Abstract
Coreceptor expression is tightly regulated during thymocyte development. Deletion of specific Cd8 enhancers leads to variegated expression of CD8αβ heterodimers in double-positive (DP) thymocytes. Here we show CD8 variegation was correlated with an epigenetic “off” state, linking Cd8 enhancer function with chromatin remodeling of the adjacent genes Cd8a and Cd8b1 (Cd8). We show the zinc finger protein MAZR, encoded by the Zfp278 gene, bound the Cd8 enhancer and interacted with the corepressor N-CoR complex in double-negative thymocytes. MAZR was down-regulated in DP and single-positive CD8+ thymocytes. Enforced expression of MAZR led to impaired Cd8 activation and variegated CD8 expression. Our results demonstrate epigenetic control of the Cd8 gene loci and identify MAZR as an important regulator of Cd8 gene expression.
Cell fate specifications during T lymphocyte differentiation are linked with spatial and temporal control of the expression of developmentally regulated genes1. Previous work has shown that epigenetic modifications are an important regulatory level to control gene expression2,3. Cd4 and Cd8 (i.e. Cd8a and Cd8b1) coreceptor gene expression have also been suggested to be epigenetically regulated4. For CD4, a T cell-specific enhancer located 13 kb upstream of the Cd4 promoter directs expression in both helper and cytotoxic T cells5, while a silencer element located in the first intron of Cd4 provides subset specificity by silencing the transcription of Cd4 in double-negative (DN) thymocytes and mature CD8+ T cells6-8. Conditional gene targeting in mice showed that the silencer element is required for establishment but not maintenance of Cd4 silencing, suggesting that the silencing of CD4 expression in mature CD8+ T cells is regulated at the epigenetic level9,10.
For CD8 (usually consisting of CD8α and CD8β heterodimers on thymic-derived T cells) at least five distinct Cd8 cis-acting elements (E8I – E8V) were identified in transgenic reporter expression assays that individually or in combination directed expression in the T cell lineage11. These studies indicate a complex regulatory network of developmental stage- and subset-specific cis-regulatory elements to achieve proper expression of the linked Cd8a and Cd8b1 genes during T cell development. To analyze the various cis-elements in more detail, mice with single and combinatorial germline deletion analysis of enhancers were generated12-15. Individual deletions of either enhancer E8I or E8II did not alter the expression pattern of CD8 during T cell development, while deletion of both enhancers impaired the expression of CD8 in double-positive (DP) thymocytes14. A population of “CD8-negative” DP thymocytes appeared that was indistinguishable from DP thymocytes by the expression of other surface markers and functional phenotype. The concurrent appearance of “CD8-negative” DP thymocytes in addition to bona fide DP cells indicated variegation of CD8 expression in the absence of E8I and E8II. Additional studies have shown that a similar phenotype is observed upon deletion of the Cd8 cis-element designated cluster II (E8V), by the combined deletion of E8II and E8III, or by altering the activity of either the BAF (Brahma-related gene/Brahma-associated factor) chromatin remodeling complex, or of members of the Ikaros family15-18. Collectively, these results suggest that Cd8a and Cd8b1 gene expression is epigenetically regulated and that some of the Cd8 cis-regulatory elements may function as recruitment sites for factors involved in chromatin remodeling to facilitate activation of the Cd8a and Cd8b1 gene loci during the DN to DP transition of T cell development.
To investigate whether deletion of E8I and E8II leads to alteration of chromatin remodeling during T cell development, we have compared the epigenetic state of the Cd8a and Cd8b1 gene loci in sorted “CD8-negative” and bona fide DP thymocytes isolated from E8I,E8II doubly deficient. Chromatin immunoprecipitation (ChIP) assays and DNA methylation analysis revealed that “CD8-negative” DP thymocytes have epigenetic modifications at the Cd8a and Cd8b1 gene complex that correspond to an “off” state of chromatin. Variegation of CD8 expression in DP thymocytes could be partially reverted by intercrossing E8I,E8II doubly deficient mice with conditional DNA methyltransferase 1 (Dnmt1)-deficient mice19, further indicating a partial “epigenetic block” of CD8 expression due to the absence of cis-regulatory elements. To study the molecular basis of the epigenetic regulation of CD8 expression in more detail, we searched for factors that bind to enhancer E8II. Yeast one-hybrid screening approaches led to the isolation of the zinc finger protein MAZR (Myc-associated Zn finger related factor20). MAZR was highly expressed in DN thymocytes and its expression was down-modulated with progressive thymocyte differentiation. We could show that Cd8 enhancers recruit MAZR to the Cd8 gene loci. MAZR interacts via its N-terminal domain, which contains a BTB-POZ motif (bric-a-bric, tramtrack, broad complex; poxvirus zinc finger) with nuclear receptor corepressor (N-CoR) complexes in DN thymocytes. Retroviral-mediated constitutive expression of MAZR during T cell development unexpectedly resulted in variegated expression of CD8 in DP thymocytes, suggesting that MAZR negatively regulates chromatin modification at the Cd8 gene loci required for the activation of CD8 expression. Thus, our results show that CD8 expression is regulated at the epigenetic level and imply that CD8 expression is actively repressed at the DN stage. Furthermore, they indicate that Cd8 enhancers have the potential to function both as negative and positive cis-acting elements by recruiting factors that block or facilitate chromatin modifications required for a transcriptional “on” state, respectively.
RESULTS
Epigenetic changes in E8I,E8II doubly deficient mice
Germline deletion of E8I and E8II leads to the appearance of a population of CD4 SP cells with low to intermediate CD3 (CD3lo) and intermediate CD5 (CD5int) surface expression phenotype characteristic of DP thymocytes14. These cells represent DP thymocytes that fail to express CD8 and therefore we refer to them as “CD8-negative” DP cells. In parallel with the appearance of “CD8-negative” DP cells the percentage of bona fide DP cells was reduced. These results demonstrate variegated expression of CD8 in DP thymocytes and point towards epigenetic regulation of the Cd8 gene complex. They also suggest that E8I and E8II may serve as recruitment sites for chromatin modifying factors during the DN to DP transition of thymocyte development. To investigate whether E8I and E8II function as recruitment elements, we analyzed epigenetic modifications at the Cd8a and Cd8b1 gene complex in CD8-expressing and “CD8-negative” DP thymocytes in E8I,E8II doubly deficient mice. To obtain a pure population of “CD8-negative” DP thymocytes devoid of any mature CD4 SP thymocytes for ChIP experiments, thymocytes from E8I,E8II doubly deficient mice backcrossed onto a Tcra−/− background were used (Fig. 1a, as previously shown14). “CD8-negative” and bona fide DP thymocytes were isolated and analyzed by ChIP experiments for differences in histone modifications, including H3 and H4 acetylation and H3 lysine 4 (H3K4) tri-methylation content at several regions within Cd8 gene loci. The non-expressing Cd8 alleles showed decreased H3 and H4 acetylation at all amplicons analyzed as compared to the expressing Cd8 alleles (Fig. 1b). In contrast, the abundance of acetylated H3 and H4 was not reduced at the Cd4 promoter or Gapdh gene regions. A similar correlation was also observed for the Cd8a and Cd8b1 promoter loci with respect to H3K4 tri-methylation, a modification associated with promoters of transcriptionally active genes21. We also performed ChIP assays with antibodies against di- or tri-methylated histone H3K9, a histone modification associated with transcriptionally inactive chromatin22. However, we found that no Cd8a and Cd8b1 gene complex specific fragments could be precipitated, indicating that H3K9 methylation is not involved in the regulation of CD8 expression during the DN to DP transition (data not shown).
Fig. 1.
Epigenetic changes at the Cd8a and Cd8b1 gene complex in E8I,E8II doubly deficient (Δ1Δ2/Δ1Δ2) thymocytes. (a) Flow cytometric analysis of CD4 and CD8 expression on thymocytes isolated from either +/Δ1Δ2 × Tcra−/− or Δ1Δ2/Δ1Δ2 × Tcra−/− mice as reported previously14. The numbers in the quadrants indicate the percentage of the corresponding thymocyte subpopulations (of total gated thymocytes). Sorting areas for “CD8-negative” (CD8-neg) and bona fide DP thymocytes for ChIP (Fig. 1b) and DNA methylation (Fig. 2) assays are indicated by rectangles. Data are representative of more than 10 mice analyzed. (b) Top: Map of the Cd8 gene loci after the deletion of E8I and E8II. Horizontal arrows indicate the transcriptional orientation of the Cd8 genes. Vertical arrows indicate the localization of DH sites that constitute clusters II, III, and IV39. The horizontal bars indicate genomic fragments used to define enhancers E8III, E8IV, and E8V as previously reported11, and Cd8a (P8a) and Cd8b1 (P8b) promoter regions. The triangles represent loxP sites left after the deletion of E8I and E8II. Only relevant BamHI (B) and EcoRI (E) sites are shown.
