Abstract
Processing bodies (PBs) and Stress Granules (SGs) are the founding members of a new class of RNA granules, known as mRNA silencing foci, as they harbour transcripts circumstantially excluded from the translationally active pool. PBs and SGs are able to release mRNAs thus allowing their translation. PBs are constitutive, but respond to stimuli that affect mRNA translation and decay, whereas SGs are specifically induced upon cellular stress, which triggers a global translational silencing by several pathways, including phosphorylation of the key translation initiation factor eIF2alpha, and tRNA cleavage among others. PBs and SGs with different compositions may coexist in a single cell. These macromolecular aggregates are highly conserved through evolution, from unicellular organisms to vertebrate neurons. Their dynamics is regulated by several signaling pathways, and depends on microfilaments and microtubules, and the cognate molecular motors myosin, dynein, and kinesin. SGs share features with aggresomes and related aggregates of unfolded proteins frequently present in neurodegenerative diseases, and may play a role in the pathology. Virus infections may induce or impair SG formation. Besides being important for mRNA regulation upon stress, SGs modulate the signaling balancing apoptosis and cell survival. Finally, the formation of Nuclear Stress Bodies (nSBs), which share components with SGs, and the assembly of additional cytosolic aggregates containing RNA –the UV granules and the Ire1 foci–, all of them induced by specific cell damage factors, contribute to cell survival.
Abbreviations: ATXN2, Ataxin-2; BicD, Bicaudal D; CBP, CREB Binding Protein; CPEB, Cytoplasmic Polyadenylation Element Binding protein; DHC, Dynein Heavy Chain; DIC, Dynein Intermediate Chain; FAK, Focal Adhesion Kinase; FUS/TLS/hnRNP P2, Fused in Sarcoma; G3BP, Ras-GAP SH3 domain binding protein; GCN2, General Control Nonderepressible-2; Grb7, Growth factor receptor-bound protein 7; HAP, hnRNP A1 interacting protein; HDAC6, Histone Deacetylase 6; HRI, Heme-Regulated Inhibitor; HSF, Heat Shock Transcription Factor; KHC, Kinesin Heavy Chain; KLC, Kinesin Light Chain; MLN51, Metastatic Lymph Node 51; NMD, Nonsense mediated decay; nSBs, Nuclear Stress Bodies; OGFOD1, 2–14 Oxoglutarate and Fe(II)-Dependent Oxygenase Domain Containing 1; PB, Processing body; PERK, Pancreatic Endoplasmic Reticulum eIF2alpha Kinase; PKR/EIF2AK2, Double stranded RNA-dependent Protein Kinase; PP1, Protein phosphatase 1; PrP, Prion protein; RBP, RNA Binding Protein; RNP, Ribonucleoparticle; Sam68, Src associated in mitosis 68 kDa; Member of STAR, Signal Transducer and Activator of RNA; SCA, Spinocerebellar Ataxia; SG, Stress Granule; SMA, Spinal Muscular Atrophy; FMRP, Fragile X Mental Retardation Protein; SMN, Survival of Motor Neuron; TDP43, TAR DNA-binding Protein 43; TRAF2, TNF receptor associated factor 2; UVGs, UV RNA Granules
Keywords: Processing body, Stress Granule, Kinesin, Dynein, Bicaudal D, Aggresome
1. Introduction
The existence of cytoplasmic granules containing translationally repressed mRNAs in germ cells, embryos and neurons is known since a long time. These macromolecular aggregates are collectively called RNA granules, and the term defines a broad spectrum of entities, ranging from neuronal RNA transport granules to specific structures for the storage of maternal mRNAs. Two additional ubiquitous granules have been recently discovered, termed “Processing Bodies” (PBs) and “Stress Granules” (SGs). PBs were initially described as cytoplasmic aggregates harbouring the RNA decay machinery [1], [2], [3], [4]. Then, work from several labs brought up the novel concept that PBs contain mRNAs that are silenced by a plethora of distinct mechanisms. Thus, cells show a variable number of PBs, depending on the amount of mRNAs under the control of silencing pathways including miRNA, RNAi, or NMD among others ([5], [6], [7], [8] reviewed in [9], [10], [11], [12]).
In addition to the numerous silencing pathways that operate in normal conditions, stress stimuli trigger several pathways leading to a global translational silencing, and this correlates with the formation of a distinct kind of mRNA silencing foci: the SGs. The formation of PBs and SGs has been recently discussed in a number of excellent reviews [9], [10], [11], [13], [14], [15]. SGs and PBs are closely linked. SGs grow in close apposition with PBs and require their presence [16], [17], [18]. In addition, SGs and PBs share a few protein components, and mRNAs can be delivered from one structure to another (reviewed in [10], [11], [12], [19]). A number of proteins stimulate the interaction between PBs and SGs, and a continuous spectrum of structures exists from PBs to SGs (reviewed in [10], [20], [21]). The cellular response to stress is highly conserved, and the formation of SGs was observed by us and other authors in trypanosomatid, yeast, mammalian, and insect cells ([10], [17], [18], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38]. SG formation in procaryotes has not been reported, but chloroplasts –organelles of bacterial ancestry– assemble similar structures [36]. SGs have also been reported in vivo, indicating that SG formation is not restricted to the stress response of cells under in vitro conditions [39], [40], [41].
We and others have also documented the presence of SGs in myelinating and neuronal cells exposed to oxidative or ER-stress, or to pro-inflammatory cytokines, all conditions associated with neurodegenerative and demyeliniating pathologies (ref [16], [39], [40], [42] and unpublished data).
The success of the stress response in helping cell survival depends on multiple mechanisms that act in concert to regulate cell metabolism, signaling pathways and gene expression at the level of transcription, translation and protein stability. Which is the relevance of SG formation to the survival response is a relevant question that we are beginning to understand, and that may have multiple answers.
2. PBs and SGs are related mRNA silencing foci
PBs are constitutive and can be further induced when a global translational silencing takes place, as it occurs upon a variety of stress insults, ranging from a raise in reactive oxygen species concentration to moderate hypoxia [6], [17], [20], [43]. Whether PBs are a cause or consequence of mRNA silencing has been a matter of debate. Current evidence indicates that mRNA silencing by miRNA, RNAi or NMD (nonsense mediated decay) can occur in the absence of visible PBs [44]. However, oligomerization of PB components appears to be required for efficient silencing [45], and several proteins present in PBs contain specific aggregation domains, many of them being conserved among different species (Table 1 ) [18], [46], [47], [48], [49], [50], [51], [52], [53]. It is important to emphasize that the recruitment of mRNAs to PBs is not simply the consequence of not being translated, but rather the effect of an active silencing mechanism. An elegant study addressing this concept was performed by Izaurralde and co-workers, showing that the translational inhibitor puromycin –which interrupts translational elongation of all transcripts and thus flooding the cytoplasm with free mRNAs–induces PBs only in the presence of active RNAi or miRNA silencing pathways [44].
Table 1.
Oligomerization domains present in PB and SG components. Oligomerization or dimerization domains relevant for foci formation were identified by deletion of distinct protein regions, or by fusion to reporter proteins. The oligomerization/dimerization domains are conserved in the species listed. The knockdown of molecules carrying the indicated aggregation domains affects foci formation in several cases. Similar putative dimerization/oligomerization domains present in additional PB components, including FMRP and CPEB, are present [46], [190], [194].
| Protein | foci | Domain | Reference |
|---|---|---|---|
| Lsm4 | PBs | C-terminal Q/N-rich (yeast) | [46], [47] |
| C-terminal RG-rich (metazoans) | |||
| EDC3 | PBs | C-terminal Yjef-N | [47], [48] |
| Gawky/GW182 | PBs | Central Q-rich | [191] |
| Ge-1/Hedls/Varicose/EDC4 | PBs | C-terminal Q/N-rich | [50], [51], [52], [53] |
| CCR4 | PBs | N-terminal Q/N | [46] |
| Dhh1p | PBs | C-terminal Q/N | [46] |
| SGs | |||
| Pop 2 | PBs | C-terminal Q/N-rich | [46] |
| G3BP | SGs | N-terminal NTF2 | [64] |
| TIA1/Pub1 | SGs | C terminal Q/N rich | [18], [192] |
| PBs | |||
| TIAR/Ngr1 | SGs | C terminal Q/N rich | [18], [192] |
| PBs | |||
| MNL51 | SGs | C terminal Q-rich | [123] |
| Pumilio 2 | SGs | N terminal Q-rich | [155] |
| Caprin | SGs | C-terminal Q-rich | [63] |
| TDP43 | SGs | C-terminal PRD Q-rich | [156], [193] |
Numerous studies in yeast, plants, trypanosomatids, insects and vertebrates describe about half a hundred proteins present in PBs. These molecules include the 5′ cap binding protein 4E, decapping enzymes and co-activators, nucleases and several RNA-binding proteins involved in NMD, miRNA-mediated silencing and general mRNA repression (reviewed in [9], [10], [11], [12]). In addition, a few splicing and mRNA export factors are also present. The presence of these factors in PBs has been studied mostly by imaging, and in most cases, they appear to display a quite uniform composition. However, many of these analyses include visualization of fluorescent chimerical proteins transiently expressed from transfected cDNAs. Extreme care should be taken when examining cells overexpressing PB components, as it was reported that alterations on the cellular stoichiometry may lead to aberrant structures, as a consequence of the intrinsic aggregative capacity of PB components, and of the titration of limiting factors [21], [50], [54]. Several reports where endogenous PB components were analyzed support the notion that heterogeneous populations of PBs are present. In mammalian cell lines, PCBP2, a facilitator of IRES-mediated translation, is present in a fraction of PBs identified by 4ET or DCP1a [55], and an important proportion of PBs lacks this protein. In the same line, a close examination of PBs in Drosophila Schneider cells reveals that Hedls, Dcp1a and XRN1 label distinct subsets of PBs, all of them being responsive to hypoxia (Fig. 1A, see also ref [43]). The heterogeneity is remarkable in mammalian neurons, where Cougot et al. have described specific foci termed dendritic P-body-like foci (dlPB). These contain the PB components DCP1a and GW182, whereas Ago2 and rck/p54 are not always present in dlPBs (Fig. 1B). Moreover, Ago2 and rck/p54 form foci that do not contain DCP1a nor GW182. In addition, unlike PBs in cell lines, dlPBs rarely contain XRN1 [56]. More recently, Bagni and collaborators reported the presence of an additional kind of dendritic foci that contain the PB component Lsm1 and exclude Dcp1a [57].
Fig. 1.

