Abstract
Introduction
Porphyromonas gingivalis has been associated with subgingival biofilms in adult periodontitis. However, the molecular mechanisms of its contribution to chronic gingival inflammation and loss of periodontal structural integrity remain unclear. The objectives of this investigation were to examine changes in the host transcriptional profiles during a P. gingivalis infection using a murine calvarial model of inflammation and bone resorption.
Methods
P. gingivalis FDC 381 was injected into the subcutaneous soft tissue over the calvaria of BALB/c mice for 3 days, after which the soft tissues and calvarial bones were excised. RNA was isolated from infected soft tissues and calvarial bones and analyzed for transcript profiles using Murine GeneChip® arrays to provide a molecular profile of the events that occur following infection of these tissues.
Results
After P. gingivalis infection, 5517 and 1900 probe sets in the infected soft tissues and calvarial bone, respectively, were differentially expressed (P ≤ 0.05) and up-regulated. Biological pathways significantly impacted by P. gingivalis infection in tissues and calvarial bone included cell adhesion (immune system) molecules, Toll-like receptors, B cell receptor signaling, TGF-β cytokine family receptor signaling, and MHC class II antigen processing pathways resulting in proinflammatory, chemotactic effects, T cell stimulation, and down regulation of antiviral and T cell chemotactic effects. P. gingivalis-induced inflammation activated osteoclasts, leading to local bone resorption.
Conclusion
This is the first in vivo evidence that localized P. gingivalis infection differentially induces transcription of a broad array of host genes that differed between inflamed soft tissues and calvarial bone.
Keywords: P. gingivalis, gene expression, calvarial tissue, bone, microarray
Introduction
Periodontitis is a chronic immuno-inflammatory infectious disease initiated by complex microbial biofilms resulting in destruction of periodontal tissues, resorption of alveolar bone, and exfoliation of teeth. Porphyromonas gingivalis, a gram-negative black pigmented anaerobe, is an important member of the pathogenic biofilm at sites of periodontal disease (18, 25). It may contribute to the disease process by a variety of mechanisms, including, elaborating numerous components that mediate its adherence to mucosal surfaces, enable its penetration into epithelial cells, inhibit host defense mechanisms, and elicit gingival tissue inflammatory destruction (25), resulting in local alveolar bone destruction (18). In vitro studies have shown that components of P. gingivalis can induce a range of proinflammatory cytokines, including IL-1α, IL-1β, TNFα, IL-6, and IL-8, which have been proposed as potential molecular effectors of connective tissue destruction and resorption of alveolar bone in periodontitis (13, 24). However, robust data on the in vivo role of these biomolecules, as well as the broader aspects of the host response in the periodontium remain to be defined.
The use of microarrays to survey transcriptional host responses after exposure to microbial pathogens has become a powerful approach to improve understanding of the molecular basis of the host response to bacterial infections which is critical for limiting tissue destruction. Host response profiling has identified transcripts uniquely affected by the pathogens, Listeria monocytogenes, Brucella abortus, and Mycobacterium tuberculosis. Recent microarray studies have determined in vitro responses of host cells to challenge with P. gingivalis or its virulence components in primary human coronary artery endothelial cells and human aortic endothelial cells (4), a stromal cell line (31), human monocytes (41), a macrophage-like human cell line U937 (33), human periodontal ligament-derived cells (20), human periodontal ligament and gingival fibroblasts (14), gingival fibroblasts from healthy and inflammatory gingival tissues (40), and human gingival epithelial cells (15). A recent review reported on transcriptional profiling of gingival epithelial cell responses to challenge by 4 different microorganisms (16). Only one previous report has described ex vivo gene expression changes in mice in response to infection with P. gingivalis and this was limited to spleen CD4 and CD8 cells (12). Thus, there remains a lack of information concerning P. gingivalis induction of gene expression when the microorganism interacts with host cells and tissues in vivo. The aim of the present study was to define changes in the transcriptome of calvarial bone and overlying soft tissues in response to localized P. gingivalis infection in mice using a calvarial model of acute inflammation and bone resorption.
Materials and Methods
Abbreviations
The following terms used herein are abbreviated as indicated: P. gingivalis (Pg), Gene Ontology (GO), Database for Annotation, Visualization, and Integrated Discovery (DAVID), Expression Analysis Systematic Explorer (EASE), leave-one-out-cross-validation (LOOCV), quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR), β-actin (ActB), interleukin-1 (IL-1), interleukin-1 receptor antagonist (IL-1RA), tumor necrosis factor alpha (TNFα), prostaglandin E receptor 3 (Ptger3), bone morphogenetic protein 2 (Bmp2), parathyroid hormone like peptide (PTHrP), tuftelin 1 (Tuft1), laminin alpha 3 chain precursor (Lama3), tissue inhibitor of proteinase 3 (Timp3), transforming growth factor beta (TGFβ), Toll-like receptor (TLR), mitogen-activated protein kinase (MAPK), matrix metalloproteinases (MMPs), cytochrome P450 family 2 gamma 1 (Cyp2g1), aldo-keto reductase family 1 member C3 (Akr1c18), metallothionein 1 (Mt1), nephronectin (Npnt), matrilin-2 (Matn2), insulin-like growth factor binding protein 4 (Igfbp4), integrin alpha-L/beta-2 (ITGAL/ITGB2), E-selectin (SELE), P-selectin (SELP), nuclear factor kappa B (NF-kB), receptor activated nuclear factor kappa ligand (RANKL), TNF-receptor associated factor (TRAF), intercellular adhesion molecule (ICAM), major histocompatibility complex (MHC), microbe-associated molecular pathogen (MAMP), and signaling adaptor protein (MyD88).
