to 10 randomly-chosen three-quarter subsets of the study group revealed that model performance was good, with the directionality of the relationships between each of the variables and survival time remaining constant.
Figure 4.
Acute infection IL-12p40, GM-CSF, IL-1α, Eotaxin and IL-7 concentrations were associated with the time taken for the study participant CD4 counts to fall below 350 cells/μl. A) Each cytokine was significantly associated with survival time (p values <0.05) and the model fitted the data well (p>χ2<0.0001). B) Kaplan Meier survival estimates of women grouped according to risk score. Risk scores were calculated for each participant using the β coefficients of each cytokine included in the Cox proportional-hazards model, and women were divided into low (0-15), medium (15-20) and high (20-25) risk groups based on risk scores. Women in the high risk group experienced rapid CD4 loss, while women in the low risk group maintained CD4 counts above 350 cells/μl for the duration of follow-up. Each dash indicates a time point at which an individual woman left the study (censored event).
Risk scores were calculated for each participant using the β-coefficients of the Cox proportional-hazards model (Fig. 4A) and observed acute infection plasma concentrations or response of each of the cytokines included in the model according to the following equation:
HIV-1-infected participants were divided into low, medium or high risk groups according to their risk scores. Women in the high risk group (n=11) experienced rapid CD4+ T cell loss to below 350 cells/μl during the study period (median time to event: 15 weeks post-infection; Fig 4B), with the exception of a single woman who left the study at 52 weeks post-infection. Women in the low risk group (n=7) maintained CD4 counts above 350 cells/μl during the study (median follow-up: 76 weeks post-infection). The Cox proportional-hazards model including acute infection IL-1α, eotaxin, IL-7, GM-CSF and IL-12p40 was a better model in terms of fit (AIC: 85.09) when compared to models including only acute infection CD4 count (AIC: 113.25), or viral load (AIC: 124.81), or the combination of both (AIC: 112.05).
Discussion
Immune activation during HIV infection has been identified as a major contributor to HIV disease progression, and is the product of inflammatory responses to HIV-encoded Toll-like Receptor ligands, microbial translocation and the homeostatic response to CD4+ T cell depletion [1, 8, 9, 28, 29]. Here we propose two models, based on a restricted set of plasma cytokines measured during acute HIV-1 infection, which are useful for the prediction of viral load set-point and CD4 decline. We show that concentrations of IL-7, IL-12p40, IL-12p70, IFN-γ and IL-15 during acute infection better predicted viral load set-point than acute infection viral load, acute infection CD4 counts, or the combination of both. Further, we show that plasma concentrations of IL-7, IL-12p40, IL-1α, eotaxin and GM-CSF during acute infection were strongly predictive of CD4 loss.
HIV viral loads in plasma and systemic CD4 counts are widely accepted predictors of HIV disease progression [16-19]. In addition, T cell proliferative capacity and activation states during early and chronic HIV infection are predictive of disease progression [1, 5, 20, 21]. Concentrations of soluble biomarkers such as TNF-RII, neopterin and β2-microglobulin during chronic infection have also been shown to predict progression to AIDS and/or CD4 decline with a degree of accuracy comparable to that of CD4 counts and viral load measurements [17, 22, 23]. While biomarkers of HIV disease progression identified during chronic infection are useful for determining rates of progression to AIDS, the ability of these markers to predict clinical course at earlier infection stages has not been tested.
We and others have shown that early HIV-1 infection is accompanied by a robust plasma pro-inflammatory cytokine response [4, 11]. Here we demonstrate that plasma immunoregulatory IL-10 and pro-inflammatory IL-1α, IL-1β, TNF-α and IP-10 were elevated in women with acute HIV-1 infection relative to pre-infection. Upregulated pro-inflammatory cytokines, IP-10 and TNF-α, were significantly associated with higher HIV viral load, suggesting that the observed inflammatory cytokine “storm” during acute infection is induced, at least in part, in response to the presence of HIV replication and products.