PBs, SGs and related RNA granules in trypanosomes, flies and mammals. A and B, PBs are heterogeneous. A. Immunofluorescence for DCP1a; Ge-1/Hedls and Pacman/XRN1 in Drosophila Schneider S2R+ cells. Double-stained foci are frequent in the case of DCP1a and Ge-1, and infrequent for DCP1a and Pacman. In all cases, single-stained foci are highly frequent. Bars: 1 μm. B. The P-body components DCP1a and rck/p54 form separate foci in hippocampal neurons, and a fraction of them partially overlap. The dendritic cytoskeleton is stained in blue (kindly provided by Luciana Luchelli, Instituto Leloir, see also [56]). Bars: 1 μm. C. ER-stress induces the transient formation of SGs (red) in mammalian cells. In a fraction of cells SGs last longer than 8 h and fuse with PBs (green) (see also [16]). Bars: 5 μm. D. Polyadenylated RNA granules are induced in T. cruzi cells exposed to nutritional stress. Left, polyA granules contain the exoribonuclease XRNa (kindly provided by Alejandro Casola and Carlos Frasch, Universidad Nacional de San Martín, Argentina). Right, polyA granules are distinct from tRNA granules, which contain 5′ halves of tRNA molecules cleaved upon stress (kindly provided by A. Cayota, Institut Pasteur de Montevideo, Montevideo, Uruguay). Bars: 1 μm.
It is assumed that all these granules contain mRNA, but this has not been tested in all of them, and thus, the possibility that they represent storage sites for specific PB components remains open. Supporting this notion, satellite granules containing truncated Ge1/Hedls are detected adjacent to PBs [7], [50]. Another structure associated to PBs and concentrating uridine-rich small nuclear ribonucleoproteins are the U bodies [58]. In this context, the heterogeneity of foci may be indicative of a maturation process where distinct factors are recruited progressively. PBs are motile, and they may come into close contact, and even fuse with each other [59], [60], [61], thus providing a way to exchange or incorporate distinct molecules. A model for PB assembly compatible with all of these observations was recently suggested [9]. According to this, silenced mRNPs are aggregated by specific dimerization or oligomerization domains (Table 1 ), which direct the formation of distinct macromolecuar complexes, likely corresponding to distinct silencing pathways. Then, homotypic interactions between protein molecules present in separate silenced mRNPs may aggregate larger foci [9]. Thus, a tempting hypothesis to test is whether foci loaded with different proteins correspond to different silencing pathways.
The stress response activates several mechanisms for translation repression, which are discussed below. Among the other pathways, the inactivation of the translation initiation factor eIF2alpha provokes the accumulation of non-functional translation initiation complexes, that include an mRNA molecule plus the 40S ribosomal subunit, the ternary complex formed by met-tRNA, eIF2 and GDP, and a number of translation initiation factors [19], [23], [26], [62]. Noteworthy, although PB number and size are enhanced upon stress, the abortive translation initiation complexes generated upon eIF2alpha phosphorylation are not recruited to pre-existing PBs, and aggregate in quite independent foci, the SGs. SGs and PBs are distinguishable in mammals and insects, but other organisms may assemble intermediate structures (Table 2, Fig. 1D and ref [10], [29], [30]). In most cases, SGs contain polyadenylated transcripts, whereas mRNAs recruited to PBs are largely deadenylated, as judged by the lack of in situ hybridization signal of oligo-dT probes [17], and by the absence of PABP [16]. Unlike PBs, SGs usually exclude components of the decapping machinery, and recruit several initiation factors and small ribosomal subunits, which are excluded from PBs (Table 2). However, in certain conditions, mammalian SGs and PBs may fuse giving place to a hybrid structure containing the PB component DCP1a and the SG component TIAR (Fig. 1C) [16].
Table 2.
Stress Granules and related foci induced upon stress. The stress-induced formation of granules containing polyadenylated RNAs is conserved through evolution, and may depend or not on the inactivation of eIF2alpha. The resulting foci may have distinct composition in different organisms. SGs induced in Drosophila by heat shock or arsenite contain classical mammalian SG components. Stress Granules from C. elegans, yeast and trypanosomes are markedly different. Bonafide Stress Granules are apparently induced in budding yeast exposed to glucose starvation or arsenite, or in fission yeast upon osmotic stress or heat shock. T. brucei respond to heat shock forming cytoplasmic SGs that contain PABP, eIF4E, eIF3 and exclude PB components, thus resembling mammalian SGs. In contrast, T. cruzi cells assemble visible granules containing polyadenylated RNA and the PB components DHH1 and XRNA when exposed to nutritional starvation (see also Fig. 1D).
| Organism | Stressor | RNA granulea | eIF2α phosphorylation | Kinase | Components included | Components excluded | References |
|---|---|---|---|---|---|---|---|
| Mammals | Arsenite or ER-stress | SG | YES | HRI or PERK respectively | PolyA(+) RNA, PABP, TIA-1/R, eIF3, G3BP, eIF4G, 40S, others. | 60S, HSP27, TTP, Dcp-1, Dcp-2, Hedls, GW182, Lsm1-7, others. | [10] |
| Heat shock | SG | YES | HSP27, polyA(+) RNA, PABP, TIA-1/R | [23] | |||
| Pateamine, hippuristanol, tiRNA, energy deprivation | SG | NO | PolyA(+) RNA, PABP, TIA-1/R, eIF3, G3BP, 40S, eIF4E, TTP (for energy deprivation), others | Hedls, 60S, Dcp1, Rck/p54 | [24], [25], [26], [27] | ||
| UV | SG | YES | PolyA(+) RNA, PABP, TIA-1/R | HSP27 | [23] | ||
| D. melanogaster | Heat shock | SG | NO | PolyA(+) RNA, FMR1, eIF4E, eIF3, PABP, Rox8 (TIA1), 18S rRNA | DCP1, RPL P0 | [28] | |
| Arsenite | SG | YES | PEK and GCN2b | PolyA(+) RNA, FMR1, eIF4E, eIF3, PABP, Rox8 (TIA1), 18S rRNA | DCP1, RPL P0 | [17], [28] | |
| T. brucei | Heat shock | SG | NO | eIF4E1 to 4, eIF2A, eIF3B, ABP1/2 | DHH1/Rck/, XRNA/XRN1 | [29] | |
| Carbon-source starvation | mRNA granules | PABP1, UBP1, polyA(+) RNA | [30] | ||||
| T. cruzi | Carbon-source starvation | mRNA granules | PABP1/2, eIF4E, TcDhh1/Rck, XRNA/XRN1, TcUBP1 to 4, 5a and 6b, polyA(+) RNA | eIF3D, TcS15(40S), TCL3 (60S) | [30] | ||
| Nutritional stress | Cytoplasmic foci | 5′ tRNA halves, 3′ tRNA halves | [31] | ||||
| S. pombe | Osmotic or heat shock | Stress-dependent foci | rRNA, eIF4E, Sum1/eIF3i, p116/eIF3b, Int6/eIF3e | [32] | |||
| S. cerevisiae | Heat shock | SG | NO | eIF3, Pab1p/PABP, eIF4G2, Rps30A (40S), Ngr1/TIAR, Pub1/TIA1, Dcp2p Dhh1/Rck | Rp125 (60S), eIF2α | [33] | |
| Glucose starvation | Pab1-containing PBs | PolyA(+) RNA, Pab1p/PABP, eIF4E, eIF4G. Partially Dcp2p | eIF3 | [37] | |||
| Glucose starvation | EGP bodies | eIF4E, eIF4G, Pab1p/PABP | eIF3b, eIF4AI, eIF2α, eIF2Bγ, | [38] | |||
| Glucose starvation | SG | YES | Gcn2 | Pub1/TIA-1, Ngr1/TIAR, Pbp1/Ataxin-2, Pab1p/PABP, eIF4GI, eIF4GII, eIF4E, Eap1/4EBP, Hrp1, Ygr250c, Gbp2 | eIF3, PeIF2α | [18] | |
| Glucose starvation or arsenite | SG | Pbp1(Ataxin-2), Pub1(TIA1), Pbp4p, Lsm12, Dhh1(Rck/p54) | [34] | ||||
| C. elegans | Heat shock/sperm depletion in female worms | RNP foci (oocytes) | RNA, MEX-3, DCP-2, CAR-1/Rap55, CGH-1/Rck, PABP, TIA1 | [35] | |||
| C. reinhardtii (chloroplast) | Oxidative stress/high light/FCCP/UV/phosphate deprivation | cpSG | cPABP, S21 (40S), mRNA | L12 (60S), L2 (60S) | [36] |
Names given by the authors. PBs are not included.
PEK main kinase, GCN2 secondary role.
Work from numerous laboratories yielded a growing list of SG protein components, most of them identified by imaging approaches (Table 2). Updated surveys [10], [11] indicate that almost a hundred proteins, not all of them linked to mRNA metabolism but involved in signalling and apoptosis, are recruited to SGs. One third of them are also present in PBs. Several SG components are normally observed in the nucleus and accumulate in the cytosol upon stress, while others reside mostly in this compartment. One third of the SG components are translation factors or associate to them or to polysomes in several manners, all this facilitating their recruitment to SGs, or helping SG formation. Splicing factors, repressors and regulators of mRNA stability are also present in SGs. Distinct RNA Binding Proteins (RBPs) are recruited by their RNA binding domains, or by protein interaction domains, including RRMs, RGG, NTF2, and ROQ domains, among others [63], [64], [65], [66]. Several of the RBP components have specific mRNA targets, and thus, they may selectively affect the expression of key transcripts. Like PBs, mammalian SGs are heterogeneous in their composition (see below).
PBs and SGs are highly dynamic, and both foci constantly exchange RNA and proteins with the cytosol. Fluorescent Recovery After Photobleaching (FRAP) analysis of several protein components reveals a wide range of exchange rates, which can be as high as 63% recovery within 30 s for a reporter mRNA [60], whereas certain protein components are almost static, as is the case of DCP2 in PBs. A comprehensive list of turnover rates is provided in a recent review by Buchan and Parker [10].
A characteristic feature of PBs and SGs is that they can release transcripts to allow their translation ([6], [7], [20], [67], reviewed in [9], [10], [19]). In the presence of drugs that stabilize polysomes, both PBs and SGs tend to dissolve, indicating that mRNAs can move from PBs and SGs to polysomes and vice-versa. When analyzed simultaneously, PBs appear less dynamic than SGs [20], [60], [68]. Distinct aggregates containing Dcp1a or Xrn1, putatively containing maternal mRNAs are present in Drosophila embryos, and show differential sensitivity to polysome-stabilizing drugs [69]. In mammalian neurons, PBs are less dynamic than PBs from cell lines [56]. However, FRAP analysis indicates that the turnover of DCP1a in neuronal PBs is dramatically enhanced by synaptic stimulation, indicating that PB dynamics and the release of mRNAs to allow their translation are controlled by neuronal activity, which is known to regulate local protein synthesis at the post-synapse [70], [71], [72], [73], [74], [75], [76], [77], [78], [79].
3. Transient SG assembly
SGs form during acute stress and their presence correlates with the transient translational silencing (Fig. 2 ). SG formation is fast, and it does not require transcription [80] indicating that they harbour mRNAs from pre-existing polysomes. In this section, we will review the mechanisms underlying SG aggregation and dissolution.