Animals
BALB/c female mice 8–10 weeks of age (Harlan, Indianapolis, IN, USA) were routinely acclimatized for at least 1 week before use. They were infected with P. gingivalis as described below following isoflurane inhalation anesthesia. All procedures were performed in accordance with the approved guidelines of the Institutional Animal Care and Use Committee at the University of Kentucky.
Microorganism and mouse infection
P. gingivalis FDC 381 was cultured and maintained for the animal infections, which were initiated within 15–30 min of bacterial preparation, as we described previously (23) in studies showing that P. gingivalis induces an inflammatory infiltrate, osteoclast activation, and increased mRNA expression of proinflammatory cytokines in calvarial tissues (22, 42). P. gingivalis was injected at 1.5 × 109 (N = 10) into the soft tissues overlying the calvaria of the mice. Bacteria [suspended in 10 µl of reduced transport fluid (RTF)] were injected into the subcutaneous tissue over the right side of the parietal bone and anterior to the lamboid suture once daily for 3 days using a Hamilton syringe (Reno, NV). A control group (N = 10) was injected with vehicle (RTF) once daily for 3 days. Mice were euthanized 8 h after the last injection using CO2 asphyxiation and cervical dislocation. The calvarial bones and overlying soft tissues from 5 mice in each group were excised, snap frozen in liquid nitrogen, and stored at –80°C until RNA isolation. The bone samples consisted predominantly of mature lamellar bone containing osteocytes, with associated osteoblasts and osteoclasts and small amounts of bone marrow. The soft tissue consisted predominantly of the cutaneous epithelium, with subcutaneous and dermis fibroblasts/connective tissue and vascular tissue and cells, and subcuticular fat. The calvarial bones with intact overlying tissue of the remaining 5 mice were fixed in 10% formalin and paraffin embedded for histological assessment of bone changes, as we described previously (42).
RNA isolation and mouse GeneChip hybridization
Total RNA was isolated from the frozen calvarial tissue and calvarial bone from each mouse (P. gingivalis infected and control animals, N = 5 in each group) with Trizol reagent (Invitrogen, CA). Quantification of RNA yield was done by spectrophotometric analysis and the absorbance at 260 and 280 nm was checked to determine the RNA concentration and purity. Quality of the RNA was determined by gel electrophoresis. For GeneChip analyses, RNA samples were further purified with Qiagen RNeasy columns (Qiagen, Valencia, CA). Equal amounts of RNA from samples were labeled and hybridized on a GeneChip following the protocol described in the GeneChip Expression Analysis Technical manual (Affymetrix, Santa Clara, CA). Briefly, reverse transcription was performed on 8 µg of total RNA using SuperScript reverse transcriptase enzyme (Invitrogen, CA) and oligo-(dT)24 primer containing a T7 RNA polymerase promoter. RNA samples were not pooled. The biotin-labeled cRNA was then synthesized by in vitro transcription, fragmented and hybridized on mouse GeneChip MG-MOE430A (Affymetrix, Santa Clara, CA). After hybridization, the GeneChip arrays were stained and scanned in an Affymetrix GCS 3000 7G Scanner. The probe sets represent genes or DNA sequences within genes. Some genes are represented by more than one probe set in the microarray, and hence probe sets are not uniquely correlated to genes. However, for ease of discussion, we use the terms ‘probe sets’ and ‘genes’ interchangeably (11).
Microarray data analysis
The microarray data was normalized and a model-based apparent gene expression matrix was generated using the algorithms implemented in dChip (28). For unsupervised analysis the dataset was analyzed as a whole including arrays hybridized with material derived from both calvarial soft tissue and calvarial bone, and in separate analyses with either calvarial soft tissue or calvarial bone derived specimens. In all cases an initial variation filter was applied to identify probe sets whose hybridization signal intensity varied the most across the respective datasets. For this purpose probe sets were selected that displayed a coefficient of variation greater than 1. The signal intensities of these probe sets were then mean centered and variance normalized to 1 on a probe set-wise basis. The variance normalized data were then subjected to hierarchal clustering using the algorithms implemented in dChip (10, 28).
For supervised analyses the model-based expression matrix was imported into BRB Array tools for high-level analysis. The expression matrix was log transformed and statistical analyses were preformed in log space. Initially the dataset was divided into tissue and treatment classes. Differences between the various treatment tissue classes were determined using a modified f-test with a random variance model and a significance threshold of P < 0.05. To assess the ability of probe sets significant at P < 0.05 to distinguish between the treatment classes, leave-one-out-cross-validation (LOOCV) studies were preformed in which each array was left out in turn, a classification model was built, and the ability of the classifier to correctly identify the array left out (not used in building the classification model) was determined using several prediction models (nearest neighbor analysis, nearest-centroid analysis and other models implemented in BRB Array tools) (11, 15–17). In cases where the misclassification rate was above zero the significance of the classifier’s performance was assessed using Monte Carlo simulations with 100 random permutations of the dataset. The performance of the classifier was considered significant at the P ≤ 0.05 threshold (17).
Genes were categorized into Gene Ontology (GO) and annotated using NetAffx©, an analysis web interface from Affymetrix and DAVID EASE (8, 19). The mean fold change of expression of P. gingivalis infection to uninfected (control) gene expression was calculated using the mean of the 5 replicates. The Gene Ontology database organizes genes into hierarchical categories based on biological process, molecular function and subcellular location. Onto-Express (microarray tool developed at the Intelligent Systems and Bioinformatics Laboratory, Wayne State University) constructs functional profiles; using Gene Ontology terms, for biological process, cellular component and molecular function (9). Gene ontology trees were populated using Pathway Express (21), available at http://vortex.cs.wayne.edu/projects.htm.