Bottom: PCR analysis of chromatin-immunoprecipitated DNA from sorted “CD8-negative” DP (−) and DP (+) thymocytes. Six amplicons denoted by the bars spanning the Cd8 gene cluster are shown. The Cd4 promoter region (P4) and the Gapdh locus were amplified as controls. Input DNA was used undiluted, or at 1:3 and 1:10 dilutions to ensure PCR quantification within a non-saturated amplification range. Relative abundance of histone H3 or H4 acetylation, and H3K4 trimethylation ((Me)3-H3K4) at the corresponding gene loci in CD8 expressing DP thymocytes were set to 1 (=100%). ChIP assays for AcH3 and AcH4 were repeated at least three times with three independent samples, while assays for (Me)3H3K4 were repeated twice.
As thymocytes develop from the DN to the DP stage progressive CpG demethylation of DNA at the Cd8 gene loci occurs concomitantly with the initiation of co-receptor expression23,24. This finding suggests that CpG demethylation is one of the epigenetic changes required for (or correlating with) the proper initiation of CD8 expression. To determine whether expressing and non-expressing Cd8 alleles display differences in CpG methylation, DNA from “CD8-negative” and bona fide DP thymocytes from E8I,E8II × Tcra−/− mice was isolated. DNA methylation was assayed by treating the DNA with sodium bisulfite followed by PCR amplification as described previously19. While CD8-expressing DP thymocytes contained clusters of non-methylated CpG dinucleotides upstream of the Cd8a and Cd8b1 promoter proximal regions, the respective CpG clusters were almost completely methylated in “CD8-negative” DP thymocytes (Fig. 2).
Fig. 2.
DNA methylation of the promoter, exon, and intron regions of the Cd8a and Cd8b1 genes. Genomic DNA from DP and “CD8-negative” DP thymocytes was treated with sodium bisulfite and amplified by PCR as indicated by black thin lines (after ref. 23,24). Positions are indicated relative to the initation codon. Individual PCR products were cloned and sequenced, each line representing one clone. Four clones of each of three independently prepared thymocytes for each group were sequenced. Unmethylated CpG sites are indicated by open circles, whereas methylated CpG sites are indicated by filled circles.
Loss of Dnmt1 leads to partial reactivation of CD8
To investigate whether the failure to activate the Cd8 gene loci properly in E8I,E8II doubly deficient mice in “CD8-negative” DP thymocytes was caused by the sustained CpG methylation of the Cd8a and Cd8b1 gene complex, E8I,E8II doubly deficient mice were intercrossed with conditional Dnmt1-deficient mice. These mice have a “floxed” Dnmt1 gene and express Cre-recombinase under the tissue-specific Lck proximal promoter, which becomes active early at DN2 stage of thymocyte development19. Deletion of Dnmt1 in DN thymocytes could partially rescue the variegated expression phenotype of E8I,E8II doubly deficient mice as revealed by the reduction of “CD8-negative” DP cells (Supplementary Fig. 1 online). This result highlights the role of Cd8 enhancers in mediating Cd8 gene loci-specific chromatin modifications.
MAZR binds to E8II in vivo
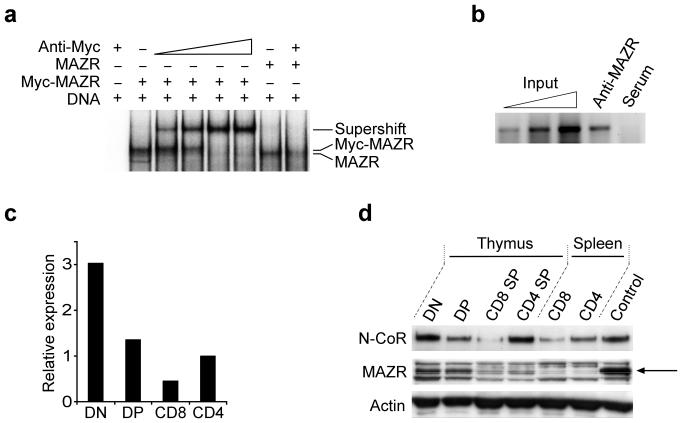
The results described above indicated that E8I and E8II may serve as recruitment sites for factors that regulate chromatin. By using a combination of bioinformatics approaches and luciferase reporter gene transactivation assays, a 154 bp E8II core enhancer containing two regulatory elements (RE) was identified within the 4.4 kb genomic E8II fragment (I.B. and W.E., manuscript in preparation). Deletion of RE-1 (nucleotides 123-154) resulted in a complete loss of enhancer activity (data not shown). Yeast one-hybrid screens (YOH) using RE-1 as bait and a GAL4-activation domain (AD) 1200M murine thymoma cell line cDNA fusion library as prey led to the isolation of a cDNA fragment that encoded the murine zinc finger protein MAZR. MAZR is a 641 amino acid long transcription factor that is encoded by the Zfp278 gene. In addition to the N-terminal BTB-domain it contains seven C2H2 zinc fingers20. The YOH clone insert encoded the first 409 aa including the BTB-domain and three complete zinc finger domains (Supplementary Fig. 2a online). MAZR binds to guanine-rich sequences20, which were also present within RE-1 (Supplementary Fig. 2b online). Binding of MAZR to RE-1 was confirmed in vitro by electrophoretic mobility shift assays (EMSA) using a 5′ Myc-tagged version of MAZR (Fig. 3a).
Fig. 3.
Enhancer E8II binding properties and expression pattern of MAZR. (a) Electrophoretic mobility shift assay (EMSA) showing in vitro binding of MAZR or Myc-MAZR to E8II RE-1 (Supplementary Fig. 2b online). Open wedge indicates addition of increasing amounts of anti-Myc. Data are representative of two independent experiments. (b) Chromatin immunoprecipitation (ChIP) assay showing in vivo interaction of MAZR with RE-1 in DN thymocytes. Primer pairs used for PCR correspond to amplicon 6 (Supplementary Table 2 online). Input DNA was used undiluted, or at 1:3 and 1:10 dilutions. Data are representative of seven independent experiments (c) Real-time qPCR quantification of MAZR expression in various thymocyte subsets. MAZR (Zfp278) transcript abundance was normalized to that of Hprt1 transcripts. (d) Immunoblot analysis showing MAZR and N-CoR expression levels in sorted thymocyte subpopulations and peripheral splenic CD8+ and CD4+ T cells. Whole cell lysates from 3 × 106 cells were used for each population. Control, protein lysate generated from thymocytes that were transduced with MAZR retrovirus (see Methods). The arrow indicates the position of MAZR. Actin protein abundance was used to confirm equal loading. Data in c and d are representative of two independent experiments.
To determine whether MAZR binds also to E8II in vivo, ChIP experiments were performed. Therefore, polyclonal rabbit antibodies were generated against a glutathione S-transferase (GST)-MAZR fusion protein (aa 146-293) (see Supplementary Fig. 3 online for anti-MAZR serum specificity). Since MAZR expression was highest in DN thymocytes (Fig. 3c and Fig. 3d), anti-MAZR serum was used to immunoprecipitate MAZR–DNA complexes from formaldehyde-treated DN thymocytes. MAZR–DNA complex precipitations with the anti-MAZR serum led to a selective enrichment of E8II sequences compared to mock precipitations (pre-immune serum), indicating in vivo binding of MAZR to E8II (Fig. 3b).