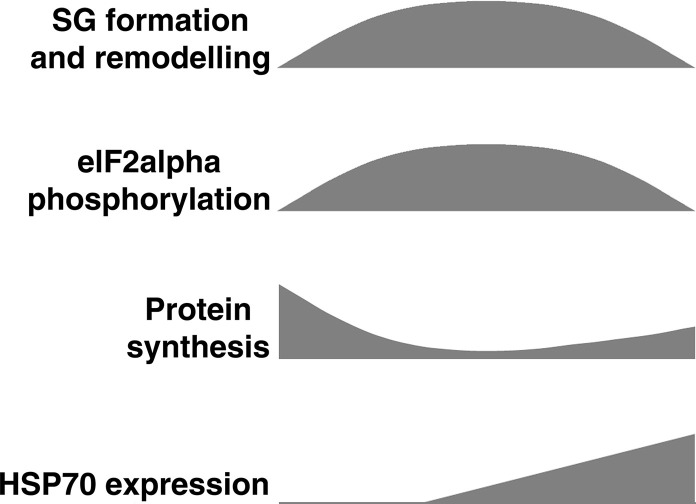
Fig. 2.
Comparative time-course of SG formation, eIF2alpha phosphorylation, protein synthesis, PB induction and heat shock protein expression upon stress induction. Maximal SG formation, eIF2alpha phosphorylation and protein synthesis inhibition occur quite simultaneously, between 1 and 2 h upon oxidative stress in mammalian or insect cells, or around 2–4 h upon ER-stress, respectively [16], [17]. All trough during the response, SG grow, undergo fusion and fission and remodelate. They can incorporate or lose components during the response (see text). Two hours after oxidative stress induction, the foci begin to dissolve synchronously and they completely vanish 1 h later. A similar time-course, with the time of maximal SG formation at around 2 h and a slower dissolution phase is observed upon ER-stress induction. SG dissolution occurs with similar time-course either in the presence or absence of oxidative or ER-stress inductors, or upon booster applications [16], [17]. SGs are induced rapidly by inhibitors of translation initiation, and do not dissolve unless the drug is removed (Loschi and Boccaccio unpublished). eIF2alpha phosphorylation reaches maximal levels and may go back down basal levels during SG dissolution. Protein synthesis shuts off at the time of maximal SG formation and then partially recovers during SG disassembly. This correlates with HSP70 expression, which keeps accumulating beyond SG disassembly. Synthesis of heat shock proteins lasts for several hours, whereas recovery of normal protein synthesis takes a longer time. PBs are induced by cellular stress, then they may return to basal conditions, move to the perinucleus or vanish, and their components can be incorporated to SGs [16], [50], [122]. Paralleling SG formation in the cytoplasm, the formation of Nuclear Stress Bodies (nSBs) occurs at specific foci in the nucleus (see text). Like SGs, nSBs are transient and remodellate during the response.
3.1. Transient translational silencing upon stress
The acute response to stress requires a rapid cellular adaptation before protective proteins begin to accumulate at functional levels. Accordingly, the initial steps are largely mediated by post-translational modifications. In most cases, translational silencing upon stress is triggered by phosphorylation of eIF2alpha by specific kinases, although additional mechanisms exist. There are four distinct eIF2alpha kinases in vertebrates, whereas other organisms may have a lower number [28], [81], [82], [83], [84], [85]. These kinases are activated by distinct stress stimuli, which promote dimerization and trans-autophosphorylation. EIF2alphaK1/HRI (Heme-Regulated Inhibitor) is activated by heat shock or arsenite –a known inductor of oxidative stress– [86], [87]. HRI is present in SGs, and a recent report suggests that HRI is positively regulated by G3BP and OGFOD1, two SG-resident proteins [88]. OGFOD1 is homologous to prolyl-hydroxilases and speculatively, it may hydroxylate a proline residue in HRI, thus stimulating HRI kinase activity.
PERK (Pancreatic Endoplasmic Reticulum eIF2alpha Kinase)/PEK/EIF2alphaK3 is an ER-transmembrane protein. Under normal conditions the endoplasmic reticulum-resident chaperone BiP/GRP78 is associated to the PERK luminal domain. Eventually BiP/GRP78 is targeted to unfolded proteins that may accumulate in the ER, releasing PERK and thus allowing dimerization and activation of the PERK kinase domain that faces the cytosol. GCN2 (General Control Nonderepressible-2) is associated to polysomes in a multiprotein complex. Recognition of uncharged tRNA by the GCN2 Histidil-tRNA sintetase (HisRS) domain triggers activation [82], [83]. Finally, EIF2AK2/PKR (double stranded RNA-dependent Protein Kinase) is restricted to metazoans and is activated by double stranded RNA, thus protecting cells from viral infections. PKR can be activated locally [89], and hypothetically, virus can induce SGs at sites of virus replication by increasing the local concentration of phosphorylated eIF2alpha. However, viral infection, eIF2alpha phosphorylation and SG formation do not always correlate, as viruses have evolved intricate mechanisms to evade the cellular defense response. They can block SG formation or moreover, benefit from it (Table 3 ).
Table 3.
Viral infections may induce or block SG formation. SGs may assemble as a defense against certain viruses, to limit the infection by sequestration and/or cleavage of translation factors, proteins and transcripts required for viral replication, and SG assembly apparently involves eIF2alpha phosphorylation [80], [195], [196], [197], [201], [202], [205], [206]. However, a number of DNA and RNA viruses block SG formation, and infected cells exposed to oxidative or ER-stress fail to aggregate SGs [196], [198], [199], [200], [204]. The 3′(−) end of flaviviruses and certain viral proteins bind TIAR and TIA1, thus inhibiting SG formation [198]. SGs can contribute positively to viral replication, by sequestration of antiviral proteins and mRNAs, or by nucleation of core particles and viral RNA, thus helping viral factories [201], [202]. Translation of alphavirus transcripts occurs in the presence of SGs and phosphorylated eIF2alpha, thanks to an adaptation called translation enhancer element [196]. Poliovirus infection triggers the formation of SGs that gradually loss PABP, G3BP and eIF4G, which are cleaved during the first hours of the viral cycle. Whether SG formation helps or inhibits poliovirus replication is unknown [80], [197], [207].
| Virus | eIF2alpha phosphorylation | SG induction (marker) | SG inhibition (stressor; marker) | References | |
|---|---|---|---|---|---|
| Positive stranded RNA | Mouse hepatitis Coronavirus | Yes | Yes (TIAR) | Unknown | [195] |
| Semliki Forest Virus | Yes | Yes (TIA-1, TIAR, eIF3) | Yes (arsenite; TIA-1) | [196] | |
| Poliovirus | Unknown | Yes (TIA-1, polyA+ mRNA, Sam68, G3BP, eIF4G, PABP) | No (heat shock; Sam68, Hsp27) | [80], [197] | |
| West Nile and Dengue virus | No | No (TIAR) | Yes (arsenite; TIAR) | [198] | |
| HIV-1 | No | No (PABP1, Staufen1) | Yes (arsenite; PABP1, Staufen1) | [199] | |
| Double stranded RNA | Rotavirus (Reoviridae) | Yes | No (TIA-1, eIF4E, S6, PABP) | Yes (arsenite; TIA-1, eIF4E, S6) | [200] |
| Reoviridae | Yes | Yes (TIA-1, TIAR, G3BP, eIF4G, eIF4E, eIF3, 7F4) | Unknown | [201], [202] | |
| DNA | Herpex Simplex virus 1 | Unknown | No (TIA1/R) | Unknown | [203] |
| Human Cytomegalovirus | Unknown | No (eIF4G) | Yes (thapsigargin; eIF4G) | [204] |
Phosphorylation of eIF2alpha leads to the accumulation of non-functional translation initiation complexes, which aggregates in SGs. However, SGs are induced in heat-shocked Drosophila, T. brucei and S. cerevisiae cells, without involving eIF2alpha phosphorylation [28], [29], [33] (Table 2). Whether translation initiation complexes or related structures containing 40S subunits accumulate in those cases is unknown. In a large proportion, translational silencing upon heat shock occurs upstream of 40S recruitment, and involves eIF4G inactivation by hsp25/hsp27, all these proteins being present at SGs [90]. Speculatively, distinct silenced mRNPs can build up SGs, as long as protein–protein interactions can be established between them, as suggested for PBs (review in [9]).
An additional mechanism recently discovered for translation repression during stress involves 5′ halves of tRNA molecules. In Giardia, Tetrahymena, mammals, plants and fungi, the anticodon loop of several tRNAs is cleaved by members of the Ribonuclease A or T2 families [91], [92], [93], [94]. Cleavage of tRNAs is enhanced by stress and the released tRNA 5′ halves inhibit translation by an unknown mechanism, likely involving mRNA degradation by guide tRNA 5′ halves [25], [92], [93], [94], [95], [96], [97], [98], [99]. The fly Dnmt2 methylase inhibits tRNA cleavage by methylating the anticodon loop [100]. Dnmt2 is recruited to SGs, and Dnmt2 mutants show a reduced survival upon stress, suggesting that excessive tRNA cleavage is noxious. Of interest, mammalian Angiogenin and yeast Rny1p –two RNAses that mediate tRNA cleavage– are secreted and may be internalized [93], [94], thus suggesting a strategy to communicate neighbouring cells that a stressor is present, and coordinating a tissular response. Translation inhibition by 5′ tRNA halves –also termed tiRNAs– induces SG formation in mammals [25], adding to the list of eIF2alpha-phosphorylation-independent mechanisms for SG induction (Table 2). A recent work in trypanosomes indicates that tiRNAs form cytoplasmic granules apparently distinct from SGs ([31], Fig. 1D, right). Given the striking differences between species, whether tRNA fragments and cognate silenced mRNAs are present in yeast or mammalian SGs remains open.
SGs are also induced upon DNA damage by UV irradiation [23], [101]. Whether this is mediated by eIF2alpha inactivation is unknown. Finally, SGs form without apparently requiring eIF2alpha phosphorylation upon exposure to mitochondrial poisons [26], or by translational inhibitors that affect ribosome scanning at different levels, namely hippuristanol, pateamine or edeine, but not by puromycin or inhibitors of 60S recruitment [16], [22], [24], [102] (Table 2). This indicates that SGs harbour mRNAs arrested at specific points during initiation. In this line of evidence, we and other authors have reported that the repressor proteins Smaug and CPEB, which block translation initiation by disrupting the 4G–4E interaction and thus preventing 40S recruitment, form silencing foci related to SGs when overexpressed [21], [103]. All this suggests that, like in the case of PBs, several repression mechanisms can target mRNAs to SGs.