Quantitative real-time qRT-PCR analysis
Selected genes that showed significant differential expression in microarray analysis were confirmed for expression by qRT-PCR analysis (LightCycler FastStart DNA Master SYBR Green I, Roche). Selected up-regulated genes from both calvarial soft tissue and bone [cytochrome P450, family 2, gamma 1 (Cyp2g1), aldo-keto reductase family 1, member C3 (Akr1c18), metallothionein 1 (Mt1), nephronectin (Npnt), matrilin-2 (Matn2), insulin-like growth factor binding protein 4 (Igfbp4)] were evaluated. Total RNA (250 ng) from pooled control and P. gingivalis infection samples were reverse transcribed to cDNA using the transcriptor first strand cDNA synthesis kit (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions. Briefly, total RNA was mixed with the anchored-oligo (dT)18 primer (2.5 µM) in a final volume of 13 µl and denatured for 10 min at 65°C and immediately cooled on ice. Then, 5x transcriptor RT reaction buffer, protector RNase inhibitor, deoxynucleotide mix, and transcriptor reverse transcriptase were added to the following final concentrations of 1x, 20U, 1mM each, and 10U, respectively, in a final volume of 20 µl. Transcriptor cDNA synthesis reactions were carried out for 30 min at 55°C, and at 85°C for 5 min for enzyme inactivation. These cDNAs were used for qRT-PCR analyses. QRT-PCR was performed using 2.0 µL of cDNA in a 20-µL reaction for 10 min at 95°C and 40 cycles of 10 s at 95°C, annealing at 60°C followed by amplification at 72°C for 8–10 s. For the determination of the melting curve, the temperature was increased 0.1°C every 1 s from 55°C to 95°C. All PCRs were repeated two or three times. Using the relative quantification application in the LightCycler analysis software, the fold change of the different gene expressions were calculated based on the expression of the target gene versus the expression of the house-keeping gene ActB of the same sample. Mouse β-actin (ActB) gene was used as an internal control based on its constant level of expression across groups in microarray analysis. Standard curves were generated for each gene. To determine the specificity of DNA quantification, a melting curve was generated for each gene.
Relative Standard Curves
Constructing a standard curve with serial dilutions of known template concentration for each target was not feasible. Therefore, dilutions (1:50, 1:100, 1:250, and 1:500) of cDNA of each sample type (control soft tissue and calvarial bone, infected soft tissue and calvarial bone) for a given target gene was used to construct a relative standard curve for that target along with ActB. Template concentrations for reactions in the relative standard were given arbitrary values of 1 (undiluted) 0.02, 0.01, 0.004, and 0.002. The undiluted cDNA from the sample was analyzed as unknown. The fold change in gene expression of the different target genes were calculated as a ratio of the relative expression of the target gene to the relative expression of ActB of the same sample. Each PCR reaction (20 µl) contained 2 µl of the respective cDNA dilution, 1X LightCycler FastStart DNA Master SYBR Green I mix, 0.5 µM primers and 4 mM MgCl2. The amplification was performed under the following conditions: a pre-incubation step of 1 min at 95°C, followed by 45 cycles of 10 s at 95°C, 5 s at 60°C and 8 – 10 s at 72°C (temperature ramp was 20°C/s). Amplification was followed by melting curve analysis using the program run for one cycle at 95°C with 0-second hold, 55°C with 15-second hold, and 95°C with 0-second hold at the step acquisition mode at 0.1°C/s. A negative control without cDNA template was run with every assay to assess the overall specificity. Unless otherwise mentioned, each assay was repeated once or twice. A relative value for the target concentration in each reaction was determined on the basis of the dilution using the LightCycler software, version 4. The concentration of ActB was used to control for input RNA because it is considered a stable housekeeping gene (34). The ActB concentration was determined once for each cDNA sample and used to normalize all other genes tested from the same cDNA sample. The relative change in gene expression was recorded as the ratio of normalized target concentrations in the undiluted cDNA.
Bone histology
The calvariae were fixed in 10% phosphate-buffered formalin and decalcified in 14% EDTA (42). The anterior half of frontal bones and most of the occipital bones were trimmed off, and the parietal bones were cut coronally into two pieces. These half calvariae were then embedded in paraffin with the edges cut coronally placed at the bottom of the cassettes, and four nonconsecutive levels were cut, providing eight coronal sections through each calvaria. Sections (4 µm thick) were stained with hematoxylin and eosin. Histomorphometric analysis of osteoclasts was carried out on two sections from calvaria from each animal. This was accomplished by counting number of osteoclasts throughout the length of the calvarial bone in each section. These bone sections contained the largest number of bone marrow spaces and thus the greatest length of bone surface was available for assessment of resorption.
Antibody analysis
Serum from mice infected with P. gingivalis or control uninfected was used to determine IgG antibody titers to P. gingivalis using a standard ELISA protocol (23), and provided an additional marker of infection. Briefly, P. gingivalis was grown in liquid culture as described above, cells harvested by centrifugation, and the pellets washed with sterile PBS (23). The cells were treated overnight with 0.5% buffered formal saline (FK cells), washed, and diluted to OD600nm=0.3 as coating antigen. Purity and sterility was confirmed using blood agar plating and the antigens stored at −20°C. Diluted infected mice serum (1:100) was incubated in wells of antigen coated microtiter plates for 2 h on a rotator at room temperature, along with a purified mice IgG (Sigma) standard to create a gravimetric curve for antibody quantization. After washing, affinity purified goat anti-mouse IgG (Fc specific) or goat anti-mouse IgM (µ-chain specific) conjugated to alkaline phosphatase (1:5000) was added to the plates and incubated for 2 h. Subsequently, rabbit anti-goat IgG alkaline phosphatase (Sigma) was added to the wells, incubated for 2 h, washed, and the assay developed with p-nitrophenolphosphate (Sigma). The assay reactions were stopped by the addition of 3M NaOH and analyzed at OD405 nm using a Bio-Rad Microplate Reader. The infected mice serum antibody level was quantified using known standard curves. Antigen-coated wells in the absence of serum provided a background negative control. Mean serum IgG antibody levels were derived from triplicate determinations for each mouse serum.