We found that higher concentrations of IL-12p70, IL-12p40 and IFN-γ were associated with lower viral set-point, while IL-12p40 and GM-CSF were associated with prolonged maintenance of CD4 counts above 350 cells/μl. Production of these cytokines is partly regulated by a positive feedback loop, with IFN-γ and GM-CSF promoting IL-12p70 production and IL-12p70 in turn stimulating IFN-γ and GM-CSF secretion [30-33]. IL-12p70 and IFN-γ promote Th1 differentiation, favouring cell-mediated immunity and inhibiting Th2 responses [34-36]. IFN-γ has previously been identified as a correlate of better disease prognosis in HIV infection, and was positively associated with CD8+ T cell and activated NK cell counts [37, 38]. In SIV-infected macaques, IL-12p70 treatment during acute infection was associated with decreased viral loads, increased CD8+ NK and T cells, reduced naïve CD4+ T cells expressing homing markers, retention of HIV-specific CTL and prolonged survival [39]. IFN-γ, GM-CSF and IL-12p40 are also principal macrophage-inducing cytokines, promoting their production, recruitment and/or activation [40-43].
In this study, elevated IL-7 and IL-15 concentrations were associated with higher viral load set-point, while IL-7, IL-1α and eotaxin were associated with greater CD4 loss. Eotaxin and IL-1α are chemotactic for T cells, potentially recruiting targets for HIV infection [44, 45]. IL-1α has additionally been found to strongly induce NF-κB activation, which binds to HIV-long terminal repeat sequences and in so doing may directly upregulate HIV replication [12, 46]. IL-7 and IL-15 are the principal regulators of CD4+ and CD8+ T cell homeostasis [47, 48]. Picker and colleagues have proposed a model whereby the balance between CD4+ central memory (TCM) and effector memory T cells (TEM) dictates the rate of HIV disease progression [49]. TEM home to effector sites and serve as the primary targets for HIV infection and destruction, but it appears to be their longer-lived TCM precursors, which replenish these populations and decay more gradually, that determine the tempo of disease progression. Higher IL-7 levels are associated with lymphopenic states during HIV infection [9, 50]. IL-7 selectively induces proliferation of naïve T cells and TCM cells and it has been proposed that, at high levels, IL-7 may disrupt the normal naïve/memory differentiation pathway by inducing memory-like characteristics on naïve cells. Exhaustion or excessive differentiation could reduce the longevity of this population, its ability to self-renew, expand and differentiate upon antigen stimulation [48]. IL-15 can induce antigen-independent proliferation and differentiation of TEM from TCM [48, 51]. Thus, elevated IL-15 levels during early infection may accelerate the loss of TEM, thereby depleting TCM more rapidly. Additionally, increased IL-15 levels during acute SIV infection led to an upregulation of CD4 expression on memory CD4+ T cells which increased in their susceptibility to SIV infection [52]. We recently demonstrated that a greater destruction of the CD8+ TCM compartment and accumulation of CD8+ TEM correlated with a higher viral set-point [53]. This may lead to exhaustion of CD8+ resources required for the control of HIV and other infections.
IL-15 has also been implicated in polyclonal B cell activation in HIV infection [54]. Polyclonal B cell activation and differentiation, together with the destruction of germinal centres, has recently been described in acute HIV infection [55], likely resulting in the characteristically ‘delayed’ antibody response to HIV. Early dysregulation of the B cell response due to elevated IL-15 levels may lead to reduced viral control, as reflected in higher set-point viral loads. Thus, memory CD4+ T cell dysfunction and depletion, CD8+ T cell exhaustion and B cell dysfunction may partly be driven by elevated levels of IL-7 and IL-15 from the earliest stages of infection, setting the course for accelerated disease progression.
We found that anti-inflammatory IL-10 was significantly elevated during acute HIV-1 infection, was correlated directly with acute infection viral loads before adjustment for multiple comparisons, and was associated with greater risk of CD4+ T cell loss in a univariate Cox survival analysis. Although IL-10 reduces HIV replication in macrophages [56], this cytokine may contribute to HIV persistence by suppressing effector T cell responses [57-59]. In support, it has been demonstrated that serum IL-10 levels increase with disease progression in HIV-infected individuals [60]. Additionally, regulatory T cells, an important source of IL-10 [61], were shown to correlate inversely with the magnitude of SIV-specific CTL responses during acute SIV infection, and may contribute to viral persistence [62].
In conclusion, we demonstrate the potential to use plasma cytokine concentrations during acute HIV-1 infection to predict subsequent disease progression. Two clusters of cytokines were more strongly predictive of viral load set-point and CD4+ T cell loss than either acute infection CD4 counts, viral loads or both combined. The identification of cytokine biomarkers which are (1) indicative of early immune activation, (2) predictive of subsequent HIV disease prognosis, and (3) can be measured directly in plasma samples from individuals with acute/early HIV infection, may inform approaches for evaluating the ability of therapeutic HIV vaccines and microbicides to control HIV infection.