The stress response is self-regulated, and the phosphorylation of eIF2alpha triggered upon stress induction facilitates the translation of proteins that mediate eIF2alpha dephosphorylation. Briefly, the mRNA encoding GADD34/PPP1R15a, the regulatory subunit of protein phosphatase 1 (PP1), a key phosphatase for eIF2alpha reactivation that is found in SGs (Loschi and Boccaccio, unpublished), contains several uORFs (upstream Open Reading Frames) and therefore, its translation is enhanced upon eIF2alpha inactivation [81], [82] . Accordingly, SG disassembly correlates with eIF2alpha dephosphorylation, which occurs even in the continuous presence of stressors, or moreover, upon booster applications (Fig. 2) (ref [16] and Loschi et al. unpublished observations). Translation inhibition triggered by stressors that do not induce eIF2alpha phosphorylation is also reversible. In all cases, SG disassembly correlates with a partial recovery of translation [16], [17], which corresponds mostly to the synthesis of HSPs and other protective molecules (Fig. 2, [41], MAD and GLB, unpublished).
3.2. Polysome disassembly
When initiation is blocked and translationally active polysomes continue to elongate, a progressive loss of ribosomes occurs. Polysome “run off” upon stress induction is negatively regulated by the double-stranded RNA-binding protein Staufen, which apparently provokes polysome stalling [16]. In both mammals and Drosophila, Staufen knockdown enhances SG formation while its overexpression impairs SG assembly (ref [17]; ML and GLB unpublished). It was recently suggested that ER-associated mRNAs may be retained in stalled polysomes thus escaping SGs [104]. A polysome stalling also occurs during mitosis, also opposing SG formation [105]. In both cases, polysome structural integrity helps translation recovery. What factors govern the stability of ER-bound polysomes is unknown, but the presence of Staufen in ER membranes and associated polysomes [16], [22], [106], [107] suggests that this protein may be involved. Finally, Anderson and co-workers recently reported that hypusination of eIF5A stimulates elongation during stress, thus accelerating polysome run off and SG formation [108]. How Staufen and eIF5A hypusination are regulated during SG assembly and dissolution remains to be elucidated.
3.3. Molecular motors govern SG assembly and disassembly
SG formation is a gradual process. Initially, numerous small aggregates begin to form. Soon afterwards, these particles coalesce into larger granules so that at the peak of the response, fewer and bigger SGs are observed [17], [20]. The rapid and coordinated assembly of SGs and their subsequent dissolution suggest that SG components are actively transported by molecular motors. Subcellular transport of silenced mRNPs has been described extensively in several cell types and organisms. Briefly, mRNPs frequently move bidirectionally, driven by dynein and kinesin motor molecules, which are recruited by specific proteins, many of them conserved in insects, amphibians and mammals ([74], see [70], [75], [76], [109], [110] for recent reviews). Members of the myosin V family are also involved in mRNA transport in both yeast and vertebrates [111], [112].
The participation of motor molecules in SG formation is supported by the effect of cytoskeleton disrupting drugs. The absence of microfilaments leads to the formation of scattered and quite small SGs [17], suggesting that unknown myosins nucleate SGs. In the same line of evidence, it was shown that a yeast myosin V, termed Myo2p, mediates PB disassembly [113]. SGs form close to PBs and PB integrity facilitates SG formation [16], [18]. Speculatively, myosins may regulate the flow of RNPs from PBs and from the cytosol to SGs. When microtubules are disrupted, the process progresses to the point that SGs nucleate and grow in size but fail to approach to the nucleus [17], [114], [115], [116] suggesting that the coalescence of the initial accretions is interrupted. These aberrant SGs contain TIA1, G3BP, eIF3 and a number of RBPs [17], [114], [115], but apparently lack a number of components such as CCAR-1, CUG-BP and HuR [114], [115], that would be incorporated by a microtubule-dependent transport.
We and others have recently identified the microtubule-dependent retrograde motor subunits DHC1 (Dynein Heavy Chain) and DIC (Dynein Intermediate Chain), and the adaptor BicD1 (Bicaudal D1) as key components of motor complexes involved in SG assembly [17], [117], [118]. We also found that the anterograde Kinesin 1b heavy chain/KIF5B and KLC1 (Kinesin Light Chain 1) are specifically required for disassembly [17]. Relevantly, the KLC-like molecule PAT1, which is involved in KLC-independent transport of maternal mRNAs [119], is not required for the kinesin-mediated dispersion [17]. The role of these molecules is conserved in flies, where Dynein, Bicaudal D, Kinesin Heavy Chain and Kinesin Light Chain govern SG assembly and disassembly, respectively (reference [17], and ML and GLB, unpublished).
Several non-mutually exclusive mechanisms for motor recruitment are likely. Kinesin and dynein adaptors can contact protein components or mRNPs, in a manner similar to that for localized mRNAs (see [70], [109]). In addition, P0, a protein from the large ribosome subunit, which is excluded from SGs, interacts with kinesin motors [120], speculatively helping dispersion of RNPs containing complete ribosomes.
3.4. Aggregation
As is the case for PBs, SG integrity depends on the self-aggregation of resident proteins which in most cases contain prion-related domains (Table 1). As mentioned, SG assembly is a multi-step process [17], [114], [115], and it has been suggested that importin alpha contributes to the first steps of SG aggregation, likely by mediating the formation of macromolecular complexes [121]. Then, a number of SG components help to recruit other factors by specific protein–protein interactions. Thus, homotypic and heterotypic interactions are instrumental in the assembly of these structures, which do not contain membranes and can be as large as 6 μm. Protein aggregation is usually controlled by chaperones, and Hsp70, which starts to accumulate almost immediately upon stress induction (Fig. 2), mediates SG dissolution [122]. Regulation by chaperones that are titrated by unfolded proteins is a common theme in the stress response. This mechanism allows the activation of the transcription factor HSF, the ER-resident proteins Ire1, PERK and ATF6 and apparently affects this novel arm of the stress response, SG formation.
4. SGs show a variable composition
Almost a hundred translation factors and modulators, nucleases, splicing factors and a few other molecular functions, are obligate SG components. However, SG composition may be subtly different according to the nature of the stress stimulus, and it may also change progressively during the response. In addition, SGs are not homogeneous, and some components can concentrate in microdomains [123], [124].
In mammalian cells, TTP is present in PBs and in FCCP-induced SGs, but absent from arsenite-induced SGs. The recruitment of TTP to SGs is inhibited by p38 and the downstream kinase MK2 [27]. More recently, it was shown that transportin/karyopherin β2, an importin which is present in both SG and PBs, associates to TTP promoting its shuttling to SGs [125]. In all cases, the exclusion of TTP from SGs inhibits the degradation of ARE-containing target mRNAs [27], [125].
Mammalian cells exposed to heat shock incorporate hsp27 in SGs and this chaperone is absent from arsenite-induced SGs [23], [90]. Among other functions, Hsp27 contributes to translational silencing by blocking 4G and thus disrupting 4F complexes [90]. Likely, the presence of these molecules in SGs is linked to translation repression. In addition, hsp27 is also involved in the degradation of ARE-containing mRNAs [126]. Several other molecules show differential recruitment to SGs in mammals, but their physiological relevance is not always clear. Heat shock-induced SGs do not include the EJC component MLN51, which is present in arsenite-induced SGs [123], [127]. Calreticulin, an ER-resident chaperone which undergoes arginylation and retrotranslocation from the ER to SGs upon alteration of intracellular Ca2+ levels, is absent from UV-induced SGs [128]. Besides being a protein chaperone, calreticulin binds RNA affecting stability, and both functions may be linked to its recruitment to SGs.
The composition of SGs can vary during the response. A striking example was reported for the endonuclease PMR1, which is normally associated to polysomes. PMR1 interacts with TIA1 and is recruited to arsenite-induced SGs in a rather mature phase [129]. The incorporation of an endonuclease at late stages is compatible with the idea that SGs serve as reparation sites to reactivate translation, and that mRNAs that fail to be reused for translation are destroyed. The sequential incorporation of components is highlighted when SG maturation is impaired by microtubule-disrupting drugs (see above; [114], [115]). Likewise, virus-induced SGs may show a progressive change in composition, as a consequence of viral activity. Initially, poliovirus-induced SGs contain G3BP, PABP and 4G. However, during later phases of the infection these molecules are cleaved and not longer detected in SGs, which still contain polyadenylated RNA and TIA1 (Table 3, [80]). Finally, SGs may incorporate PBs or PB components at later stages (Fig. 1, Fig. 2). Differences in the composition of SGs triggered by distinct stimuli, as well as changes observed along the response undoubtedly reflect distinct physiological conditions that may affect mRNA metabolism and other functions that remain to be unveiled.
5. Which mRNA species are present in SGs?
As discussed above, distinct kinds of silenced mRNPs are targeted to SGs. However, which mRNA species are present in SGs is unknown. The hsp70 mRNA appears to be excluded from SGs, and this is consistent with its high translation rate during stress [14]. It has been speculated that most transcripts silenced by stress will be targeted to SGs. However, recent evidence indicates that additional factors are involved. The presence of TIA1 binding elements consisting of an U-rich RNA 30–37 nucleotide-long bipartite element that forms loops of variable size plus a bent stem [130], facilitates mRNA targeting to SGs [104]. Besides TIA1 and TIAR, there are other proteins present in SGs that bind specific RNA motifs, and that induce SG formation (reviewed in [10]). Thus, additional RNA elements are expected to direct messengers to SGs. Unexpectedly, transcripts triggered to SGs by TIA1 are excluded from SGs when engineered to encode a transmembrane domain that target the corresponding polysomes to the ER. Apparently, ER-associated polysomes are more resistant to stress-induced disruption, likely due to a slower elongation [104]. Escape of ER-associated mRNAs from SGs was shown in an oxidative stress model [104]. It remains to be investigated whether this also occurs under ER-stress. This condition requires alleviation of protein loading at the ER, and can trigger the decay of hundreds of mRNAs associated to ER membranes by a specific pathway that involves the endonuclease Ire1, and that is therefore termed regulated Ire1-dependent decay (RIDD) [131], [132]. RIDD is triggered in both Drosophila and mammals upon activation of Ire1 by unfolded proteins.
A few mRNAs have been confirmed to be present in SGs [122], [133] and a few others were shown to be excluded. SG isolation has remained elusive and thus, protein and mRNA composition is currently evaluated by imaging approaches. Important information is expected to be gathered by biochemical approaches in the future.
6. mRNA silencing foci regulation
It is predicted that the composition and dynamics of the mRNA silencing foci, as well as the interaction among them, will be modulated by the stress response. Indeed, post-translational modifications affecting recruitment, RNA binding or enzymatic activity of several protein components were described [27], [64], [134], [135], [136], [137].
In a pioneering work, Anderson and co-workers performed a systematic survey of genes involved in the assembly of these structures [138]. In a genome-wide RNAi screen, a hundred cell functions were identified as important for SG and/or PB formation. Among others, the O-GlcNAc post-translational modification of ribosomal proteins and other targets is apparently required for SG assembly. O-glycosilation is reversible and frequently reciprocal with phosphorylation, and both modifications may regulate protein self-aggregation [139].