Statistical analyses
Microarray data were analyzed as described above. P values of 0.05 or less were considered significant. The qRT-PCR data from two independent experiments were combined and results were expressed as means ± SD.
RESULTS
Ontology of gene expression changes in murine calvarial soft tissue and bone
The mouse gene chip MOE430A contains 22,690 probe sets, and 11,391 and 1,677 probe sets (P < 0.001) were detected above background in soft tissue and calvarial bone, respectively. Mean gene expression levels of 6,452 and 2341 probe sets were significantly different in soft tissue and bone in response to the P. gingivalis infection (P < 0.05). In addition, expression of 5,517 probes was up-regulated and 935 probes were significantly down-regulated in calvarial soft tissue infection with P. gingivalis (data not shown). In contrast, expression of 1,900 probes was up-regulated and 441 probes were down-regulated significantly in calvarial bone at the site of infection with P. gingivalis (data not shown). P. gingivalis acute infection stimulated changes in a wide array of genes (6452), particularly in the calvarial soft tissue and more than 139, 40, and 15 genes were greater than 10-, 25-, and 50-fold up-regulated, respectively (data not shown). In contrast, all of the 1900 genes up-regulated in bone to P. gingivalis acute infection were less than 5-fold altered. These results also demonstrate a low level of similarity (overlap) in up- and down-regulated genes within the tissues examined.
In order to mine the array data for biologically relevant information, an ontology analysis of known metabolic pathways was performed using individual comparisons of the P. gingivalis infected tissue or bone samples to the corresponding baseline uninfected state. A pair-wise comparison was imposed by the ontology algorithms described above. For this analysis, signal intensities were renormalized across all samples, and supervised analyses were repeated as described above. To populate biological pathways to the maximal extent and thus enhance their predictive power, a significance level of P < 0.05 was adopted. 6452 genes were differentially expressed in the tissue samples, whereas 2341 genes were differentially expressed in bone samples compared to uninfected controls. The ability of probe sets significant at P < 0.05 to correctly identify between treatment groups was confirmed by LOOCV analysis (Supplementary data: Table 1). The classifiers performed flawlessly and correctly predicted the treatment group with 100% accuracy. Probe sets at the P < 0.05 significance level were analyzed by the Pathway Express (PE) algorithm. Pathways significantly impacted by P. gingivalis in bone tissue types included, Antigen Processing and Presentation (Fig. 1), Toll-like Receptor Signaling (Fig. 2 & 3), Cell Adhesion (immune system) Molecules (Fig. 4) and Cytokine-Cytokine Receptor Interaction (Fig. 5). Table 1 shows soft-tissues and calvarial bone pathways generated in this analysis that were predominantly affected in order of their impact factors. P. gingivalis significantly impacted 25 pathways in tissue and 2 in bone with an impact factor above 10. Moreover, Toll-like receptor, B cell receptor, cytokine-cytokine receptor, and MAPK signaling pathways are other major pathways impacted in calvarial soft tissue. Cell adhesion molecules and phosphatidylinositol signaling system pathways were significantly impacted or overlapped in both the calvarial tissue and bone. Interestingly, few classical proinflammatory genes, e.g. IL-1, TNF, IL-6, were altered by the localized P. gingivalis infections using these criteria. However, various genes related to bone remodeling activities, e.g. Ptger3 (prostaglandin E receptor 3), Bmp1 (bone morphogenetic protein 1), Bmp2, Bmp4, PTHrP (parathyroid hormone-related protein), Tuft1 (tuftelin 1), Ptger4 (prostaglandin E receptor 4), and tissue extracellular matrix, eg. Lama3 (laminin alpha 3 chain precursor), Timp1 (tissue inhibitor of proteinase 1), Timp3, were up-regulated in the localized inflamed tissue. These data also emphasize that this localized infection by P. gingivalis appeared to up-regulate genes in the overlying infected tissue that could negatively impact physiologic and remodeling processes in the underlying bone. A few inflammatory/innate immune/acute phase response genes, including serum amyloid A, S100A9 calcium binding protein, and chemokine ligand 2, (e.g. S100a9, S100a8, Saa3, Cp, and Ccl2) were highly up-regulated in inflamed soft tissues.