Downregulation of MAZR during T cell development
MAZR is expressed in multiple tissues including fetal liver, bone marrow and thymus20. To determine the expression pattern of MAZR in various thymocyte subsets, real-time quantitative PCR (qPCR) analysis and immunoblot analysis was performed on sorted DN, DP, CD8 SP and CD4 SP thymocyte populations. MAZR was expressed more abundantly in DN thymocytes as compared to DP cells (Fig. 3c and Fig. 3d). MAZR expression was further reduced in CD8 SP thymocytes. Almost no MAZR expression was detected in peripheral T cells (Fig. 3d).
MAZR induces variegation of CD8 expression
MAZR was originally isolated as an interacting protein of the B cell and neuronal transcriptional repressor Bach2. MAZR can also transactivate the Myc and Fgf4 promoter in transient reporter expression assays20. Since MAZR interacted with E8II, we investigated whether it regulates Cd8a and Cd8b1 gene expression. Therefore, MAZR was ectopically expressed in thymocytes using retroviral-mediated gene transduction of hematopoietic stem cells followed by bone marrow (BM) transplantation into irradiated recipients (see Supplementary Fig. 4a online for the constructs used). Enforced expression of MAZR resulted in a small increase in the percentage of CD4 SP thymocytes leading to an approximately two-fold increase of the CD4/CD8 ratios in GFP+ thymocytes as compared to GFP− thymocytes (2.0 ± 0.7, n = 5) (Fig. 4b and data not shown). In contrast, GFP+ thymocytes transduced with the parental MIG-R vector had no increase in the percentage of CD4 SP thymocytes, resulting in similar CD4/CD8 ratios in GFP+ and GFP− thymocytes (1.1 ± 0.1, n = 5).
Fig. 4.
Forced expression of MAZR in thymocytes induces variegation of CD8 expression. (a) Two-color flow cytometry of MAZR transduced wild-type thymocytes. Representative dot plots show CD4 versus CD8 expression. Numbers in the dot plot quadrants indicate the percentage of the corresponding thymocyte subpopulations of total gated GFP− (top) and GFP+ thymocytes (bottom), respectively. (b) Expression of CD3 and CD5 on CD4 SP (left), DP (middle) and CD8 SP (right) thymocytes (as defined by the gates in Fig. 4a). Top, expression of CD3 (left) and CD5 (right) in MIG-R-transduced thymocytes. Bottom, expression of CD3 (left) and CD5 (right) in MAZR-transduced thymocytes. Expression patterns for GFP+ and GFP− populations are indicated. Numbers in the CD4 SP-gated histograms indicate the percentage of cells within the indicated regions (i.e. CD3lo and CD5int, respectively) of total gated thymocytes. Data are representative of at least 5 experiments.
To investigate the increase in the percentage of CD4 SP thymocytes in more detail, the developmental stage of MAZR-transduced CD4 SP cells was determined. Mature CD4 SP thymocytes have high surface expression of CD3 and CD5, while immature DP thymocytes are CD3loCD5int 25, a phenotype that was also observed in MIG-R transduced CD4 SP and DP thymocytes, respectively (Fig. 4b). In contrast, the GFP+ CD4 SP population displayed two distinct cell subsets in thymocytes expressing MAZR constitutively. In addition to mature CD3hiCD5hi CD4 SP cells, a CD3loCD5int CD4+CD8− subset was detected in all mice reconstituted with the MAZR-transduced BM (Fig. 4b and Fig. 5a, left panel). Furthermore, CD4+CD3lo cells developed in chimeric Rag2−/− mice reconstituted with MAZR-transduced Tcra−/− BM (Supplementary Fig. 5 online). This finding suggests that these cells develop before the onset of positive selection. The appearance of the “CD8-negative” DP subset was more pronounced when gated only on cells expressing high amounts of GFP, which correlated with higher MAZR expression (Fig. 5a). This result demonstrated a dose-dependent influence of MAZR on the expression of CD8. The phenotype in MAZR expressing thymocytes is thus reminiscent of the appearance of “CD8-negative” DP thymocytes and the CD8 variegation phenotype observed in E8I,E8II doubly deficient mice or in cluster II (E8V)-deficient mice, thus linking MAZR to the regulation of CD8 expression.
Fig. 5.
MAZR-induced variegation in wild-type, E8I- and E8II-deficient thymocytes (a) Representative GFP expression profile of thymocytes transduced with MIG-R and MAZR. Gating areas for GFP− (−) and GFP+ (+) cells (thin line rectangles), and high (hi) GFP-expressing cells (thick line rectangles) are shown. Scatter plots show the percentage of CD3lo (top) and CD5int (bottom) cells within the CD4 SP subset of MIG-R and MAZR transduced wild-type (wt; left), E8I-deficient (Δ1/Δ1; middle) or E8II-deficient (Δ2/Δ2; right) thymocytes, respectively. Each circle represents one mouse. Horizontal bars indicate average value. (b) Left, representative histograms of GFP+ thymocytes indicating gating areas for CD4+CD8-CD3lo, CD4+CD8-CD3hi and DP thymocytes, respectively, that were used to determine expression as shown in the right panel. Right, expression of the zinc finger transcription factor Th-POK in MAZR-trasnduced thymocytes. Semi-quantitative RT-PCR of Zbtb7b (Th-POK) mRNA levels in CD4+CD3lo, DP, and CD4+CD3hi populations of MAZR-transduced E8I-deficient thymocytes. Open wedges indicate undiluted sample, 1:3, and 1:10 dilutions, respectively. M, marker; −, RT(−) control. Hprt1 mRNA was used as quantification control.
Variegation of CD8 in the absence of E8I or E8II
Enforced expression of MAZR might negatively regulate Cd8a and Cd8b1 gene expression through interaction with E8II. In contrast to combined deletions, deletion of either E8I or E8II alone had only a marginal effect on the expression of CD8 during thymocyte development12-14. Thus, a potential repressive role of MAZR acting at E8II could be counterbalanced by compensatory factors acting at E8I. To test this possibility, E8I-deficient BM cells were transduced with MAZR-expressing retrovirus and transplanted into irradiated E8I-deficient mice. The percentage of “CD8-negative” DP thymocytes (CD4+CD3loCD5int) upon forced MAZR expression increased substantially in E8I-deficient thymocytes as compared to wild-type thymocytes (Fig. 5a, middle). To confirm that the increased CD4+CD3lo cells in MAZR-transduced E8I-deficient BM chimeric mice had a DP phenotype (and were not mature CD4 SP thymocytes that for unknown reasons are CD3lo and CD5int), CD4+CD3lo, DP and CD4+CD3hi cells were sorted and the expression of Th-POK (also known as cKrox) was determined. Th-POK is induced in developing CD4 SP thymocytes upon T cell receptor stimulation of DP cells and is a master regulator for CD4 lineage differentiation26,27. While Th-POK was expressed in CD4+CD3hi cells, it was not expressed in DP or in CD4+CD3lo cells (Fig. 5b). This finding indicated that the CD4+CD3lo cells are not mature CD4 SP cells and thus showed again the presence of “CD8-negative” DP cells upon forced expression of MAZR.
To determine whether E8II is the only recruitment site and therefore required for MAZR function, E8II-deficient BM cells were transduced with MAZR retrovirus and transplanted into irradiated E8II-deficient mice. Increased variegation was also observed upon forced expression of MAZR in E8II-deficient thymocytes (Fig. 5a, right). This finding indicates that in addition to E8II other Cd8 cis-regulatory elements were able to recruit MAZR to the Cd8a and Cd8b1 gene complex.