SG formation also appears to be regulated by protein acetylation. HDAC6 (Histone Deacetylase 6) is required for SG assembly, likely by modifying tubulin and other cellular components, thus affecting subcellular transport [117]. Another post-translational modification found in SG proteins is ubiquitination [117]. This may tag them for degradation, providing an additional level of SG modulation. Supporting this notion, the inhibition of proteosome activity or knockdown of the E3-ubiquitin ligase EDD induces SG formation [122]. In addition, ubiquitination may contribute to signalling by unknown pathways.
Several other signaling molecules have been found in SGs. In addition to HDAC6 [117] and OGT [138], SGs contain the catalytic subunit of PP1 (ML and GLB, unpublished), likely involved in eIF2alpha dephosphorylation. The stress-activated JNK and the upstream kinase MKK7 are recruited to SGs induced upon oxidative stress by a specific scaffold, termed WDR62, and pharmacological inhibition of JNK reduces SG and PB size and number [140]. More recently, the small GTPase RhoA and its downstream kinase ROCK1 were shown to mediate SG assembly, and both molecules are present in SGs in their active forms [124]. The RhoA/ROCK1 pathway regulates cytoskeleton dynamics and the JNK pathway, all this likely contributing to SG formation. In addition to this signaling molecules, the protein kinase-A (PKA) scaffold AKAP350A [114], and IP5K (Ins(1,3,4,5,6)P5 2-kinase), which synthesize InsP6, are also recruited to SGs [141]. Speculatively, this may provoke a local increase of signaling molecules that may play a role in SG dynamics.
STE20 is another regulatory kinase that affects SG formation. In a recent report, Parker and co-workers showed that yeast Dcp2 is phosphorylated by STE20 upon stress induction, and Dcp2 phosphorylation is required for SG formation [135]. Dcp2 phosphorylation inhibits the decapping of a number of mRNAs [135], likely favoring the flux of mRNAs from PBs to SGs. SG dissolution is regulated by Focal Adhesion Kinase (FAK). FAK hyperphosphorylates the SG promoting protein Grb7 (Growth factor receptor-bound protein 7), thus inducing SG disassembly [134].
Finally, the antagonistic anterograde and retrograde transports of mRNP components are expected to be modulated during the response at the level of motor activity or recruitment, thus allowing transient SG assembly [17]. Interestingly, a number of stress-activated protein kinases, collectively known as SAPKs, including p38 and JNK, are known to modulate molecular motors differentially affecting anterograde and retrograde transports (see [142] for a recent review). Further regulatory mechanisms affecting protein self-aggregation, subcellular transport and other functions relevant to SG dynamics are expected to be discovered.
7. Relevance to mRNA regulation and cell survival
The formation of SGs upon a variety of stress stimuli (Table 2) and viral infections (Table 3) highlights their relevance to the cell survival response [121], [143], [144], [145], [146] which appears to involve multiple mechanisms.
7.1. mRNA regulation
It was initially speculated that SGs may contribute to the global translational silencing by sequestration of mRNAs and translation factors. However, current evidence seems to indicate that this is not the case. Several authors reported that SG disruption by distinct molecular approaches is not accompanied by impaired silencing. Among other studies, the disruption of retrograde transport [17]; the inhibition of protein acetylation [117]; the knockdown of key factors for O-glycosilation [138]; or the knock out of yeast pub1, pbp1 or eIF4G in S. cerevisiae [18], all these dramatically impairing SG assembly, do not affect translational silencing. Collectively these observations support the notion that aggregation of microscopically visible SGs is not required to keep translation off, and that SGs are the consequence and not the cause of the translational shut off upon stress. It has been suggested that SGs are sites where translation initiation occurs (discussed in [17], [147]), and moreover, that they may function in the reparation of defective initiation complexes [10]. Translation upon stress largely depends on uORF and IRES [81], [82] and interestingly, eIF3, which promotes translation reinitiation of uORF-containing transcripts [148] is recruited to SGs and, moreover, it is required for SG formation [138]. Similarly, the IRES trans-activating factor PCBP2 is recruited to SGs [55], all this supporting that translation initiation of specific mRNAs may occur in SGs. In accordance with a role in reparation of translation initiation factors and complexes, mammalian SGs contain eIF2alpha, that can be present in its phosphorylated form, and the exchange factor eIF2B ([16], [17], [115], [147]). In yeast, both factors are found in specific foci in both normal or stress conditions termed eIF2B bodies, and eIF2B bodies are sensitive to polysome stabilization by cycloheximide [149], [150], [151]. Thus, submicroscopic SGs may exist under normal conditions. These primordial structures would grow and incorporate additional components if the amount of arrested initiation complexes increases. Also supporting these hypotheses, stress-induced SGs disassemble when eIF2alpha is dephosphorylated (Fig. 2, [16], [17]) but SGs induced by translation initiation blockers persist until the drug is removed (ML and GLB, unpublished observations). Also in this line, FRAP analysis of several RNA Binding Proteins and reporter mRNAs (reviewed in [10]) indicates that they transit in and out of SGs quickly and that are not stably retained in SGs, which is also compatible with the notion that mRNAs are released as soon as they are ready for translation.
Whether translation of selected mRNA species is affected by SGs is unknown. Current knowledge indicates that SGs do affect the stability of a number of mRNAs, including p21 mRNA, with consequences on cell survival [122].
7.2. Sequestration of pro-apoptotic molecules
Additional cell survival mechanisms linked with SG formation are likely to occur, and an emerging example is the sequestration of pro-apoptotic molecules. The TNF receptor associated factor 2 (TRAF2) was the first case reported [152]. TRAF2 facilitates apoptosis by two independent pathways: TNFR activation, and caspase activation upon ER-stress induction [153]. TRAF2 interacts with eIF4G and is therefore retained in SGs, thus avoiding apoptosis [152]. More recently, two key molecules that activate the p38/JNK apoptotic pathway, namely RACK1 and ROCK1 were shown to be localized in SGs [124], [146], thus favoring cell survival. For a sequestration–inhibition mechanism to be efficient, the turnover of those molecules in SGs should be relatively slow. This seems to be the case for FAST, an apoptotic inhibitor present in SGs with a relative low turnover [62]. Whether this assumption is valid for other examples remains to be investigated. The retention of key molecules in SGs to control apoptosis is reminiscent of the regulatory role of the nucleolus and other nuclear structures specifically induced upon stress (see below). Thus, the sequestration of key molecules in large RNA–protein structures appears to be a conserved cell strategy present in both the cytosol and the nucleus.
7.3. SGs in neuronal health
Normal neuron physiology largely depends on the presence of the so-called neuronal RNA granules, which are the functional units for transport and translation regulation. Apparently, neuronal RNA transport granules are distinct from PB and dlPB, although they can share some components and get in close contact, likely allowing a flow of mRNAs and proteins [56], [154]. There is also an apparent relationship between neuronal RNA granules and SGs, as both contain polyadenylated mRNAs, ribosomal subunits and a number of common RBPs, including SMN, Staufen, Smaug, FMRP (Fragile X Mental Retardation Protein), Pumilio, ZBP1, CPEB, TDP43 and FUS/TLS/hnRNP P2, among others [21], [22], [103], [111], [127], [133], [155], [156], [157], [158], [159]. The presence of common RBP components suggests that similar mRNA regulatory pathways operate in neuronal RNA granules and SGs. RNA repression in neuronal granules is mediated by several mechanisms, which include polyA-tail length regulation and miRNA-mediated silencing, in addition to the inhibition of initiation provoked by a plethora of RNA Binding Proteins (reviewed by [71], [81]). Relevantly, eIF2alpha inactivation by GCN2 regulates mRNA expression in the CNS, but whether this involves granule formation is unknown ([160], [161], [72], [75], [76], [77], [78], [79]). Strikingly, a number of the above RBPs, namely SMN, CPEB, Smaug, Pumilio, Staufen, TDP-43 and FMRP modulate SG formation [16], [21], [103], [127], [155], [156], [158], [162], and thus, they may govern the dynamics of neuronal RNA granules. Whether neuronal RNA granules behave as mRNA silencing foci and remodellate upon stress is poorly described [22], [155]. Noteworthy, neurons are frequently exposed to excitotoxic stimuli, and their survival will depend on an efficient stress response. Strikingly, a number of the above RBPs that affect SG formation are also associated to neuronal defects. Staufen 1, which negatively modulates SG formation [16], affects neuronal function in flies and mammals [163], [164], and modulates Spinocerebellar Ataxia 8 (SCA 8) noncoding RNA, thus affecting the outcome of the disease [165]. Whether dysregulation of SG formation by defective Staufen contributes to the neuronal deficit is unknown. In the same line, SMN, an RBP that nucleates SGs, is altered in Spinal Muscular Atrophy (SMA). Interestingly, the SMN variant preferentially expressed in SMA fails to be recruited to SGs and SG formation is reduced in SMA cells [166]. Mutant FMRP triggers a specific neurodegenerative condition, termed Fragile X Mental Retardation Syndrome. Several mRNAs are deregulated upon FMRP mutation [167] and this seems to be causative of the syndrome. In addition, Didiot et al. [127] reported recently that FMRP K.O. or FMRP mutants show a severe impairment in SG formation. Finally, the SG and PB component Ataxin-2 (ATXN2), and its yeast orthologue pbp1 are required for SG formation [18], [168]. ATXN2 is mutated in the polyglutamine-associated disorder Spinocerebellar Ataxia type 2 (SCA2). Whether formation of dysfunctional SGs by defective SMN, FMRP or ATXN2 contributes to pathology onset or perpetuation remains to be investigated.
SGs can also be important in the context of unfolded protein diseases. In many aspects, SGs resemble aggresomes and unfolded protein aggregates frequently present in neurodegenerative pathologies. Among other remarkable similarities (Table 4 ), SG formation is mediated by specific protein–aggregation domains similar to those of prion protein or polyglutamine expansions, and their dissolution requires molecular chaperones. In addition, both SGs and aggresomes are actively assembled by the retrograde motor dynein. Like unfolded protein aggregates, SGs contain ubiquitinated proteins and are enhanced by inhibitors of protein degradation machineries. A striking difference between these structures is that SGs are transient and highly dynamic, whereas aggresomes and related protein aggregates are quite static. Taking into account all this, it is tempting to speculate whether SGs and intracellular protein aggregates may interact. Remarkably, aggregates of mutant huntingtin formed in cell lines may include TIA1 [169]. In a related study, Roucou and co-workers, reported that prion protein (PrP) forms aggresomes that sequester polyadenylated RNA, and interferes with SG formation and hsp70 expression, thus affecting cell survival [170]. More recently, attention has been focused on TDP43 and FUS/TLS, two RNA-binding proteins that interact and form aggregates in several neurodegenerative conditions [156], [159], [171], [172], [173]. TDP 43 contains an aggregation domain (Table 1), and it was recently shown to be present in intracellular aggregates in Amyotrophic Lateral Sclerosis, Alzheimer's disease and frontotemporal dementia.