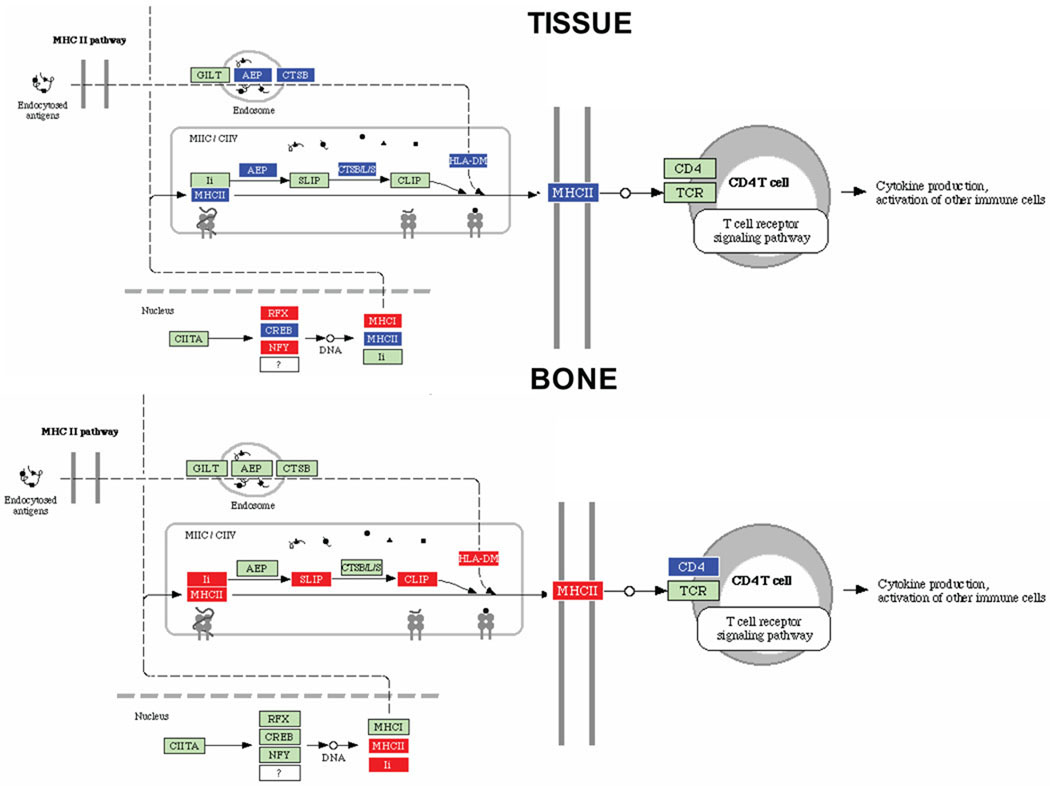
Fig. 1.
MHC class II antigen processing pathways containing P. gingivalis differentially regulated genes in calvarial tissue and calvarial bone at P < 0.05, adopted from Pathway Express and using the Kyoto Encyclopedia of Genes and Genomes nomenclature. Red indicates up regulation, blue indicates repressed, and green indicates no change in gene expression. An arrow indicates a molecular interaction resulting in activation of T cell receptor signaling pathway, and a line without an arrowhead indicates a molecular interaction resulting in inhibition.
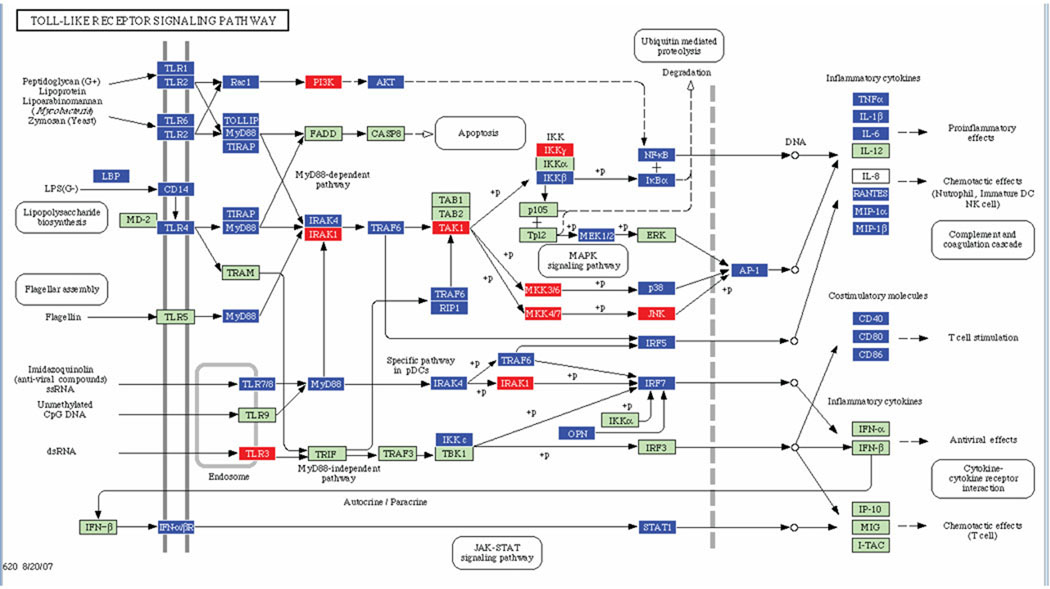
Fig. 2.
Toll-like receptor signaling pathways containing P. gingivalis differentially regulated genes in calvarial tissue at P ≤ 0.05, adopted from Pathway Express and using the Kyoto Encyclopedia of Genes and Genomes nomenclature. Red indicates up regulation, blue indicates repressed, and green indicates no change in gene expression. +P indicates phosphorylation, and −P indicates dephosphorylation. An arrow indicates a molecular interaction resulting in proinflammatory, chemotactic effects, and down-regulation of antiviral and T cell chemotactic effects.
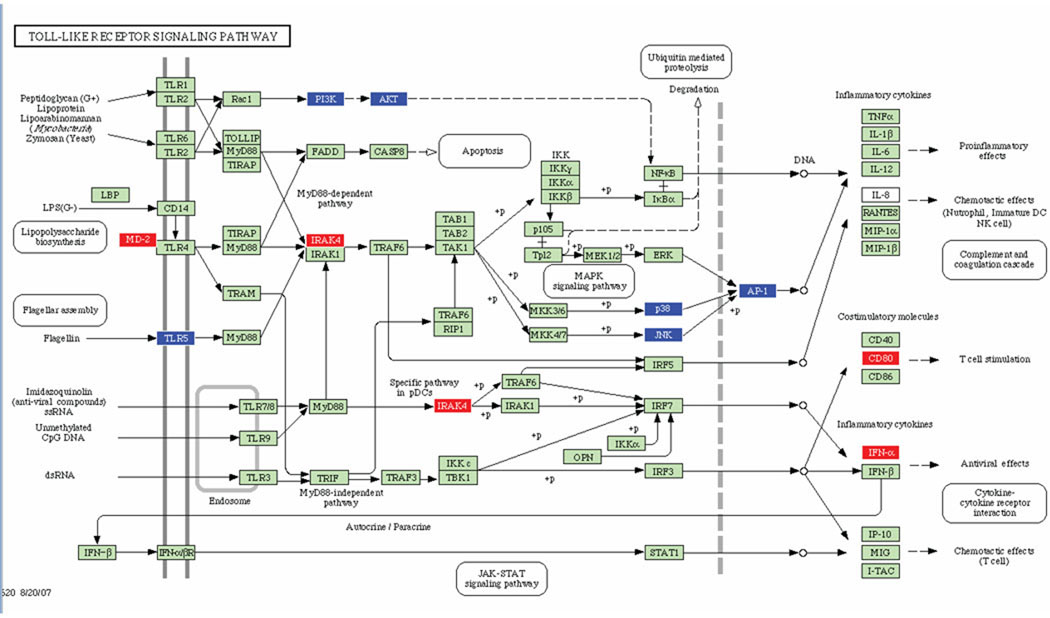
Fig. 3.