Multiple MAZR binding sites at the Cd8 gene loci
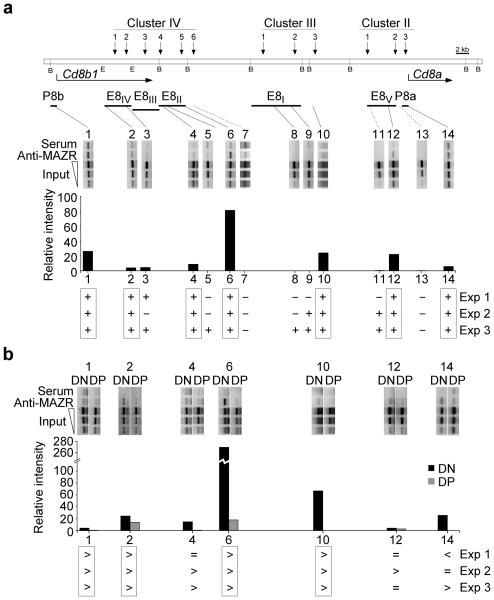
Nucleotide sequence analysis based on the MAZR binding motif within E8II RE-1 and the published MAZR consensus binding site revealed at least 11 additional potential binding sites within the five known Cd8 cis-regulatory elements. ChIP experiments were performed to determine whether MAZR interacts with these sites. MAZR–DNA complex precipitations from DN thymocytes with the polyclonal rabbit anti-mouse MAZR serum led to a selective enrichment of at least 7 of the 11 PCR fragments containing the binding sites compared to fragments without MAZR binding sites or to mock (pre-immune serum) precipitations (Fig. 6a). Thus, MAZR is recruited to multiple sites within the Cd8 gene loci in DN thymocytes. To test whether MAZR was recruited to these sites in DP cells, comparative ChIP experiments between DN and DP thymocytes were performed. At least 4 of the 7 amplicons were less amplified in DP thymocytes as compared to DN cells, indicating reduced MAZR binding at the Cd8a and Cd8b1 gene complex in DP cells (Fig. 6b).
Fig. 6.
MAZR is recruited to multiple sites within the Cd8a and Cd8b1 gene complex. (a) Top, schematic map of the Cd8a and Cd8b1 gene complex40. Horizontal arrows indicate the transcriptional orientation of the Cd8a and Cd8b1 genes. Vertical arrows indicate the localization of DH sites that constitute clusters II, III, and IV39. The horizontal thick bars below DH sites indicate genomic fragments used to define enhancers E8I, E8II, E8III, E8IV, and E8V as previously reported11, and Cd8a (P8a) and Cd8b1 (P8b) promoter regions. All BamHI (B) but only relevant EcoRI (E) sites are shown. Middle, PCR analysis of chromatin-immunoprecipitated DNA from sorted primary DN thymocytes. 14 amplicons spanning the Cd8 gene cluster are shown. Solid lines indicate PCR amplicons containing potential MAZR binding sites (1, 2, 3, 4, 5, 6, 8, 9, 10, 12 and 14), while dotted lines indicate control PCR amplicons (7, 11 and 13) within or outside of known enhancers that contain no potential MAZR binding sites. (b) Top, PCR analysis of chromatin-immunoprecipitated DNA from sorted primary DN and DP thymocytes. The 7 PCR amplicons (1, 2, 4, 6, 10, 12 and 14) showing MAZR binding in DN thymocytes (Fig. 6a) are shown. The input DNA in a and b was used undiluted, or at 1:3 and 1:10 dilutions to ensure PCR quantification within a non-saturated amplification range. Signal intensities at 1:3 input dilutions were used to calculate relative signal intensities for each amplicon. One representative out of three independent experiments is shown (Experiment 1 for a and experiment 3 for b). The relative densitometric quantification values (arbitrary units) of PCR signal intensities of precipitated DNA relative to the input fraction for each amplicon were calculated as described in the Methods section. The summary for three independent experiments is shown below. Boxed amplicons indicate results observed in all three ChIP experiments. (a) + : indicates binding; − : no binding. (b) > : indicates more MAZR binding in DN cells; < : less MAZR binding in DN cells; = : equal MAZR binding in DN and DP cells.
MAZR interacts with the N-CoR corepressor
The BTB domains of most known transcription factors are able to interact with N-CoR and/or SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) corepressors28-31. This interaction is dependent on critical conserved amino acid residues28,31 that are also present within the BTB domain of MAZR (Supplementary Fig. 4c online). To test whether the BTB domain of MAZR interacts with N-CoR as well, co-immunoprecipitation experiments were performed. HEK293T cells were transfected with N-CoR and either wild-type MAZR or a mutant form of MAZR (MAZRmut) containing point mutations within the conserved amino acid residues of the BTB domain required for corepressor interaction28. MAZR interacted with N-CoR in a BTB domain-dependent manner, since only wild-type but not MAZRmut co-immunoprecipitated with N-CoR (Fig. 7a). More importantly, interaction between MAZR and N-CoR was also be observed in primary DN thymocytes (Fig. 7b). Thus, these findings point to a potential molecular mechanism of how MAZR negatively regulates the expression of CD8.
Fig. 7.
BTB domain-dependent interaction of MAZR and N-CoR. (a) HEK293T cells overexpressing N-CoR and either MAZRmut (lane 1) or MAZR (lane 2) were lysed and N-CoR was immunoprecipitated using anti-N-CoR. Immunoblots were performed using either anti-MAZR serum or anti-N-CoR. Input shows anti-MAZR immunoblot on total cell lysates. (b) Lysates of 107 sorted DN thymocytes (lane 1), 107 total thymocytes (lane 2 and 3) were immunoprecipitated with anti-N-CoR (lanes 1 and 2). For mock IP (lane 3) thymocyte extracts were incubated with protein G beads only. Immunoblots were performed using either anti-MAZR serum or anti-N-CoR. Immunprecipitation data shown with DN and total thymocyte populations are representative of three independent experiments each.
The BTB domain alone does not induce CD8 variegation
MAZR binds to E8II and other Cd8 cis-regulatory elements, suggesting a direct regulation of CD8 expression. However, forced expression of MAZR could also sequester endogenous factors from the Cd8a and Cd8b1 gene complex, resulting in an indirect influence of MAZR on CD8 expression. Thus, additional BM transplantation experiments with truncated forms of MAZR (see Supplementary Fig. 4b online for MAZR truncations) were performed to discriminate whether CD8 expression is directly or indirectly regulated by MAZR. None of the C-terminal truncations of MAZR gave variegated expression of CD8 (Fig. 8), even though the truncated molecules retained their capacity to localize to the nucleus and were still able to bind N-CoR (data not shown). These data indicate that MAZR regulates CD8 expression directly by binding to the Cd8 gene loci.
Fig. 8.
The BTB domain alone is not sufficient to induce variegated expression of CD8 in DP thymocytes. Scatter plot showing percentage of CD3lo cells within GFP−, GFP+, and GFPhi CD4 SP thymocytes of MIG-R, BTBMAZR, and MAZRΔZF transduced wild-type thymocytes. Representative gating areas for GFP expression are as in Fig. 5a. Each circle represents one mouse. Horizontal bars indicate average value.
DISCUSSION
It has been shown previously that the combined deletion of Cd8 enhancers E8I and E8II leads to variegated expression of CD8 in DP thymocytes14. Here we demonstrate that CD8 variegation correlated with an epigenetic “off” state of the Cd8a and Cd8b1 gene complex in cells that failed to express CD8, thus linking Cd8 enhancers to Cd8 gene loci-specific chromatin modification pathways. We further show that the zinc finger protein MAZR interacts in DN thymocytes with CD8 enhancers and with N-CoR corepressor complexes. Expression of MAZR was down-regulated during thymocyte development, and forced expression of MAZR caused impaired activation and variegated expression of CD8 in DP thymocytes. Our results demonstrate epigenetic control of the Cd8 gene loci and identify MAZR as an important regulator of Cd8 gene expression. The results also imply that CD8 is actively repressed at the DN stage, and indicate that CD8 enhancers can function either as negative or positive cis-acting elements by recruiting factors that block or facilitate chromatin modifications required for an epigenetic “on” stage.
Combined deletion of E8I and E8II causes variegation of CD8 expression, resulting in the development of a subset of DP thymocytes that does not express CD8 (ref. 14). These “CD8-negative” DP thymocytes showed lower histone H3 and H4 acetylation over the whole Cd8 gene loci. Furthermore, these cells showed lower H3K4 tri-methylation and sustained DNA methylation at the Cd8a and Cd8b1 promoter region compared to bona fide DP thymocytes. Therefore, all epigenetic patterns analyzed indicate that the Cd8a and Cd8b1 gene complex is locked in an epigenetic “off” state in “CD8-negative” DP thymocytes. However, thymocytes that failed to activate the Cd8 gene loci due to deletion of cis-regulatory elements have not completely lost the potential to express CD8. Conditional deletion of Dnmt1 at the DN2 stage leads to a partial re-activation of CD8 expression and therefore to a reduction of CD8 variegation. It has been shown that deletion of Dnmt1 at the DN2 stage is accompanied by demethylation of the Cd8 gene loci (but not induction of CD8 expression in TCRαβ-lineage DN cells)19. Thus, removal of negative epigenetic marks at the Cd8 gene loci at the proper developmental stage can at least partially rescue impaired CD8 expression in DP thymocytes. This observation highlights the need for Cd8 cis-regulatory elements to recruit factors that facilitate epigenetic modifications required for initiating CD8 expression.