Table 4.
Similarities between SGs and unfolded protein aggregates. SGs and aggresomes share components and their assembly is mechanistically linked, suggesting a role for SGs in unfolded protein diseases (see text).
| Stress Granules | Unfolded protein aggregates |
|---|---|
| Similarities | |
| Induced by several stressors | Present in several pathologies (ER-stress and/or oxidative stress involved) [208] |
| Induced by proteasome or autophagy inhibitors | Dissolution requires proteosome activity or autophagy. [209] |
| Aggregation modulated by HSP70 | Miss-folded protein aggregation modulated by chaperones. |
| Contain ubiquitinated proteins | Contain ubiquitinated proteins. [210], [211] |
| Contain O-glycosilated proteins | Contain O-glycosilated proteins [212] |
| Microtubule and dynein-dependent | Microtubule and dynein-dependent. [213], [214] |
| Differences | |
| Highly dynamic | Quite static [214] |
| Transient | Long-lived |
| Protective | Pathogenic or protective |
Pathological TDP43 aggregates are distinct from SGs, but TDP43 fragments are able to induce, and are recruited to SGs [156], [159], [171], [172], [173]. Among several possibilities, it remains open whether the eventual formation of SGs containing TDP43 fragments might initiate the irreversible aggregation of pathogenic TDP43.
A role for SGs in TLS/FUS-related pathologies is strongly suggested by the fact that the pathologic aggregates include the SG components PABP and eIF4G [159]. Moreover, TLS/FUS pathogenic mutants induce SGs and pathogenicity correlates with their recruitment to SGs [159], [174]. A model has been proposed where cellular stress initiates aggregation of FUS/TLS, which will be facilitated by specific mutations [159], and perpetuated during the disease. Vice-versa, whether the presence of TDP43 or TLS/FUS inclusions that retain SG components interferes with SG formation and function, thus affecting the stress response and cell survival is an open question.
8. Other RNA granules induced upon stress
8.1. Ire1 foci
A new type of stress-induced foci distinct from SGs and PBs was recently described in yeast cells by Walter and co-workers [175], [176]. These foci contain the ER-associated endonuclease Ire1, and a substrate mRNA encoding the transcription factor HAC1. Briefly, Ire1 initiates the splicing of the translationally repressed HAC1 mRNA, which occurs in the cytoplasm, allowing its translation and the consequent expression of protective genes. Endonucleolytic cleavage of HAC1 transcripts requires Ire1 oligomerization. Two protein surfaces in Ire1 mediate homotypic interactions, allowing the formation of visible foci that contain tens of Ire1 molecules. Ire1 aggregation does not require HAC1 mRNA, although the contribution of other mRNA substrates, such as the transcripts degraded by RIDD remains open.
8.2. Yeast UV RNA Granules
Exposure of cells to UV provokes RNA damage. Gaillard and Aguilera [177] reported that in yeast, damaged transcripts accumulate in specific granules termed UV RNA Granules (UVGs), which are distinct from PB, EGP bodies, aggresomes and SGs. Strikingly, UVGs are not induced by other stressors and are not mRNA silencing foci, as they are insensitive to polysome-stabilizing drugs. UVGs protect cells from damaged RNA that could be deleterious if engaged in translation.
8.3. Nuclear Stress Bodies
The exposure of primate cells to heat, heavy metals, aminoacid deprivation or proteasome inhibitors provokes the transient formation of highly packed ribonucleoprotein complexes in the nucleus, termed Nuclear Stress Bodies (nSBs) (see [178] for a recent review). nSBs contain HSF (Heat Shock Transcription Factor) [179] and polyadenylated transcripts from satellite III repeats, which are transcribed by RNA polymerase II during the stress response [180], [181]. The formation and maintenance of nSBs depend on ongoing transcription, and in addition to Pol II, nSBs contain several protein factors involved in transcription and RNA processing, such as HAP (hnRNP A1 interacting protein) [182], hnRNP M (heterogeneous nuclear Ribonucleoprotein M) [183], CBP (CREB Binding Protein) and acetylated histones [180], and the splicing factors SF2/ASF, SRp300, and Sam68 (KH domain-containing, RNA-binding, signal transduction-associated protein 1) [182]. However, only a small fraction of satellite transcripts are spliced, and none of them are found in the cytoplasm [180]. It is speculated that the recruitment of transcription and splicing factors to nSBs reduces the availability of these molecules in splicing speckles and in the nucleoplasm, thus affecting transcription and splicing of a number of messenger RNAs [182]. Like SGs, nSBs are induced shortly upon stress induction, and both foci decay after reaching a maximum at around 1–3 h [180]. Interestingly, a number of splicing factors present in SGs –namely TIA1/TIAR, hnRNP A1, Sam68– are also recruited to nSBs [182]. The entrapment of these molecules in both nuclear and cytoplasmic macromolecular aggregates may reinforce splicing regulation under stress.
9. Concluding remarks
The spontaneous formation of microscopically visible aggregates that concentrate molecules involved in a given pathway is not exclusive of mRNA metabolism. An et al. reported the formation of the so-called “purinosome” in mammalian cells, which include the enzymes for purine biosynthesis and assemble when purine levels decrease [184]. Similar structures were recently reported in yeast cells [185]. The purine biosynthetic enzyme Ade4, and the glutamine synthetase reversibly concentrate in discrete foci upon starvation, and dissolve in the presence of the specific end-product metabolite. Like in the case of the mRNA silencing foci described in this review, aggregation of these factors apparently depends on the presence of self-aggregation domains [185]. Also in yeast cells, Ashe and co-workers reported the presence of discrete foci, termed eIF2B bodies, that concentrates eIF2 and the cognate guanine-exchange factor eIF2B [149], [150]. The integrity and dynamics of eIF2B bodies, which are distinct from PBs and SGs, is important for eIF2 recycling [151]. In all these cases, the aggregation of enzymes and factors that act successively may optimize the process, channeling substrates through the pathway and minimizing diffusion to the cytosol.
In addition, aggregates may represent storage sites, that may help to protect unemployed molecules from decay [185], [186], [187]. Aggregation is also a strategy to protect the cell from deleterious molecules, and both unfolded proteins and damaged RNA can be packed off in specific compartments [177].
Additional significance for the formation of macromolecular structures with complex composition may be linked to the sequestration of key molecules. In this way, the inhibition or release of distinct factors, such as the pro-apoptotic proteins recruited to SGs, can be regulated in a coordinate manner, and from a few spots in the cell cytoplasm. The existence of specialized foci in the nucleus including splicing speckles, Cajal bodies, and nucleolus, among others, has been known for long. These nuclear machineries have a primary function in RNA biogenesis and processing, and may also serve as a depot for controlling the availability of specific molecules.
Finally, aggregation of distinct RNA-binding proteins was shown to be instrumental for their function. Oligomerization is required for Ire1 activity [175]. Translation of certain IRES is helped by oligomerization of the trans-acting factor PCP2 [188], which is present in both PBs and SGs. Similarly, aggregation mediated by a prion-related domain present in CPEB is relevant to mRNA regulation in neurons, with a dramatic impact on neuronal activation [189], [190].
PBs and SGs are complex structures that come in subtly different flavors, and they appear to be involved in several pathways that control mRNA metabolism, cell signalling and survival. Further investigation on their dynamics and composition would contribute to understand the functional relevance of these intriguing structures.
Acknowledgements
We are grateful to Dr. L. J. Martinez Tosar (FCEyN-UBA, Argentina) for critical reading of the manuscript, Dr. A. Cassola and Dr. C. Frasch, (Universidad Nacional de San Martín, Argentina), Dr. A. Cayota (Institut Pasteur de Montevideo, Montevideo, Uruguay), and L. Luchelli (Instituto Leloir, Bs. As., Argentina) for kindly providing the figure panels. This work was supported by the following grants to GLB: X834 from UBA, Argentina; PIP 6173 from Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET); PICT 38006 and PICT 1965 from Agencia Nacional de Promoción Científica y Tecnológica, (ANPCyT), Argentina and 1R03 TW 006037–01A1, NIH, USA. GLB and MGT are investigators of CONICET; M.L. is recipient of the “Cardini Fellowship” from the Instituto Leloir, and of a fellowship from CONICET.