Supplementary Material
Acknowledgements
LR performed all laboratory work, analysis, modelling and prepared the manuscript; JP developed the hypothesis, performed the analysis and prepared the manuscript; CW designed the cohort, is protocol co-chair for CAPRISA 002 Acute Infection Study, developed the hypothesis, and prepared the manuscript; FL developed the model, performed the analysis and prepared the manuscript; LB performed some of the laboratory work and developed the hypothesis; KM designed and managed the cohort, is protocol co-chair for CAPRISA 002 Acute Infection Study, heads the clinical aspects of the study, performed clinical analysis and prepared the manuscript; WB developed the hypothesis and prepared the manuscript; FVL developed and managed the cohort; GW and JDS performed some of the laboratory work and contributed to manuscript preparation and writing; QAK and SAK conceptualized the CAPRISA cohort and prepared the manuscript; SAK developed the hypothesis. The authors would like to acknowledge the following people for their contribution to this work: the Centre for the AIDS Programme of Research in South Africa (CAPRISA), and especially the members of the Acute Infection Study Team; and the participants of the Acute Infection Study, without whom the work would not have been possible. This work was supported by grants from the Comprehensive International Program of Research on AIDS (CIPRA) of the Division of AIDS (DAIDS), National Institute of Allergy and infectious Disease (NIAID), National Institutes of Health (NIH), US Department of Health and Human Services (DHHS) (grant# U19 AI51794); the Center for HIV-AIDS Vaccine Immunology (CHAVI) by grants from the National Institute of Allergy and Infectious Disease (NIAID), National Institutes of Health (NIH) and the US Department of Health and Human Services (DHHS) (AI51794), and the Poliomyelitis Research Foundation (PRF) of South Africa. JP is a recipient of a Wellcome Trust Intermediate Fellowship in Infectious Diseases. LB was supported by the Columbia University-Southern African Fogarty AIDS International Training and Research Programme (AITRP) and the Fogarty Ellison Programme funded by the Fogarty International Center, National Institutes of Health (grant# D43TW00231). LR was supported by the South African Medical Research Council (MRC), PRF, KW Johnstone Research and Benfara.
This work was supported by grants from the Comprehensive International Program of Research on AIDS (CIPRA) of the Division of AIDS (DAIDS), National Institute of Allergy and infectious Disease (NIAID), National Institutes of Health (NIH), US Department of Health and Human Services (DHHS) (grant# U19 AI51794); the Center for HIV-AIDS Vaccine Immunology (CHAVI) by grants from the National Institute of Allergy and Infectious Disease (NIAID), National Institutes of Health (NIH) and the US Department of Health and Human Services (DHHS) (AI51794), and the Wellcome Trust. The NIH requires that published results arising from NIH funds be made available to the public within 12 months of publication. We therefore request that the authors of this manuscript retain the right to provide a copy of the final manuscript to the NIH upon acceptance for Journal publication, for public archiving in PubMed Central as soon as possible but no later than 12 months after publication by AIDS.