Differences in the epigenetic pattern resulting from deletion of cis-regulatory elements at the Cd8 gene loci imply that E8I and E8II (and probably other cis-regions) recruit factors that induce activating chromatin modifications. These modifications alter the repressive state present at early stage DN thymocytes and therefore it was expected that factors that bind to Cd8 enhancers positively influence the expression of CD8. However the observation that forced expression of MAZR causes variegation and therefore negatively influences CD8 expression indicates a more complex regulatory activity of Cd8 cis-acting elements. Depending on the developmental stage, Cd8 enhancers recruit either negative or positive regulators of CD8 expression. Members of the BAF chromatin remodelling complex and the Ikaros gene family have been shown to act as positive regulators16,18. Recently it has been suggested that Runx3 (recruited by E8I) is another positive acting factor required for CD8 expression after positive selection in CD8 SP thymocytes32. In contrast, MAZR appears to function as a negative regulator of CD8 expression in DN thymocytes. It is not known whether the same or distinct sites within these enhancers are required for the recruitment of positive or negative acting factors.
ChIP assays have shown that MAZR interacts in vivo with several CD8 enhancers in DN thymocytes. Multi-site recruitment has also been observed for members of the Ikaros gene family. Several functional binding sites for Ikaros were shown to overlap with hypersensitivity sites within E8I and E8V 16. Thus, recruitment of either negative or positive acting regulatory factors to multiple sites with the Cd8a and Cd8b1 gene complex seems to ensure the proper and coordinated regulation of CD8 expression. To understand the complex regulatory pattern, it will be important to determine the developmental order of recruitment of MAZR and other factors by using ChIP assays combined with Cd8 gene loci-specific DNA microarray analysis (“ChIP on CHIP”).
We could demonstrate that MAZR interacted in a BTB domain dependent manner with N-CoR upon overexpression in cell lines. More importantly, this interaction was also observed in primary DN thymocytes. N-CoR and the related SMRT corepressor molecules are components of complexes containing proteins involved in epigenetic modifications of chromatin templates (like Sin3A, NURD, SWI/SNF-like BAF and histone deacetylases) and it has been shown that N-CoR is recruited by several transcription factors33,34. The association with histone deacetylases has been suggested as part of the mechanism of how N-CoR interacting transcription factors are able to repress transcription. Thus, enhancer-mediated recruitment of MAZR and MAZR–N-CoR interactions may provide a molecular mechanism by which epigenetic modifications corresponding to a repressive state are introduced at the Cd8 gene loci in DN thymocytes. However, N-CoR-deficient mice have a block at the DN stage of thymocyte development33, indicating that the absence of N-CoR alone is not sufficient for allowing CD8 expression in DN thymocytes. Progression to the DP stage correlates with a down-modulation of both N-CoR and MAZR, leading to a reduction of MAZR at the Cd8a and Cd8b1 gene complex, and thus allowing positive regulators to overcome the repressive state and to activate CD8. If high concentrations of MAZR due to forced expression are maintained, the balance between repressing and activating complexes at the Cd8 gene loci is disturbed, thus leading to variegated expression of CD8 and the appearance of “CD8-negative” DP thymocytes. This hypothesis is supported by the observation that the degree of MAZR-induced CD8 variegation is increased in the absence of E8I or E8II that serve also as recruitment sites for positive acting factors. Nevertheless, one could argue that due to the high amounts of MAZR upon forced expression endogenous factors are sequestered from the Cd8 gene loci, thereby leading to CD8 variegation. However, we consider this scenario unlikely since forced expression of a form of MAZR that lacks the C-terminal zinc fingers (MAZRΔZF) did not induce variegation. This truncated version retained a properly folded BTB domain that could bind N-CoR, localized to the nucleus, and should therefore be able, like the full-length form, to sequester endogenous factors from the Cd8 gene loci. Thus, these data together with result from ChIP assays strongly indicate that MAZR acts directly at the Cd8a and Cd8b1 gene complex.
Finally, our results imply active repression of CD8 expression in DN thymocytes. Recruitment of MAZR and N-CoR corepressor complexes via Cd8 enhancers may therefore prevent CD8 expression in DN cells. Since it has been shown that premature transgenic expression of CD8 in DN2 and DN3 cells causes a partial developmental block leading to reduced numbers of DP thymocytes35, active repression of CD8 expression by MAZR in DN thymocytes may be essential for β-selection and proper T cell development.
Taken together, we propose that CD8 enhancers regulate CD8 expression by controlling the balance between closed and open state of chromatin at the Cd8 gene loci. Dependent on the developmental stage, they recruit either negative- or positive-acting regulators of CD8 expression. MAZR appears to function as a negative regulator of CD8 expression in DN thymocytes. Therefore, the gradual down-regulation of endogenous MAZR expression during thymocyte development may be a prerequisite for the proper initiation of CD8 expression.
METHODS
Cell lines and Culture
HEK293T and Phoenix-E cell lines were grown in Dulbecco's modified Eagle's medium, both supplemented with 10% fetal calf serum (FSC) and antibiotics.
Mice
C57BL/6J were purchased from Harlan. Conditional LckCre-Tg × Dnmt1fl/fl mice, E8I-deficient and E8II-deficient mice, and Tcra−/− × E8I,E8II doubly deficient mice have been described12,14,19. All mice were bred and maintained in the animal facility of the Medical University of Vienna. All animal experiments were done according to protocols approved by the Federal Ministry for Education, Science and Art.
Bisulfite sequencing analysis
The bisulfite-based methylation analysis was performed as described19. The sample was amplified by two sequential PCR reactions. The primers used were described previously19 or are listed in Supplementary Table 1 online. The gel-purified PCR products were cloned into the pCR2.1-TOPO TA cloning vector (Invitrogen), and 12 individual clones were sequenced for each group.
Chromatin immunoprecipitation (ChIP)
107 DP, “CD8-negative” DP, or DN thymocytes (purity of each population >90%) were treated with 0.3% formaldehyde in PBS for 5 min at 37 °C, followed by addition of glycine (0.25 M final concentration). After washing twice with PBS, cells were lysed on ice and sonicated to obtain 0.5-1 kb sheared chromatin fragments. Subsequent ChIP steps were performed according the protocol from Upstate Biotechnology. 5 μl of anti-AcH3 (#06-599; Upstate Biotechnology), 10 μl of anti-AcH4 (#06-866; Upstate Biotechnology), 8 μl of anti-(Me)3H3K4 (#07-473; Upstate Biotechnology), 3 μl anti-tri- or anti-di-methyl-H3K9 (kindly provided by T. Jenuwein, Research Institute of Molecular Pathology, Vienna, Austria) 10 μl of anti-MAZR rabbit serum or rabbit pre-immune serum were used per reaction. The presence of DNA target sequences was assessed by PCR. The PCR was carried out for 3 min at 95 °C, followed by 40 cycles of 30 sec at 95 °C, 45 sec at 58 °C, and 1 min at 72 °C. PCR products were resolved on a 2% agarose gel and quantified using Lumiimager analyzer and Lumianalyst software package (Roche). Signal intensities at 1:3 input dilutions were used to calculate relative signal intensities for each amplicon. The relative values (arbitrary units) for each amplicon are shown below the PCR blots and were calculated according to ref. 32 : ([intensity antibody precipitates] – [intensity mock precipitates])/(intensity input). The primers used for the various PCR reactions are listed in Supplementary Table 2 online.
Retroviral constructs and virus production
Full-length or truncated forms of murine MAZR were cloned into the MIG-R retroviral vector (kindly provided by H. Singh, University of Chicago, IL). High-titer viral preparations were generated by transient transfection of retroviral constructs by standard calcium-phosphate precipitation onto the ecotropic producer cell line Phoenix-E. Viral supernatants were harvested 2 and 3 days post-transfection, filtered through 0.45 μm filters and directly used for the infection of hematopoietic stem cells.