References
- 1.Bashkirov V.I. J. Cell Biol. 1997;136(4):761. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Dijk E. EMBO J. 2002;21(24):6915. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ingelfinger D. RNA. 2002;8(12):1489. [PMC free article] [PubMed] [Google Scholar]
- 4.Sheth U., Parker R. Science. 2003;300(5620):805. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teixeira D. RNA. 2005;11(4):371. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brengues M., Teixeira D., Parker R. Science. 2005;310(5747):486. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharyya S.N. Cell. 2006;125(6):1111. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 8.Franks T.M., Lykke-Andersen J. Genes Dev. 2007;21(6):719. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franks T.M., Lykke-Andersen J. Mol. Cell. 2008;32(5):605. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchan J.R., Parker R. Mol. Cell. 2009;36(6):932. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulkarni M., Ozgur S., Stoecklin G. Biochem. Soc. Trans. 2010;38(Pt 1):242. doi: 10.1042/BST0380242. [DOI] [PubMed] [Google Scholar]
- 12.Eulalio A., Behm-Ansmant I., Izaurralde E. Nat. Rev. Mol. Cell Biol. 2007;8(1):9. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 13.Kedersha N., Anderson P. Meth. Enzymol. 2007;431:61. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 14.Anderson P., Kedersha N. J. Cell Sci. 2002;115(Pt 16):3227. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- 15.Anderson P., Kedersha N. Trends Biochem. Sci. 2008;33(3):141. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Thomas M.G. J. Cell Sci. 2009;122(Pt 4):563. doi: 10.1242/jcs.038208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loschi M. J. Cell Sci. 2009;122(Pt 21):3973. doi: 10.1242/jcs.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchan J.R., Muhlrad D., Parker R. J. Cell Biol. 2008;183(3):441. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson P., Kedersha N. J. Cell Biol. 2006;172(6):803. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kedersha N. J. Cell Biol. 2005;169(6):871. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilczynska A. J. Cell Sci. 2005;118(Pt 5):981. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- 22.Thomas M.G. Mol. Biol. Cell. 2005;16(1):405. doi: 10.1091/mbc.E04-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kedersha N.L. J. Cell Biol. 1999;147(7):1431. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dang Y. J. Biol. Chem. 2006;281(43):32870. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- 25.Emara M.M. J. Biol. Chem. 2010;285(14):10959. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kedersha N. Mol. Biol. Cell. 2002;13(1):195. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoecklin G. EMBO J. 2004;23(6):1313. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farny N.G., Kedersha N.L., Silver P.A. RNA. 2009;15(10):1814. doi: 10.1261/rna.1684009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kramer S. J. Cell Sci. 2008;121(Pt 18):3002. doi: 10.1242/jcs.031823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cassola A., De Gaudenzi J.G., Frasch A.C. Mol. Microbiol. 2007;65(3):655. doi: 10.1111/j.1365-2958.2007.05833.x. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Silva M.R. Mol. Biochem. Parasitol. 2010;171(2):64. doi: 10.1016/j.molbiopara.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Dunand-Sauthier I. Mol. Biol. Cell. 2002;13(5):1626. doi: 10.1091/mbc.01-06-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grousl T. J. Cell Sci. 2009;122(Pt 12):2078. doi: 10.1242/jcs.045104. [DOI] [PubMed] [Google Scholar]
- 34.Swisher K.D., Parker R. PLoS ONE. 2010;5(4):e10006. doi: 10.1371/journal.pone.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jud M.C. Dev. Biol. 2008;318(1):38. doi: 10.1016/j.ydbio.2008.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uniacke J., Zerges W. J. Cell Biol. 2008;182(4):641. doi: 10.1083/jcb.200805125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brengues M., Parker R. Mol. Biol. Cell. 2007;18(7):2592. doi: 10.1091/mbc.E06-12-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoyle N.P. J. Cell Biol. 2007;179(1):65. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Degracia D.J. Neuroscience. 2006;146(2):562. doi: 10.1016/j.neuroscience.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kayali F. Neuroscience. 2005;134(4):1223. doi: 10.1016/j.neuroscience.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 41.Moeller B.J. Cancer Cell. 2004;5(5):429. doi: 10.1016/s1535-6108(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 42.Nakano S. Neurology. 2005;65(3):420. doi: 10.1212/01.wnl.0000171341.76482.15. [DOI] [PubMed] [Google Scholar]
- 43.Dekanty A. PLoS Genet. 2010;6(6):e1000994. doi: 10.1371/journal.pgen.1000994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eulalio A. Mol. Cell. Biol. 2007;27(11):3970. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tritschler F. Proc. Natl. Acad. Sci. USA. 2009;106(51):21591. doi: 10.1073/pnas.0909871106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reijns M.A. J. Cell Sci. 2008;121(Pt 15):2463. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Decker C.J., Teixeira D., Parker R. J. Cell Biol. 2007;179(3):437. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tritschler F. Mol. Cell. Biol. 2007;27(24):8600. doi: 10.1128/MCB.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eulalio A. Nucleic Acids Res. 2009;37(9):2974. doi: 10.1093/nar/gkp173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu J.H. RNA. 2005;11(12):1795. doi: 10.1261/rna.2142405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu J. Plant Cell. 2006;18(12):3386. doi: 10.1105/tpc.106.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eulalio A. Genes Dev. 2007;21(20):2558. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jinek M. RNA. 2008;14(10):1991. doi: 10.1261/rna.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fenger-Gron M. Mol. Cell. 2005;20(6):905. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 55.Fujimura K., Kano F., Murata M. RNA. 2008;14(3):425. doi: 10.1261/rna.780708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cougot N. J. Neurosci. 2008;28(51):13793. doi: 10.1523/JNEUROSCI.4155-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.di Penta A. J. Cell Biol. 2009;184(3):423. doi: 10.1083/jcb.200807033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu J.L., Gall J.G. Proc. Natl. Acad. Sci. USA. 2007;104(28):11655. doi: 10.1073/pnas.0704977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Z. J. Cell Sci. 2004;117(Pt 23):5567. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- 60.Mollet S. Mol. Biol. Cell. 2008;19(10):4469. doi: 10.1091/mbc.E08-05-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aizer A. Mol. Biol. Cell. 2008;19(10):4154. doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kedersha N. J. Cell Biol. 2005;169(6):871. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Solomon S. Mol. Cell. Biol. 2007;27(6):2324. doi: 10.1128/MCB.02300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tourriere H. J. Cell Biol. 2003;160(6):823. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Mazroui R. Hum. Mol. Genet. 2003;12(23):3087. doi: 10.1093/hmg/ddg335. [DOI] [PubMed] [Google Scholar]
- 66.Athanasopoulos V. FEBS J. 2010;277(9):2109. doi: 10.1111/j.1742-4658.2010.07628.x. [DOI] [PubMed] [Google Scholar]
- 67.Kedersha N. J. Cell Biol. 2000;151(6):1257. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leung A.K., Calabrese J.M., Sharp P.A. Proc. Natl. Acad. Sci. USA. 2006;103(48):18125. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin M.D. Dev. Biol. 2008;322(2):276. doi: 10.1016/j.ydbio.2008.07.033. [DOI] [PubMed] [Google Scholar]
- 70.Holt C.E., Bullock S.L. Science. 2009;326(5957):1212. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Besse F., Ephrussi A. Nat. Rev. Mol. Cell Biol. 2008;9(12):971. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- 72.Sutton M.A., Schuman E.M. Cell. 2006;127(1):49. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 73.Schuman E.M., Dynes J.L., Steward O. J. Neurosci. 2006;26(27):7143. doi: 10.1523/JNEUROSCI.1796-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Steward O. Neuron. 1997;18(1):9. doi: 10.1016/s0896-6273(01)80041-6. [DOI] [PubMed] [Google Scholar]
- 75.Steward O. Neuron. 2002;36(3):338. doi: 10.1016/s0896-6273(02)01006-1. [DOI] [PubMed] [Google Scholar]
- 76.Steward O., Schuman E.M. Neuron. 2003;40(2):347. doi: 10.1016/s0896-6273(03)00635-4. [DOI] [PubMed] [Google Scholar]
- 77.Zalfa F., Achsel T., Bagni C. Curr. Opin. Neurobiol. 2006;16(3):265. doi: 10.1016/j.conb.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 78.Bagni C., Greenough W.T. Nat. Rev. Neurosci. 2005;6(5):376. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- 79.Costa-Mattioli M. Neuron. 2009;61(1):10. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Piotrowska J. J. Virol. 2010;84(7):3654. doi: 10.1128/JVI.01320-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gebauer F., Hentze M.W. Nat. Rev. Mol. Cell Biol. 2004;5(10):827. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Holcik M., Sonenberg N. Nat. Rev. Mol. Cell Biol. 2005;6(4):318. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 83.Wek R.C., Jiang H.Y., Anthony T.G. Biochem. Soc. Trans. 2006;34(Pt 1):7. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 84.Vonlaufen N. Cell. Microbiol. 2008;10(12):2387. doi: 10.1111/j.1462-5822.2008.01210.x. [DOI] [PubMed] [Google Scholar]
- 85.Moraes M.C. Eukaryot. Cell. 2007;6(11):1979. doi: 10.1128/EC.00249-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rafie-Kolpin M., Han A.P., Chen J.J. Biochemistry. 2003;42(21):6536. doi: 10.1021/bi034005v. [DOI] [PubMed] [Google Scholar]
- 87.McEwen E. J. Biol. Chem. 2005;280(17):16925. doi: 10.1074/jbc.M412882200. [DOI] [PubMed] [Google Scholar]
- 88.Wehner K.A., Schutz S., Sarnow P. Mol. Cell. Biol. 2010;30(8):2006. doi: 10.1128/MCB.01350-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ben-Asouli Y. Cell. 2002;108(2):221. doi: 10.1016/s0092-8674(02)00616-5. [DOI] [PubMed] [Google Scholar]
- 90.Cuesta R., Laroia G., Schneider R.J. Genes Dev. 2000;14(12):1460. [PMC free article] [PubMed] [Google Scholar]
- 91.Fu H. FEBS Lett. 2009;583(2):437. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 92.Thompson D.M. RNA. 2008;14(10):2095. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thompson D.M., Parker R. J. Cell Biol. 2009;185(1):43. doi: 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yamasaki S. J. Cell Biol. 2009;185(1):35. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li Y. Nucleic Acids Res. 2008;36(19):6048. doi: 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee S.R., Collins K. J. Biol. Chem. 2005;280(52):42744. doi: 10.1074/jbc.M510356200. [DOI] [PubMed] [Google Scholar]
- 97.Zhang S., Sun L., Kragler F. Plant Physiol. 2009;150(1):378. doi: 10.1104/pp.108.134767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jochl C. Nucleic Acids Res. 2008;36(8):2677. doi: 10.1093/nar/gkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Elbarbary R.A. PLoS ONE. 2009;4(6):e5908. doi: 10.1371/journal.pone.0005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schaefer M. Gene Dev. 2010 Aug 1;24(15):1590. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pothof J. Cell Cycle. 2009;8(21):3462. doi: 10.4161/cc.8.21.9835. [DOI] [PubMed] [Google Scholar]
- 102.Mokas S. Mol. Biol. Cell. 2009;20(11):2673. doi: 10.1091/mbc.E08-10-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Baez M.V., Boccaccio G.L. J. Biol. Chem. 2005;280(52):43131. doi: 10.1074/jbc.M508374200. [DOI] [PubMed] [Google Scholar]
- 104.Unsworth H. FASEB J. 2010 [Google Scholar]