REFERENCES
- 1.Deeks SG, Kitchen CMR, Liu L, Guo H, Gascon R, Narvaez AB, et al. Immune activation set point during early HIV infection predicts subsequent CD4 + T-cell changes independent of viral load. Blood. 2004;104:942–947. doi: 10.1182/blood-2003-09-3333. [DOI] [PubMed] [Google Scholar]
- 2.Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, et al. CD4+ T Cell Depletion during all Stages of HIV Disease Occurs Predominantly in the Gastrointestinal Tract. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehandru S, Poles MA, Tenner-Racz K, Horowitz A, Hurley A, Hogan C, et al. Primary HIV-1 Infection Is Associated with Preferential Depletion of CD4+ T Lymphocytes from Effector Sites in the Gastrointestinal Tract. J Exp Med. 2004;200:761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stacey AR, Norris PJ, Qin L, Haygreen EA, Taylor E, Heitman J, et al. Induction of a striking systemic cytokine cascade prior to peak viraemia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol. 2009;83:3719–3733. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hazenberg MD, Otto SA, van Benthem BHB, Roos MTL, Coutinho RA, Lange JMA, et al. Persistent immune activation in HIV-1 infection is associated with progression to AIDS. AIDS. 2003;17:1881–1888. doi: 10.1097/00002030-200309050-00006. [DOI] [PubMed] [Google Scholar]
- 6.Lane HC, Masur H, Edgar LC, Whalen G, Rook AH, Fauci AS. Abnormalities of B-cell activation and immunoregulation in patients with the acquired immunodeficiency syndrome. N Engl J Med. 1983;309:453–458. doi: 10.1056/NEJM198308253090803. [DOI] [PubMed] [Google Scholar]
- 7.Alter G, Malenfant JM, Delabre RM, Burgett NC, Yu XG, Lichterfeld M, et al. Increased Natural Killer Cell Activity in Viremic HIV-1 Infection. J Immunol. 2004;173:5305–5311. doi: 10.4049/jimmunol.173.8.5305. [DOI] [PubMed] [Google Scholar]
- 8.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 9.Catalfamo M, Mascio MD, Hu Z, Srinivasula S, Thaker V, Adelsberger J, et al. HIV infection-associated immune activation occurs by two distinct pathways that differentially affect CD4 and CD8 T cells. Proc Natl Acad Sci USA. 2008;105:19851–19856. doi: 10.1073/pnas.0810032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norris PJ, Pappalardo BL, Custer B, Spotts G, Hecht FM, Busch MP. Elevations in IL-10, TNF-α and IFN-γ from the Earliest Point of HIV Type 1 Infection. AIDS Res Hum Retroviruses. 2006;22:757–762. doi: 10.1089/aid.2006.22.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bebell LM, Passmore JA, Williamson C, Mlisana K, Iriogbe I, van Loggerenberg F, et al. Relationship between Levels of Inflammatory Cytokines in the Genital Tract and CD4+ Cell Counts in Women with Acute HIV-1 Infection. J Infect Dis. 2008;198:710–714. doi: 10.1086/590503. [DOI] [PubMed] [Google Scholar]
- 12.Osborne L, Kunkel S, Nabel GJ. Tumor necrosis factor-alpha and interleukin-1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappa B. Proc Natl Acad Sci USA. 1989;86:2336–2340. doi: 10.1073/pnas.86.7.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin RH, Hwang YW, Yang BC, Lin CS. TNF receptor-2-triggered apoptosis is associated with the down- regulation of BclxL on activated T cells and can be prevented by CD28 costimulation. J Immunol. 1997;158:598–603. [PubMed] [Google Scholar]
- 14.Swingler S, Mann A, Jacque JM, Brichacek B, Sasseville VG, Williams K, et al. HIV-1 Nef mediates lymphocyte chemotaxis and activation by infected macrophages. Nat Med. 1999;5:997–1003. doi: 10.1038/12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mascola JR, Nabel GJ. Vaccines for the prevention of HIV-1 disease. Curr Opinion Immunol. 2001;13:489–495. doi: 10.1016/s0952-7915(00)00246-6. [DOI] [PubMed] [Google Scholar]
- 16.Goujard C, Bonarek M, Meyer L, Bonnet F, Chaix ML, Deveau C, et al. CD4 cell count and HIV DNA level are independent predictors of disease progression after primary HIV type 1 infection in untreated patients. Clin Infect Dis. 2006;42:709–715. doi: 10.1086/500213. [DOI] [PubMed] [Google Scholar]