Toll-like receptor signaling pathways containing P. gingivalis differentially regulated genes in calvarial bone at P ≤ 0.05. Red indicates up regulation, blue indicates repressed, and green indicates no change in gene expression. +P indicates phosphorylation, and −P indicates dephosphorylation. An arrow indicates a molecular interaction resulting in proinflammatory, chemotactic effects, T. cell stimulation, and down-regulation of antiviral and T cell chemotactic effects.
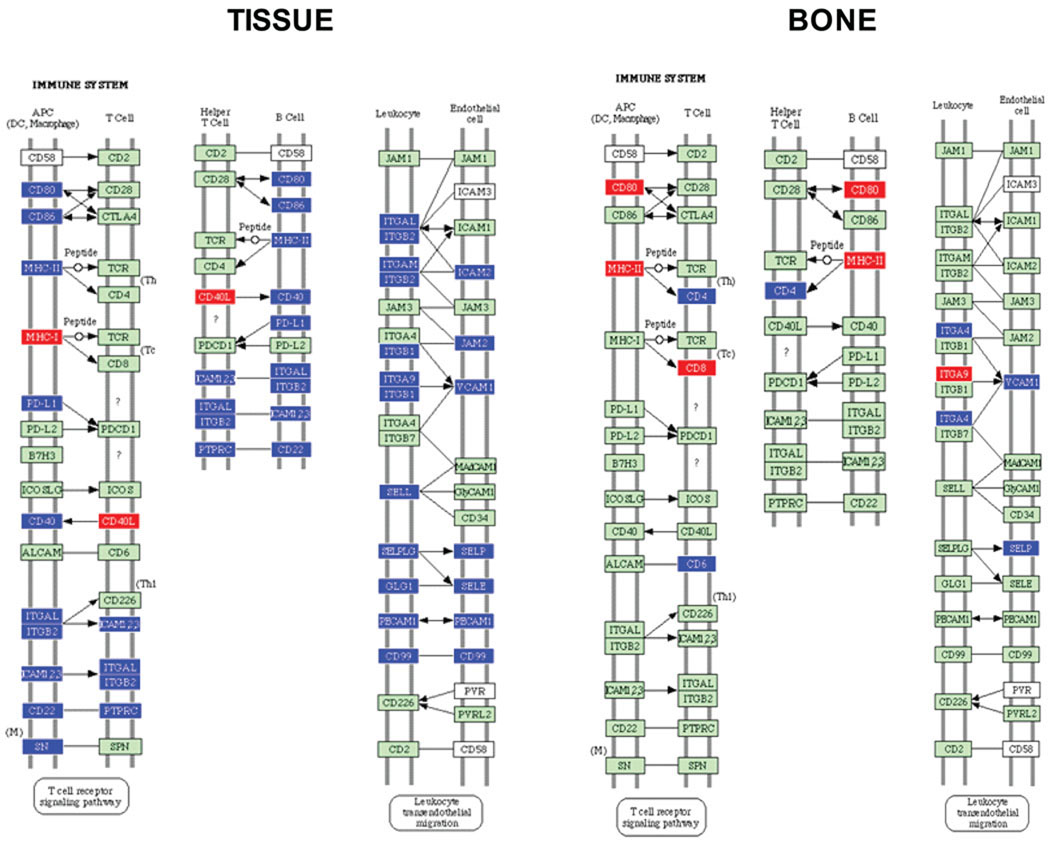
Fig. 4.
Cell adhesion (immune system) molecules pathway containing P. gingivalis differentially regulated genes in calvarial tissue at P ≤ 0.05, adapted from Pathway Express and using the Kyoto Encyclopedia of Genes and Genomes nomenclature. Red indicates up-regulation, blue indicates repressed, and green indicates no change in gene expression. An arrow indicates a molecular interaction resulting in activation, and a line without an arrowhead indicates a molecular interaction resulting in inhibition.
Fig. 5.