Retroviral infection of hematopoietic stem cells (HSC)
Transduction and transplantation of BM cells was done according to published protocols36 with minor modifications. Briefly, C57BL/6J donor mice were injected intraperitoneally with 5-fluorouracil (5-FU; Sigma) at 10 mg/ml 4 days prior to BM harvest. After red blood cell lysis, BM cells were pre-stimulated for 24h in DMEM (supplemented with 10% FCS, penicillin/streptomycin and L-Glutamine) in the presence of recombinant murine SCF (5 U/ml), IL-6 (104 U/ml) and IL-3 (6 U/ml) (Peprotech).
For retroviral transductions, 1 - 2 × 107 activated BM cells were transferred to a non-TC 10 cm plate (Sterilin) pre-coated with RetroNectin® (Takara INC). Infection was done according to the manufacturers instructions by incubation with 7 ml of viral supernatant supplemented with SCF, IL-3 and IL-6 at concentrations indicated above for 2-3 rounds. BM cells were harvested 48 h post infection, washed three times with PBS and injected at 0.5 - 3 × 106 cells/ mouse into the tail veins of lethally irradiated C57BL/6J recipient mice. γ-irradiation was performed at 8.25 Gy using a Hille TH-150 instrument. Mice were given acidified drinking water supplemented with 25 μg/ml neomycin and 25,000 U/ml polymyxin B sulfate for 1 week.
Isolation of nuclear protein extracts
Nuclear protein extracts were prepared as described37. Briefly, HEK293T cells (total of 1 × 107 cells) were harvested and washed once with PBS. The cells were pelleted (280 × g for 5 min) and resuspended in 1 ml of cold buffer A (10 mM HEPES; pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol (DTT), 0.2 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitors; Roche). Following a 10 min incubation on ice, 50 μl of 10% NP-40 was added, and the cells were lysed for additional 10 min on ice. The nuclei were pelleted (280 × g for 4 min) and resuspended in 100 μl buffer C (20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 420 mM NaCl, 25 % glycerol, 0.2 mM EDTA, 0.5 mM DTT and 0.2 mM PMSF) for the extraction of nuclear proteins. After 30 min incubation on ice, the extract was pelleted at 16,000 × g in a microcentrifuge for 3 min at 4 °C. The supernatant was snap frozen in liquid N2 and stored at −80 °C until use.
Electrophoretic mobility shift assay (EMSA)
Duplex oligonucleotides were labeled with T4 polynucleotide kinase and γ-[32P]-ATP. Labeled oligonucleotides (2.5 × 104 c.p.m.) were incubated in the binding reaction buffer (10 mM Tris-HCl; pH 7.5, 50 mM NaCl, 0.5 mM EDTA, 0.5 mM DTT, 1 mM MgCl2, 4 % glycerol, 1μg bovine serum albumin (BSA), 1 μg poly dI:dC) with 10 μg of nuclear protein extract HEK293T cell line transfected with Myc-tagged MAZR construct. MAZR cDNA was cloned in-frame into EcoRI restriction site of pCDNA3-Myc plasmid (kindly provided by R. de Martin, Medical University of Vienna, Austria). The binding reaction was carried out in a total volume of 15 μl for 30 min at 30°C. For supershift experiments, 0.1, 0.5, 1 or 3 μg of anti-Myc monoclonal antibody (OP10L, Calbiochem) was added to the reactions, respectively. Protein-DNA complexes were separated on a 4% non-denaturing polyacrylamide/bisacrylamide (19:1) gel (0.5 × TBE) at 10 mA for 2-3 h at 4 °C. Afterwards the gel was dried and exposed to a Storage PhosphoImager Screen (Molecular Dynamics). The following radiolabeled duplex oligonucleotide was used for EMSA (only the sequence of one oligonucleotide strand is indicated: 5′-AGGTGTGCTGCCCCCAGGTCCACCCGCAGGAGGAGAGGGGGCT-3′.
Generation of a rabbit anti-MAZR serum
A MAZR fragment (corresponding to aa 146-293) was cloned into the pGEX-5X-1 vector and the GST-MAZR fusion plasmid was transformed into Escherichia coli strain BL-21. 800 μg of GST-MAZR146-293 fusion protein was batch purified using glutathione sepharose 4B according to manufacturer's instructions (Amersham Biosciences). The GST fusion protein was used for rabbit immunizations.
Flow cytometric analysis and antibodies
Thymus was removed from euthanized animals and placed into 60 mm tissue culture dishes containing staining buffer (phosphate-buffered saline supplemented with 2% FCS and 0.1% sodium azide). Single cell suspensions were made by passing the tissue through a 70 μm nylon cell strainer. The cell suspensions were washed once with staining buffer and 0.5 - 5 × 105 cells were incubated on ice with Fc-block (Pharmingen) for 5 min and subsequently with the appropriate antibodies for 30 min. Afterwards, the cells were washed once with staining buffer and analyzed or incubated with secondary antibodies on ice for 30 min. The following antibodies were used for the stainings: PE-anti-mCD8α (CT-CD8a), TC-anti-mCD4 (CT-CD4), Cy5-anti-mCD3ε (500A2) and Cy5-anti-mCD8α (CT-CD8a) from Caltag, PE-anti-mCD5 (53-7.3) from Pharmingen. Cells were analyzed using Becton Dickinson FACSCalibur flow cytometer and Cellquest Pro software (Becton Dickinson).
Cell sorting
Thymocytes were stained with FITC-anti-CD8, TC-anti-CD4, and PE-anti-CD5 antibodies to sort CD4−CD8− (DN), CD4+CD8+ (DP), CD4+ (CD4 SP), and CD8+CD5hi (CD8 SP) thymocyte subpopulations using FACSAria cell sorter (Becton Dickinson). For the isolation of peripheral CD4+ and CD8+ T cells, splenocytes were briefly treated with red blood cell lysis buffer, washed, and stained with FITC-anti-CD4 and FITC-anti-CD8, respectively. Subsequently, the cells were incubated with anti-FITC magnetic beads (Miltenyi) and purified over a MACS LS Column to >97% purity according to the manufacturer's instructions.
Yeast one-hybrid screen
The screen was carried out using the Matchmaker One-Hybrid System according to the manufacturer's instructions (Clontech Laboratories). Three tandem repeats of the E8II RE-1 were inserted into the pHISi-1 vector and used as bait to screen for the interacting proteins. The cDNA expression library was cloned into pACT2 vector.
cDNA synthesis, RT-PCR and real-time qPCR
Total RNA was isolated from sorted thymocyte subsets using TRI reagent (Sigma) and cDNA synthesis was performed using random hexamer and oligo-dT primers and SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's protocol. Cycling conditions for RT-PCR were 2 min at 95 °C, followed by 35 cycles of 30 sec at 95 °C, 45 sec at 60 °C, and 1 min at 72°C. Real-time qPCR was carried out using SYBR green and the LightCycler detection system (Roche). The real-time qPCR conditions were 5 sec at 95 °C, 5 sec at 65 °C and 15 sec at 72 °C over a total intervall of 55 cycles. The following primers were used for MAZR: 5′-GCAGGTGCACACTTCTGAGCGAC-3′, 5′-GCATTTCTGGCCTTCTCGGTTACA-3′; for Hprt1: 5'-GATACAGGCCAGACTTTGTTG-3′, 5′-GGTAGGCTGGCCTATAGGCT-3′; for Th-POK: 5′-CACCTTCCACCTCTCTCTGGCTCGA-3′, 5′-TTGCTTGGCGTGGTGTCCCA-3′.