- 17.Mellors JW, Munoz A, Giorgi JV, Margolick JB, Tassoni CJ, Gupta P, et al. Plasma Viral Load and CD4+ Lymphocytes as Prognostic Markers of HIV-1 Infection. Ann Intern Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 18.de Wolf F, Spijkerman I, Schellekens PT, Langendam M, Kuiken C, Bakker M, et al. AIDS prognosis based on HIV-1 RNA, CD4+ T-cell count and function: markers with reciprocal predictive value over time after seroconversion. AIDS. 1997;11:1799–1806. doi: 10.1097/00002030-199715000-00003. [DOI] [PubMed] [Google Scholar]
- 19.Lepri AC, Katzenstein TL, Ullum H, Phillips AN, Skinhøj P, Gerstoft J, et al. The relative prognostic value of plasma HIV RNA levels and CD4 lymphocyte counts in advanced HIV infection. AIDS. 1998;12:1639–1643. doi: 10.1097/00002030-199813000-00011. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, Cumberland WG, Hultin LE, Prince HE, Detels R, Giorgi JV. Elevated CD38 antigen expression on CD8+ T cells is a stronger marker for the risk of chronic HIV disease progression to AIDS and death in the Multicenter AIDS Cohort Study than CD4+ cell count, soluble immune activation markers, or combinations of HLA-DR and CD38 expression. J Acquir Immune Defic Syndr. 1997;16:83–92. doi: 10.1097/00042560-199710010-00003. [DOI] [PubMed] [Google Scholar]
- 21.Giorgi JV, Hultin LE, McKeating JA, Johnson TD, Owens B, Jacobson LP, et al. Shorter Survival in Advanced Human Immunodeficiency Virus Type 1 Infection Is More Closely Associated with T Lymphocyte Activation than with Plasma Virus Burden or Virus Chemokine Coreceptor Usage. J Infect Dis. 1999;179:859–870. doi: 10.1086/314660. [DOI] [PubMed] [Google Scholar]
- 22.Fahey JL, Taylor JMG, Manna B, Nishanian P, Aziz N, Giorgi JV, et al. Prognostic significance of plasma markers of immune activation, HIV viral load and CD4 T-cell measurements. AIDS. 1998;12:1581–1590. doi: 10.1097/00002030-199813000-00004. [DOI] [PubMed] [Google Scholar]
- 23.Zangerle R, Steinhuber S, Sarcletti M, Dierich MP, Wachter H, Fuchs D, et al. Serum HIV-1 RNA Levels Compared to Soluble Markers of Immune Activation to Predict Disease Progression in HIV-1-Infected Individuals. Int Arch Allergy Immunol. 1998;116:228–239. doi: 10.1159/000023949. [DOI] [PubMed] [Google Scholar]
- 24.van Loggerenberg F, Mlisana K, Williamson C, Auld SC, Morris L, Gray CM, et al. Establishing a Cohort at High Risk of HIV Infection in South Africa: Challenges and Experiences of the CAPRISA 002 Acute Infection Study. PLoS ONE. 2008;3:e1954. doi: 10.1371/journal.pone.0001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siawaya JF, Roberts T, Babb C, Black G, Golakai HJ, Stanley K, et al. An Evaluation of Commercial Fluorescent Bead-Based Luminex Cytokine Assays. PLoS ONE. 2008;3:e2535. doi: 10.1371/journal.pone.0002535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Columb MO, Sagadai S. Multiple comparisons. Curr Anaesth Crit Care. 2006;17:233–236. [Google Scholar]
- 27.Lee JS, Dixon DO, Kantarjian HM, Keating MJ, Talpaz M. Prognosis of chronic lymphocytic leukaemia: a multivariate regression analysis of 325 untreated patients. Blood. 1987;69:929–936. [PubMed] [Google Scholar]
- 28.Meier A, Alter G, Frahm N, Sidhu H, Li B, Bagchi A, et al. MyD88-Dependent Immune Activation Mediated by Human Immunodeficiency Virus Type 1-Encoded Toll-Like Receptor Ligands. J Virol. 2007;81:8180–8191. doi: 10.1128/JVI.00421-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Centlivre M, Sala M, Wain-Hobson S, Berkhout B. In HIV-1 pathogenesis the die is cast during primary infection. AIDS. 2007;21:1–11. doi: 10.1097/QAD.0b013e3280117f7f. [DOI] [PubMed] [Google Scholar]