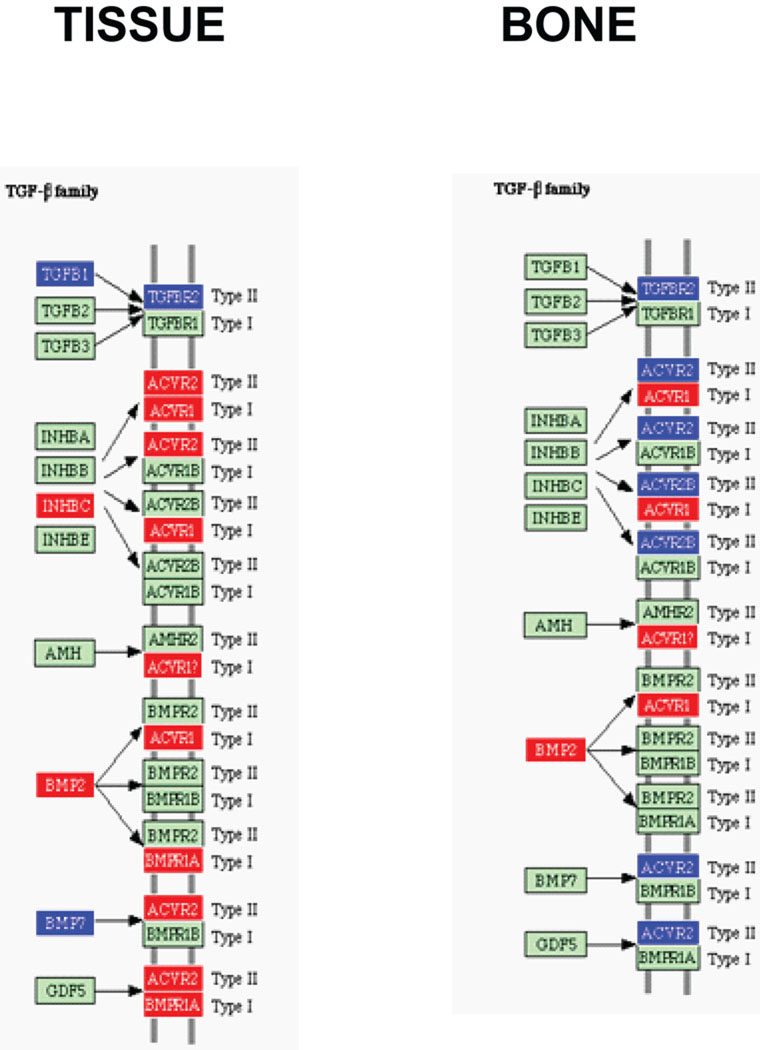
TGF-β cytokine family receptor interaction (Cytokine-cytokine receptor interaction) pathway containing P. gingivalis differentially regulated genes in calvarial tissue and bone at P ≤ 0.05. Red indicates up-regulation, blue indicates repressed, and green indicates no change in gene expression. An arrow indicates a molecular interaction resulting in activation.
Table 1.
Ontology analysis of calvarial tissue and bone gene pathways impacted by infection with P. gingivalis a
| Impacted pathwayb | Impact Factorc | No. of input genes/no. of pathway genesd |
|---|---|---|
| Calvarial tissue | ||
| Adherens junction | 47.991 | 18/74 |
| Cell adhesion molecules | 41.085 | 36/157 |
| Antigen processing and presentation | 36.401 | 17/99 |
| Circadian rhythm | 27.498 | 3/14 |
| Toll-like receptor signaling pathway | 27.299 | 45/101 |
| B cell receptor signaling pathway | 24.705 | 31/66 |
| Apoptosis | 19.262 | 32/81 |
| Cytokine-cytokine receptor interaction | 18.667 | 63/242 |
| Leukocyte transendothelial migration | 18.487 | 39/115 |
| Natural killer cell mediated cytotoxicity | 17.046 | 37/127 |
| Fc epsilon RI signaling pathway | 14.506 | 28/78 |
| Phosphatidylinositol signaling system | 14.256 | 12/70 |
| Insulin signaling pathway | 13.955 | 40/137 |
| Regulation of actin cytoskeleton | 13.618 | 52/206 |
| Chronic myeloid leukemia | 13.599 | 27/77 |
| MAPK signaling pathway | 13.085 | 64/258 |
| ErbB signaling pathway | 13.08 | 28/87 |
| Jak-STAT signaling pathway | 12.531 | 39/151 |
| Focal adhesion | 12.308 | 49/192 |
| T cell receptor signaling pathway | 11.354 | 28/105 |
| GnRH signaling pathway | 11.088 | 30/95 |
| Acute myeloid leukemia | 10.871 | 21/58 |
| Glioma | 10.663 | 21/65 |
| Hematopoietic cell lineage | 10.643 | 27/85 |
| Pancreatic cancer | 10.042 | 23/73 |
| Calvarial bone | ||
| Cell adhesion molecules | 112.874 | 12/157 |
| Phosphatidylinositol signaling system | 33.315 | 4/70 |
Kyoto Encyclopedia of genes and genome pathways (http://www.genome.jp/kegg/).
The impact factor measures the pathways most affected by changes in gene expression in calvarial tissue and bone by considering the proportion of differentially regulated genes, the perturbation factors of all pathway genes, and the propagation of these perturbations throughout the pathway. Only pathways with an impact factor greater than 10 are included in this table.
Number of regulated genes in a pathway/total number of genes currently mapped to this pathway.
Histology
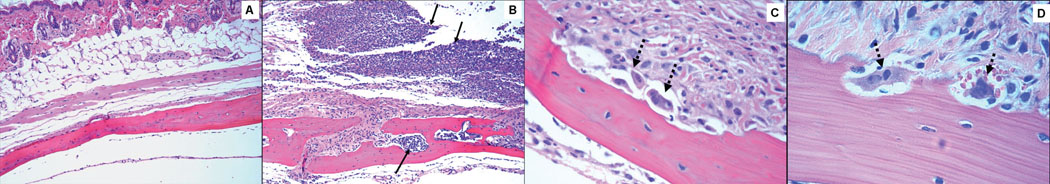
No edema and minimal inflammation were present in the soft tissue over the calvaria of the control uninfected mice (Fig. 6A). Calvarial soft tissue swelling occurred at the injection site within 24 h of the first injection and increased in size following 48 and 72 h in almost all of the mice injected with P. gingivalis, but not with control uninfected mice. None of the P. gingivalis infected mice developed local abscesses, ulceration of the overlying skin, or any evidence of spread of infection to other sites. Histological examination revealed moderate edema and an intense mixed inflammatory cell infiltrate of variable intensity in the calvarial tissues and this large infiltrate consisted of polymorphonuclear leukocytes, lymphocytes, and macrophages (Fig. 6B). Microabscesses and areas of soft tissue necrosis were also evident in sections and where the necrosis abutted the parietal bone. Osteoclasts were also present in some mice on the upper periosteal surface, consistent with more aggressive resorption. In addition, significantly increased numbers of osteoclasts were seen inside the calvaria compared to uninfected controls (data not shown), and these caused an increase in the size of the bone marrow spaces due to increased endosteal bone resorption (Fig. 6C). P. gingivalis cells were not seen in Gram-stained calvarial sections from mice with moderate soft tissue inflammatory infiltrates. Thus, we assumed that the bacteria were removed by host immune cells during the 8 h between the last injection and euthanasia.