Generation of full-length MAZR and site-directed mutagenesis
Full-length Zfp278 cDNA (encoding for 641 aa) was generated by combining the Zfp278 cDNA sequence from the YOH clone (encoding for 409aa) with a cDNA fragment encoding the C-terminal MAZR sequence. The Zfp278 3′ cDNA sequence was PCR amplified from thymocyte cDNA using 5′-GCAGGTGCACACTTCTGAGCGAC-3′ and 5′-ACAGGTACCGAATTCGTCGACGGGACACAGCATGTCTCACTTC-3′ as primers and inserted as XhoI fragment into the Zfp278 YOH clone. BTBMAZR and MAZRΔZF truncated forms of MAZR were amplified as XhoI fragments by PCR using Myc-tagged Zfp278 cDNA as template. The following primers were used: 5′-AGACTCGAGGCTGCCACCATGGAACAAAAG-3′ with either 5′-AGACTCGAGTCATTAAGATCTCGATGACCGAC-3′ for BTBMAZR or 5′-AGACTCGAGTCATTATGGAAGGATGCCTGC-3′ for MAZRΔZF. Mutations into the BTB domain of MAZR (MAZRmut) were introduced by site directed mutagenesis using two-step overlap PCR. The following primers were used: for 1st step PCR (1) 5′-AGACTCGAGGCTGCCACCATGGAACAAAAG-3′, (2) 5′-GAGTGATTGGTTCAGGTTGTGCGACATCTCCGTAC-3′ and (3) 5′-TCGCACAACCTGAACCAATCACTCAAAAACGGCGG-3′, (4) 5′-ACAGGTACCGAATTCGTCGACGGGACACAGCATGTCTCACTTC-3′; for 2nd step PCR products from the 1st step and primers (1) and (4). Mutated sequences are underlined. The final PCR product was digested with XhoI restriction enzyme and inserted instead of the wild-type sequence.
Transfection, immunoprecipitation and immunoblot analysis
HEK293T cells were seeded on 10 cm tissue culture plates and transfected with 30 μg FLAG-tagged N-CoR (kind gift of K. Jepsen and M. Rosenfeld, University of California, San Diego, CA) and 20 μg of the various MAZR expression plasmids. 48 h post-transfection, the cells were harvested, washed with PBS, and lysed in lysis buffer (20 mM Tris-HCl, pH 8.0, 138 mM NaCl, 10 mM EDTA, 100 mM NaF, 1% NP-40, 10% glycerol) containing protease inhibitors (Roche). Lysates were snap-frozen in liquid N2 and stored at −80 °C until use. After thawing cell debris was removed by 10 min centrifugation, and lysates were incubated with anti-N-CoR with gentle rotation over night at 4 °C. Upon addition of 30% (Vol/Vol) protein G beads (Roche) and 1 h rotation at 4 °C, the immunocomplexes were pelleted, washed, and the beads were resuspended in Laemli buffer. Polyacrylamide gel electrophoresis and immunoblotting was performed according to standard procedures38. N-CoR immunoprecipitations and immunoblots were performed with polyclonal anti-N-CoR (C-20; Santa Cruz). Immunoblot analysis for MAZR and actin expression was performed with rabbit anti-MAZR serum or rabbit anti-actin (A2066, Sigma), respectively.
Supplementary Material
Loss of Dnmt1 leads to a partial reactivation of CD8 expression in variegated DP cells. Left, representative flow-cytometry histograms showing the percentage of CD3lo cells within the gated CD4 SP thymocytes of floxed Dnmt1 alleles on E8I,E8II doubly deficient background (Dnmt1fl/fl × Δ1Δ2/Δ1Δ2). Floxed Dnmt1 alleles were deleted using LckCre transgenic mice. Upper and lower histograms correspond to LckCre− (Cre−) and LckCre+ (Cre+) littermates. Right, scatter plots showing the percentages of CD3lo thymocytes within the gated CD4 SP cells. The genotype of mice is either Dnmt1fl/fl or Dnmt1fl/fl × Δ1Δ2/Δ1Δ2. Each circle represents one mouse. + and − indicates the presence or absence of the LckCre transgene. Horizontal bars indicate average value for each genotype.
(a) Schematic map of MAZR showing the N-terminal BTB domain and the seven C2H2 Zn-finger domains. The clone isolated from Yeast One-Hybrid screens (YOH clone) is shown below. Numbers next to the maps indicate amino acid residues. (b) Sequence of RE-1 used as bait in YOH screens. Putative MAZR binding sites are shown. The approximate location of RE-1 within E8II downstream of the Cd8b1 gene is indicated.
Specificity of the anti-MAZR serum. (a) Total cell lysates of untransfected (−) or Myc-MAZR-expressing (+) HEK293T cells. Immunoblots were performed with anti-Myc (left), anti-MAZR serum (middle), or with preimmune serum (right). Actin protein abundance was used as loading controls. (b) HEK293T cells overexpressing MAZR were lysed and MAZR was immunoprecipitated using anti-MAZR serum. Immunoblots were performed using anti-MAZR serum. Input (left) shows anti-MAZR immunoblot on total cell lysates.
Retroviral constructs used for the transduction of hematopoietic stem cells (a) MIG-R, control retroviral construct expressing only GFP. MAZR, retroviral construct expressing various forms of MAZR and GFP (see Methods). (b) Map of the various MAZR forms expressed. MAZR, full-length form of MAZR. MAZRΔZF, Myc-tagged version of MAZR lacking the C-terminal zinc fingers. BTBMAZR, Myc-tagged BTB domain of MAZR. Numbers indicate amino acid residues. (c) Map showing partial amino acid sequence of the BTB domain of MAZR. Triangles indicate critical residues within BTB domains required for homodimerization and interaction with corepressors28. These critical amino acid residues were exchanged to generate a mutated form of MAZR (MAZRmut). The mutations were: L28S, Q34S, R35L.
“CD8-negative” DP thymocytes develop upon forced expression of MAZR in Tcra−/− thymocytes. (a) Two-color flow cytometry of MIG-R (upper row) and MAZR (lower row) transduced Tcra−/− thymocytes on Rag2−/− background. Representative dot plots show CD4 versus CD8 expression. Numbers in the dot plot quadrants indicate the percentage of the corresponding thymocyte subpopulations of total gated GFP− (left) and GFP+ thymocytes (right), respectively. (b) Scatter plot showing the ratio of GFP+/GFP− thymocytes within the CD4+CD8− quadrant in Supplementary Fig. 5a. Each circle represents one mouse. Horizontal bars indicate average value from the mice analyzed for each construct.
ACKNOWLEDGMENTS
The authors thank V.J. Bardwell for providing N-CoR expression constructs, E. Pfeiffer for help with irradiation, and S. Sakaguchi, P. Kinross and R. Herbst for critical reading of the manuscript. Work supported by the Austrian Science Fund (FWF; research grants P14261, P16708), by the START Program (Project Y-163) of the Austrian Ministry of Education, Science and Culture (BM:BWK), by the K-plus Competence Center Biomolecular Therapeutics, and by the Federal Bank of Austria (OeNB 10530). Support for W.E. was also provided by the Austrian Program for Advanced Research and Technology (APART) of the Austrian Academy of Sciences (W.E.). C.B.W. is supported by grants HD39454 and HD18184 from the National Institutes of Health.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interest.