- 30.Kubin M, Kamoun M, Trinchieri G. Interleukin 12 Synergizes with B7/CD28 Interaction in Inducing Efficient Proliferation and Cytokine Production of Human T Cells. J Exp Med. 1994;180:211–222. doi: 10.1084/jem.180.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flesch IE, Hess JH, Huang S, Aguet M, Rothe J, Bluethmann H, et al. Early interleukin 12 production by macrophages in response to mycobacterial infection depends on interferon γ and tumor necrosis factor α. J Exp Med. 1995;181:1615–1621. doi: 10.1084/jem.181.5.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gazzinelli RT, Hieny S, Wynn TA, Wolf S, Sher A. Interleukin-12 is required for the T-lymphocyte independent induction of interferon-γ by an intracellular parasite and induces resistance in T-deficient hosts. Proc Natl Acad Sci USA. 1993;90:6115–6119. doi: 10.1073/pnas.90.13.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tripp CS, Wolf SF, Unanue ER. Interleukin 12 and tumor necrosis factor alpha are costimulators of interferon gamma production by natural killer cells in severe combined immunodeficiency mice with listeriosis, and interleukin 10 is a physiologic antagonist. Proc Natl Acad Sci USA. 1993;90:3725–3729. doi: 10.1073/pnas.90.8.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manetti R, Gerosa F, Giudizi MG, Biagiotti R, Parronchi P, Piccinni M, et al. Interleukin 12 induces stable priming for interferon-γ (IFN-γ) production during differentiation of human T helper (Th) cells and transient IFN-γ production in established Th2 cell clones. J Exp Med. 1994;179:1273–1283. doi: 10.1084/jem.179.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manetti R, Parronchi P, Giudizi MG, Piccinni MP, Maggi E, Trinchieri G, et al. Natural killer cell stimulator-y factor (NKSF/IL-12) induces Thl-type specific immune responses and inhibits the development of IL-4 producing Th cells. J Exp Med. 1993;177:1199–1204. doi: 10.1084/jem.177.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-γ. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 37.Ullum H, Lerri AC, Bendtzen K, Victor J, Gùtzsche PC, Philips AN, et al. Low production of interferon-γ is related to disease progression in HIV infection: evidence from a cohort of 347 HIV-infected individuals. AIDS Res Hum Retrovir. 1997;13:1039–1046. doi: 10.1089/aid.1997.13.1039. [DOI] [PubMed] [Google Scholar]
- 38.Bailer RT, Holloway A, Sun J, Margolick JB, Martin M, Kostman J, et al. IL-13 and IFN-g Secretion by Activated T Cells in HIV-1 Infection Associated with Viral Suppression and a Lack of Disease Progression. J Immunol. 1999;162:7534–7542. [PubMed] [Google Scholar]
- 39.Ansari AA, Mayne AE, Sundstrom JB, Bostik P, Grimm B, Altman JD, et al. Administration of recombinant rhesus interleukin-12 during acute simian immunodeficiency virus (SIV) infection leads to decreased viral loads associated with prolonged survival in SIVmac251-infected rhesus macaques. J Virol. 2002;76:1731–174. doi: 10.1128/JVI.76.4.1731-1743.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nathan CF, Murray HW, Wiebe ME, Rubin BY. Identification of interferon-γ as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J Exp Med. 1983;158:670–689. doi: 10.1084/jem.158.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metcalf D. The molecular biology and functions of the granulocyte-macrophage colony-stimulating factors. Blood. 1986;67:257–267. [PubMed] [Google Scholar]
- 42.Jana M, Dasgupta S, Saha RN, Liu X, Pahan K. Induction of tumor necrosis factor-alpha (TNF-alpha) by interleukin-12 p40 monomer and homodimer in microglia and macrophages. J Neurochem. 2003;86:519–528. doi: 10.1046/j.1471-4159.2003.01864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooper AM, Khader SA. IL-12p40: an inherently agonistic cytokine. Trends Immunol. 2007;28:33–38. doi: 10.1016/j.it.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 44.Miossec P, Yu CL. Lymphocyte chemotactic activity of human interleukin 1. J Immunol. 1984;133:2007–2011. [PubMed] [Google Scholar]
- 45.Gerber BO, Zanni MP, Uguccioni M, Loetscher M, Mackay CR, Pichler WJ, et al. Functional expression of the eotaxin receptor CCR3 in T lymphocytes co-localizing with eosinophils. Curr Biol. 1997;7:836–843. doi: 10.1016/s0960-9822(06)00371-x. [DOI] [PubMed] [Google Scholar]
- 46.Niu J, Li Z, Peng B, Chiao PJ. Identification of an Auto-regulatory Feedback Pathway Involving IL-1α in Induction of Constitutive NF-κB Activation in Pancreatic Cancer Cells. J Biol Chem. 2004;279:16452–16462. doi: 10.1074/jbc.M309789200. [DOI] [PubMed] [Google Scholar]
- 47.Fry TJ, Mackall CL. The many faces of IL-7: From lymphopoiesis to peripheral T cell maintenance. J Immunol. 2005;174:6571–6576. doi: 10.4049/jimmunol.174.11.6571. [DOI] [PubMed] [Google Scholar]