- 105.Sivan G., Kedersha N., Elroy-Stein O. Mol. Cell. Biol. 2007;27(19):6639. doi: 10.1128/MCB.00798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kiebler M.A. J. Neurosci. 1999;19(1):288. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Marion R.M. Mol. Cell. Biol. 1999;19(3):2212. doi: 10.1128/mcb.19.3.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li C.H. PLoS ONE. 2010;5(4):e9942. doi: 10.1371/journal.pone.0009942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Meignin C., Davis I. Curr. Opin. Cell Biol. 2010;22(1):112. doi: 10.1016/j.ceb.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 110.Martin K.C., Ephrussi A. Cell. 2009;136(4):719. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yoshimura A. Curr. Biol. 2006;16(23):2345. doi: 10.1016/j.cub.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 112.Heuck A. J. Cell Biol. 2010;189(3):497. doi: 10.1083/jcb.201002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chang W. RNA. 2008;14(3):491. doi: 10.1261/rna.665008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kolobova E. Exp. Cell Res. 2009;315(3):542. doi: 10.1016/j.yexcr.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fujimura K. Biochim. Biophys. Acta. 2009;1793(11):1728. doi: 10.1016/j.bbamcr.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 116.Ivanov P.A., Chudinova E.M., Nadezhdina E.S. Exp. Cell Res. 2003;290(2):227. doi: 10.1016/s0014-4827(03)00290-8. [DOI] [PubMed] [Google Scholar]
- 117.Kwon S., Zhang Y., Matthias P. Genes Dev. 2007;21(24):3381. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tsai N.P., Tsui Y.C., Wei L.N. Neuroscience. 2009;159(2):647. doi: 10.1016/j.neuroscience.2008.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Loiseau P. Development. 2010;137(16):2763. doi: 10.1242/dev.048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bisbal M. J. Biol. Chem. 2009;284(14):9489. doi: 10.1074/jbc.M808586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fujimura K. Biochim. Biophys. Acta. 2010;1803(7):865. doi: 10.1016/j.bbamcr.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 122.Mazroui R. Mol. Biol. Cell. 2007;18(7):2603. doi: 10.1091/mbc.E06-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Baguet A. J. Cell Sci. 2007;120(Pt 16):2774. doi: 10.1242/jcs.009225. [DOI] [PubMed] [Google Scholar]
- 124.Tsai N.P., Wei L.N. Cell. Signal. 2010;22(4):668. doi: 10.1016/j.cellsig.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chang W.L., Tarn W.Y. Nucleic Acids Res. 2009;37(19):6600. doi: 10.1093/nar/gkp717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sinsimer K.S. Mol. Cell. Biol. 2008;28(17):5223. doi: 10.1128/MCB.00431-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Didiot M.C. Mol. Biol. Cell. 2009;20(1):428. doi: 10.1091/mbc.E08-07-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Carpio M.A. Biochem. J. 2010;429(1):63. doi: 10.1042/BJ20091953. [DOI] [PubMed] [Google Scholar]
- 129.Yang F. Mol. Cell. Biol. 2006;26(23):8803. doi: 10.1128/MCB.00090-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lopez de Silanes I. Mol. Cell. Biol. 2005;25(21):9520. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hollien J., Weissman J.S. Science. 2006;313(5783):104. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 132.Hollien J. J. Cell Biol. 2009;186(3):323. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Stohr N. J. Cell Biol. 2006;175(4):527. doi: 10.1083/jcb.200608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tsai N.P., Ho P.C., Wei L.N. EMBO J. 2008;27(5):715. doi: 10.1038/emboj.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yoon J.H., Choi E.J., Parker R. J. Cell Biol. 2010;189(5):813. doi: 10.1083/jcb.200912019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Blackwell E., Zhang X., Ceman S. Hum. Mol. Genet. 2010;19(7):1314. doi: 10.1093/hmg/ddq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Blumenthal J. FEBS Lett. 2009;583(1):197. doi: 10.1016/j.febslet.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 138.Ohn T. Nat. Cell Biol. 2008;10(10):1224. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hu P., Shimoji S., Hart G.W. FEBS Lett. 2010 Jun 18;584(12):2526. doi: 10.1016/j.febslet.2010.04.044. [DOI] [PubMed] [Google Scholar]
- 140.Wasserman T. Mol. Biol. Cell. 2010;21(1):117. doi: 10.1091/mbc.E09-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Brehm M.A. Biochem. J. 2007;408(3):335. doi: 10.1042/BJ20070382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Morfini G.A. J. Neurosci. 2009;29(41):12776. doi: 10.1523/JNEUROSCI.3463-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lin J.C., Hsu M., Tarn W.Y. Proc. Natl. Acad. Sci. USA. 2007;104(7):2235. doi: 10.1073/pnas.0611015104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kim W.J., Kim J.H., Jang S.K. EMBO J. 2007;26(24):5020. doi: 10.1038/sj.emboj.7601920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Eisinger-Mathason T.S. Mol. Cell. 2008;31(5):722. doi: 10.1016/j.molcel.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Arimoto K. Nat. Cell Biol. 2008;10(11):1324. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- 147.Kimball S.R. Am. J. Physiol. Cell Physiol. 2003;284(2):C273. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- 148.Roy B. RNA. 2010 Apr 16;16(4):748. doi: 10.1261/rna.2056010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Campbell S.G., Hoyle N.P., Ashe M.P. J. Cell Biol. 2005;170(6):925. doi: 10.1083/jcb.200503162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Campbell S.G., Ashe M.P. Cell Cycle. 2006;5(7):678. doi: 10.4161/cc.5.7.2607. [DOI] [PubMed] [Google Scholar]
- 151.Taylor E.J. Mol. Biol. Cell. 2010;13:2202. doi: 10.1091/mbc.E09-11-0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kim W.J. Mol. Cell. Biol. 2005;25(6):2450. doi: 10.1128/MCB.25.6.2450-2462.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yoneda T. J. Biol. Chem. 2001;276(17):13935. doi: 10.1074/jbc.M010677200. [DOI] [PubMed] [Google Scholar]
- 154.Zeitelhofer M. J. Neurosci. 2008;28(30):7555. doi: 10.1523/JNEUROSCI.0104-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Vessey J.P. J. Neurosci. 2006;26(24):6496. doi: 10.1523/JNEUROSCI.0649-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Colombrita C. J. Neurochem. 2009;111(4):1051. doi: 10.1111/j.1471-4159.2009.06383.x. [DOI] [PubMed] [Google Scholar]
- 157.Bechade C. Eur. J. Neurosci. 1999;11(1):293. doi: 10.1046/j.1460-9568.1999.00428.x. [DOI] [PubMed] [Google Scholar]
- 158.Hua Y., Zhou J. Biochem. Biophys. Res. Commun. 2004;314(1):268. doi: 10.1016/j.bbrc.2003.12.084. [DOI] [PubMed] [Google Scholar]
- 159.Dormann D. EMBO J. 2010 Aug 18;29(16):2841. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Costa-Mattioli M. Nature. 2005;436(7054):1166. doi: 10.1038/nature03897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Costa-Mattioli M. Cell. 2007;129(1):195. doi: 10.1016/j.cell.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mazroui R. Hum. Mol. Genet. 2002;11(24):3007. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- 163.Dubnau J. Curr. Biol. 2003;13(4):286. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 164.Vessey J.P. Proc. Natl. Acad. Sci. USA. 2008;105(42):16374. doi: 10.1073/pnas.0804583105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kim Y.K. Cell. 2005;120(2):195. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 166.Hua Y., Zhou J. FEBS Lett. 2004;572(1–3):69. doi: 10.1016/j.febslet.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 167.De Rubeis S., Bagni C. Mol. Cell. Neurosci. 2010;43(1):43. doi: 10.1016/j.mcn.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 168.Nonhoff U. Mol. Biol. Cell. 2007;18(4):1385. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Waelter S. Mol. Biol. Cell. 2001;12(5):1393. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Goggin K. Biochim. Biophys. Acta. 2008;1783(3):479. doi: 10.1016/j.bbamcr.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 171.Nishimoto Y. J. Biol. Chem. 2010 Jan 1;285(1):608. doi: 10.1074/jbc.M109.022012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Buratti E., Baralle F.E. RNA Biol. 2010;7(4) doi: 10.4161/rna.7.4.12153. [DOI] [PubMed] [Google Scholar]
- 173.Ling S.C. Proc. Natl. Acad. Sci. USA. 2010;107(30):13318. doi: 10.1073/pnas.1008227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gal J. Neurobiol. Aging. 2010 Jul 29 [Epub ahead of print] [Google Scholar]
- 175.Aragon T. Nature. 2009;457(7230):736. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Korennykh A.V. Nature. 2009;457(7230):687. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gaillard H., Aguilera A. Mol. Biol. Cell. 2008;19(11):4980. doi: 10.1091/mbc.E08-02-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Biamonti G., Vourc'h C. Cold Spring Harb. Perspect. Biol. 2010;2(6):a000695. doi: 10.1101/cshperspect.a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sarge K.D., Murphy S.P., Morimoto R.I. Mol. Cell. Biol. 1993;13(3):1392. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Jolly C. J. Cell Biol. 2004;164(1):25. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rizzi N. Mol. Biol. Cell. 2004;15(2):543. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Denegri M. Mol. Biol. Cell. 2001;12(11):3502. doi: 10.1091/mbc.12.11.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mahl P. J. Cell Biol. 1989;109(5):1921. doi: 10.1083/jcb.109.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.An S. Science. 2008;320(5872):103. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- 185.Narayanaswamy R. Proc. Natl. Acad. Sci. USA. 2009;106(25):10147. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ray P.S., Arif A., Fox P.L. Trends Biochem. Sci. 2007;32(4):158. doi: 10.1016/j.tibs.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 187.Sagot I. Mol. Biol. Cell. 2006;17(11):4645. doi: 10.1091/mbc.E06-04-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bedard K.M., Walter B.L., Semler B.L. RNA. 2004;10(8):1266. doi: 10.1261/rna.7070304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Si K., Lindquist S., Kandel E.R. Cell. 2003;115(7):879. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- 190.Si K. Cell. 2010;140(3):421. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 191.Eulalio A., Tritschler F., Izaurralde E. RNA. 2009;15(8):1433. doi: 10.1261/rna.1703809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gilks N. Mol. Biol. Cell. 2004;15(12):5383. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Johnson B.S. Proc. Natl. Acad. Sci. USA. 2008;105(17):6439. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Banerjee P. J. Neurosci. 2010;30(19):6782. doi: 10.1523/JNEUROSCI.6369-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Raaben M. Cell. Microbiol. 2007;9(9):2218. doi: 10.1111/j.1462-5822.2007.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.McInerney G.M. Mol. Biol. Cell. 2005;16(8):3753. doi: 10.1091/mbc.E05-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.White J.P. Cell Host Microbe. 2007;2(5):295. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 198.Emara M.M., Brinton M.A. Proc. Natl. Acad. Sci. USA. 2007;104(21):9041. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Abrahamyan L.G. J. Cell Sci. 2010;123(Pt 3):369. doi: 10.1242/jcs.055897. [DOI] [PubMed] [Google Scholar]
- 200.Montero H. J. Virol. 2008;82(3):1496. doi: 10.1128/JVI.01779-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Smith J.A. J. Virol. 2006;80(4):2019. doi: 10.1128/JVI.80.4.2019-2033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Qin Q., Hastings C., Miller C.L. J. Virol. 2009;83(21):11090. doi: 10.1128/JVI.01239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Esclatine A., Taddeo B., Roizman B. J. Virol. 2004;78(16):8582. doi: 10.1128/JVI.78.16.8582-8592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Isler J.A., Maguire T.G., Alwine J.C. J. Virol. 2005;79(24):15388. doi: 10.1128/JVI.79.24.15388-15397.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Beckham C.J., Parker R. Cell Host Microbe. 2008;3(4):206. doi: 10.1016/j.chom.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mazroui R. Mol. Biol. Cell. 2006 Oct 17;17(10):4212. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mazan-Mamczarz K. Mol. Cell. Biol. 2006;26(7):2716. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Tran P.B., Miller R.J. Trends Neurosci. 1999;22(5):194. doi: 10.1016/s0166-2236(99)01409-5. [DOI] [PubMed] [Google Scholar]
- 209.Filimonenko M. J. Cell Biol. 2007;179(3):485. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Neumann M. Science. 2006;314(5796):130. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 211.Arai T. Biochem. Biophys. Res. Commun. 2006;351(3):602. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 212.Liu F. Proc. Natl. Acad. Sci. USA. 2004;101(29):10804. doi: 10.1073/pnas.0400348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Garcia-Mata R. J. Cell Biol. 1999;146(6):1239. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Johnston J.A., Ward C.L., Kopito R.R. J. Cell Biol. 1998;143(7):1883. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]