REFERENCES
- 1.Fisher AG. Cellular identity and lineage choice. Nat Rev Immunol. 2002;2:977–982. doi: 10.1038/nri958. [DOI] [PubMed] [Google Scholar]
- 2.Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- 3.Smale ST. The establishment and maintenance of lymphocyte identity through gene silencing. Nat Immunol. 2003;4:607–615. doi: 10.1038/ni0703-607. [DOI] [PubMed] [Google Scholar]
- 4.Taniuchi I, Ellmeier W, Littman DR. The CD4/CD8 lineage choice: new insights into epigenetic regulation during T cell development. Adv Immunol. 2004;83:55–89. doi: 10.1016/S0065-2776(04)83002-5. [DOI] [PubMed] [Google Scholar]
- 5.Sawada S, Littman DR. Identification and characterization of a T-cell-specific enhancer adjacent to the murine Cd4 gene. Mol Cell Biol. 1991;11:5506–5515. doi: 10.1128/mcb.11.11.5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donda A, Schulz M, Burki K, De Libero G, Uematsu Y. Identification and characterization of a human CD4 silencer. Eur J Immunol. 1996;26:493–500. doi: 10.1002/eji.1830260232. [DOI] [PubMed] [Google Scholar]
- 7.Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates Cd4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 8.Siu G, Wurster AL, Duncan DD, Soliman TM, Hedrick SM. A transcriptional silencer controls the developmental expression of the CD4 gene. EMBO J. 1994;13:3570–3579. doi: 10.1002/j.1460-2075.1994.tb06664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou YR, et al. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat Genet. 2001;29:332–336. doi: 10.1038/ng750. [DOI] [PubMed] [Google Scholar]
- 10.Leung RK, et al. Deletion of the Cd4 silencer element supports a stochastic mechanism of thymocyte lineage commitment. Nat Immunol. 2001;2:1167–1173. doi: 10.1038/ni733. [DOI] [PubMed] [Google Scholar]
- 11.Kioussis D, Ellmeier W. Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nat Rev Immunol. 2002;2:909–919. doi: 10.1038/nri952. [DOI] [PubMed] [Google Scholar]
- 12.Ellmeier W, Sunshine MJ, Losos K, Littman DR. Multiple developmental stage-specific enhancers regulate CD8 expression in developing thymocytes and in thymus-independent T cells. Immunity. 1998;9:485–496. doi: 10.1016/s1074-7613(00)80632-9. [DOI] [PubMed] [Google Scholar]
- 13.Hostert A, et al. Hierarchical interactions of control elements determine CD8α gene expression in subsets of thymocytes and peripheral T cells. Immunity. 1998;9:497–508. doi: 10.1016/s1074-7613(00)80633-0. [DOI] [PubMed] [Google Scholar]
- 14.Ellmeier W, Sunshine MJ, Maschek R, Littman DR. Combined deletion of CD8 locus cis-regulatory elements affects initiation but not maintenance of CD8 expression. Immunity. 2002;16:623–634. doi: 10.1016/s1074-7613(02)00309-6. [DOI] [PubMed] [Google Scholar]
- 15.Garefalaki A, et al. Variegated expression of CD8α resulting from in situ deletion of regulatory sequences. Immunity. 2002;16:635–647. doi: 10.1016/s1074-7613(02)00308-4. [DOI] [PubMed] [Google Scholar]
- 16.Harker N, et al. The CD8A gene locus is regulated by the Ikaros family of proteins. Mol Cell. 2002;10:1403–1415. doi: 10.1016/s1097-2765(02)00711-6. [DOI] [PubMed] [Google Scholar]
- 17.Feik N, et al. Functional and molecular analysis of the double-positive stage-specific CD8 enhancer E8III during thymocyte development. J Immunol. 2005;174:1513–1524. doi: 10.4049/jimmunol.174.3.1513. [DOI] [PubMed] [Google Scholar]
- 18.Chi TH, et al. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature. 2002;418:195–199. doi: 10.1038/nature00876. [DOI] [PubMed] [Google Scholar]
- 19.Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi A, et al. A combinatorial code for gene expression generated by transcription factor Bach2 and MAZR (MAZ-related factor) through the BTB/POZ domain. Mol Cell Biol. 2000;20:1733–1746. doi: 10.1128/mcb.20.5.1733-1746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 23.Carbone AM, Marrack P, Kappler JW. Demethylated Cd8 gene in CD4+ T cells suggests that CD4+ cells develop from CD8+ precursors. Science. 1988;242:1174–1176. doi: 10.1126/science.2460926. [DOI] [PubMed] [Google Scholar]
- 24.Hamerman JA, Page ST, Pullen AM. Distinct methylation states of the Cd8b gene in peripheral T cells and intraepithelial lymphocytes. J Immunol. 1997;159:1240–1246. [PubMed] [Google Scholar]
- 25.van Meerwijk JP, Germain RN. Development of mature CD8+ thymocytes: selection rather than instruction? Science. 1993;261:911–915. doi: 10.1126/science.8102208. [DOI] [PubMed] [Google Scholar]
- 26.He X, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 27.Sun G, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 28.Huynh KD, Bardwell VJ. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene. 1998;17:2473–2484. doi: 10.1038/sj.onc.1202197. [DOI] [PubMed] [Google Scholar]
- 29.Polo JM, et al. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med. 2004;10:1329–1335. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- 30.Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell. 2003;12:723–734. doi: 10.1016/j.molcel.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Melnick A, et al. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol Cell Biol. 2002;22:1804–1818. doi: 10.1128/MCB.22.6.1804-1818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato T, et al. Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity. 2005;22:317–328. doi: 10.1016/j.immuni.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 33.Jepsen K, Rosenfeld MG. Biological roles and mechanistic actions of co-repressor complexes. J Cell Sci. 2002;115:689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- 34.Metivier R, et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 35.Wack A, Coles M, Norton T, Hostert A, Kioussis D. Early onset of CD8 transgene expression inhibits the transition from DN3 to DP thymocytes. J Immunol. 2000;165:1236–1242. doi: 10.4049/jimmunol.165.3.1236. [DOI] [PubMed] [Google Scholar]
- 36.Pear WS, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- 37.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology Greene Publishing Associates/Wiley Interscience, New York, NY. Greene Publishing Associates/Wiley Interscience; New York: 1992. [Google Scholar]
- 39.Hostert A, et al. A CD8 genomic fragment that directs subset-specific expression of CD8 in transgenic mice. J Immunol. 1997;158:4270–4281. [PubMed] [Google Scholar]
- 40.Gorman SD, Sun YH, Zamoyska R, Parnes JR. Molecular linkage of the Ly-3 and Ly-2 genes. Requirement of Ly-2 for Ly-3 surface expression. J Immunol. 1988;140:3646–3653. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Loss of Dnmt1 leads to a partial reactivation of CD8 expression in variegated DP cells. Left, representative flow-cytometry histograms showing the percentage of CD3lo cells within the gated CD4 SP thymocytes of floxed Dnmt1 alleles on E8I,E8II doubly deficient background (Dnmt1fl/fl × Δ1Δ2/Δ1Δ2). Floxed Dnmt1 alleles were deleted using LckCre transgenic mice. Upper and lower histograms correspond to LckCre− (Cre−) and LckCre+ (Cre+) littermates. Right, scatter plots showing the percentages of CD3lo thymocytes within the gated CD4 SP cells. The genotype of mice is either Dnmt1fl/fl or Dnmt1fl/fl × Δ1Δ2/Δ1Δ2. Each circle represents one mouse. + and − indicates the presence or absence of the LckCre transgene. Horizontal bars indicate average value for each genotype.
(a) Schematic map of MAZR showing the N-terminal BTB domain and the seven C2H2 Zn-finger domains. The clone isolated from Yeast One-Hybrid screens (YOH clone) is shown below. Numbers next to the maps indicate amino acid residues. (b) Sequence of RE-1 used as bait in YOH screens. Putative MAZR binding sites are shown. The approximate location of RE-1 within E8II downstream of the Cd8b1 gene is indicated.
Specificity of the anti-MAZR serum. (a) Total cell lysates of untransfected (−) or Myc-MAZR-expressing (+) HEK293T cells. Immunoblots were performed with anti-Myc (left), anti-MAZR serum (middle), or with preimmune serum (right). Actin protein abundance was used as loading controls. (b) HEK293T cells overexpressing MAZR were lysed and MAZR was immunoprecipitated using anti-MAZR serum. Immunoblots were performed using anti-MAZR serum. Input (left) shows anti-MAZR immunoblot on total cell lysates.
Retroviral constructs used for the transduction of hematopoietic stem cells (a) MIG-R, control retroviral construct expressing only GFP. MAZR, retroviral construct expressing various forms of MAZR and GFP (see Methods). (b) Map of the various MAZR forms expressed. MAZR, full-length form of MAZR. MAZRΔZF, Myc-tagged version of MAZR lacking the C-terminal zinc fingers. BTBMAZR, Myc-tagged BTB domain of MAZR. Numbers indicate amino acid residues. (c) Map showing partial amino acid sequence of the BTB domain of MAZR. Triangles indicate critical residues within BTB domains required for homodimerization and interaction with corepressors28. These critical amino acid residues were exchanged to generate a mutated form of MAZR (MAZRmut). The mutations were: L28S, Q34S, R35L.
“CD8-negative” DP thymocytes develop upon forced expression of MAZR in Tcra−/− thymocytes. (a) Two-color flow cytometry of MIG-R (upper row) and MAZR (lower row) transduced Tcra−/− thymocytes on Rag2−/− background. Representative dot plots show CD4 versus CD8 expression. Numbers in the dot plot quadrants indicate the percentage of the corresponding thymocyte subpopulations of total gated GFP− (left) and GFP+ thymocytes (right), respectively. (b) Scatter plot showing the ratio of GFP+/GFP− thymocytes within the CD4+CD8− quadrant in Supplementary Fig. 5a. Each circle represents one mouse. Horizontal bars indicate average value from the mice analyzed for each construct.