- 48.Picker LJ, Reed-Inderbitzin EF, Hagen SI, Edgar JB, Hansen SG, Legasse A, et al. IL-15 induces CD4+ effector memory T cell production and tissue emigration in nonhuman primates. J Clin Invest. 2006;116:1514–1524. doi: 10.1172/JCI27564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okoye A, Meier-Schellersheim M, Brenchley JM, Hagen SI, Walker JM, Rohankhedkar M, et al. Progressive CD4+ central-memory T cell decline results in CD4+ effector-memory insufficiency and overt disease in chronic SIV infection. J Exp Med. 2007;204:2171–2185. doi: 10.1084/jem.20070567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Napolitano LA, Grant RM, Deeks SG, Schmidt D, De Rosa SC, Herzenberg LA, et al. Increased production of IL-7 accompanies HIV-1–mediated T-cell depletion: implications for T-cell homeostasis. Nat Med. 2001;7:73–79. doi: 10.1038/83381. [DOI] [PubMed] [Google Scholar]
- 51.Geginat J, Sallusto F, Lanzavecchia A. Cytokine-driven proliferation and differentiation of human naive, central memory, and effector memory CD4(+) T cells. J Exp Med. 2001;194:1711–1719. doi: 10.1084/jem.194.12.1711. 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eberly MD, Kader M, Hassan W, Rogers KA, Zhou J, Mueller YM, et al. Increased IL-15 Production Is Associated with Higher Susceptibility of Memory CD4 T Cells to Simian Immunodeficiency Virus during Acute Infection. J Immunol. 2009;182:1439–1448. doi: 10.4049/jimmunol.182.3.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burgers WA, Riou C, Mlotshwa M, de Assis Rosa D, Maenetje P, Brenchley JM, et al. Association of HIV-specific and total CD8+ T memory phenotypes in subtype C HIV-1 infection with viral set point. J Immunol. 2009;182:4751–61. doi: 10.4049/jimmunol.0803801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kacani L, Sprinzl GM, Erdei A, Dierich MP. Interleukin-15 enhances HIV-1-driven polyclonal B-cell response in vitro. Exp Clin Immunogenet. 1999;16:162–172. doi: 10.1159/000019108. [DOI] [PubMed] [Google Scholar]
- 55.Levesque MC, Moody MA, Hwang KK, Marshall DJ, Whitesides JF, Amos JD, et al. Polyclonal B Cell Differentiation and Loss of Gastrointestinal Tract Germinal Centers in the Earliest Stages of HIV-1 Infection. PLOS Med. 2009;6:e1000107. doi: 10.1371/journal.pmed.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akridge RE, Oyafuso LK, Reed SG. IL-10 is induced during HIV-1 infection and is capable of decreasing viral replication in human macrophages. J Immunol. 1994;153:5782–5789. [PubMed] [Google Scholar]
- 57.Brooks DG, Trifilo MJ, Edelmann KH, Teyton L, McGavern DB, Oldstone MB. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12:1301–1309. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ejrnaes M, Filippi CM, Martinic MM, Ling EM, Togher LM, Crotty S, et al. Resolution of a chronic viral infection after interleukin-10 receptor blockade. J Exp Med. 2006;203:2461–2472. doi: 10.1084/jem.20061462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D'Andrea A, Aste-Amezaga M, Valiante NM, Ma X, Kubin M, Trinchieri G. Interleukin 10 (IL-10) Inhibits Human Lymphocyte Interferon-γ-Production by Suppressing Natural Killer Cell Stimulatory Factor/IL-12 Synthesis in Accessory Cells. J Exp Med. 1993;178:1041–1048. doi: 10.1084/jem.178.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stylianou E, Aukrust P, Kvale D, Muller F, SS Froland. IL-10 in HIV infection: increasing serum IL-10 levels with disease progression – down-regulatory effect of potent anti-retroviral therapy. Clin Exp Immunol. 1999;116:115–120. doi: 10.1046/j.1365-2249.1999.00865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Couper KN, Blount DG, Riley EM. IL-10: The Master Regulator of Immunity to Infection. J Immunol. 2008;180:5771–5777. doi: 10.4049/jimmunol.180.9.5771. [DOI] [PubMed] [Google Scholar]
- 62.Estes JD, Li Q, Reynolds MR, Wietgrefe S, Duan L, Schacker T, et al. Premature Induction of Immunosuppressive Regulatory T Cell Response during Acute Simian Immunodeficiency Virus Infection. J Infect Dis. 2006;193:703–712. doi: 10.1086/500368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.