Fig. 6.
Effects of local injection of P. gingivalis on mouse calvaria. Live P. gingivalis bacteria (1.5 × 109) were injected once daily for 3 days into the s.c. tissues overlying the calvaria of mice. All photomicrographs of slides stained with hematoxylin and eosin. A. No edema and minimal inflammation present in the soft tissue over the calvaria of the control mouse (magnification × 10). B. Section from P. gingivalis infected mouse reveals enlarged bone marrow spaces with an intense inflammatory cell infiltrate (solid black arrows) and edema present in the adjacent soft tissue and within the marrow spaces (magnification × 10). C. Multiple osteoclasts (dashed black arrows) present in resorption lacunae. (magnification × 40). D. Osteoclasts (dashed black arrows) at higher magnification (× 63).
Validation of microarray gene expression
P. gingivalis-induced transcript expression results from the microarray studies were analyzed by qRT-PCR for selected genes, including Cyp2g1, Akr1c18, Mt1, Npnt, Matn2, Igfbp4 in soft tissue and in calvarial bone from aliquots of the pooled RNA samples used for the microarray (Supplementary data: Table 2). Transcripts of β-actin were used as an expression control and the qRT-PCR analyses were performed at least twice for each gene. The expression levels of Akr1c18, Mt1, Npnt, Matn2 and Igfbp4 genes by qRT-PCR were higher than microarray expression level and the expression levels of Cyp2g1 gene by qRT-PCR were lower than microarray expression levels. The direction and magnitude of expression level of the genes quantified with qRT-PCR generally did not show equivalent expression change with the microarray fold-change outcomes.
Serum IgG antibody in calvarial infections
To provide additional documentation of calvarial infection and to demonstrate an immunological response to P. gingivalis, we evaluated the levels of P. gingivalis specific IgG antibody in mice sera collected at the end of the 3 days of acute infection period. As expected, P. gingivalis infection for 3 days (acute infection) did not induce P. gingivalis-specific serum IgG antibody responses that were not different than those observed in control uninfected mice.
Discussion
Microarray technology continues to evolve as a powerful tool to study host-pathogen interactions and to expand our understanding of the molecular pathogenesis of diseases induced by pathogens (5, 7, 8, 11, 15). Here, we evaluated the gene transcription profiles of host soft tissue and bone in a calvarial bone resorption model during a localized acute infectious challenge with the oral pathogen, P. gingivalis. In the soft tissue P. gingivalis down-regulated several cytokines involved in bone resorption including IL-1β, IL-1α, IL-6, and TNFα, while in bone none of these cytokines was differentially regulated. The result are consistent with previous results in a murine model that showed P. gingivalis infection did not significantly up-regulate bone resorptive cytokines in gingival tissue (35). Moreover, under the conditions of infection in the current study, P. gingivalis did not appear to stimulate the RANKL bone resorption pathway (39), as TRAF6 and NF-κB were down-regulated in tissue and were not altered in bone. One possible explanation for this lack of increase in expression levels of osteoclastogenic cytokines is that the bones were harvested 8 hours after the last injection of bacteria following 3 days of injections and that expression levels had been elevated, but returned to normal levels by the time the bones were sampled. Further studies will be required to examine this possibility. One means by which P. gingivalis may impact bone turnover is through down-regulation of the anti-inflammatory cytokine IL-10. IL-10−/− mice exhibit a hyper inflammatory phenotype and display increased susceptibility to P. gingivalis-induced periodontal bone loss (35). In addition, down-regulation of ICAMs and P-selectin in tissue may contribute to bone loss as knockout mice deficient in cellular adhesion molecules show increased P. gingivalis-induced alveolar bone loss (2). Another possible mechanism for pro-inflammatory cytokine independent alveolar bone loss is through down-regulation of activin receptor type II (ACVR2), which occurred in bone samples. Activins are dimeric growth and differentiation factors which belong to the TGF-β superfamily (3). Activins can be stimulated by a subset of BMPs (38), and will bind and activate the SMAD family of transcriptional regulators, which control osteoblast expansion, differentiation, and bone formation (27, 29).
A major difference between the tissue and bone samples was in the impact on the antigen processing and presentation pathway. In bone, P. gingivalis up-regulated several components of the MHCII pathway, including MHCII itself, that processes endocytosed antigens and leads to expansion of CD4 positive T cells including Th cells. In addition, in bone samples IL-18 was up-regulated, which in combination with a lack of IL-12 up-regulation, is indicative of a Th2 protective response (32). By contrast, in tissue the MHC class II pathway was down-regulated which would be predicted to decrease the level of stimulation of CD4 T cells and consequently lower the levels of cytokines and B-cells. Indeed, several Th-B-cell interacting molecules were down-regulated in tissue including CD80, CD86, CD40 and CD22. Leukocyte migration through the endothelium was also potentially abrogated by reduced expression of many interacting components. Leukocyte adhesion components that were down-regulated included integrin alpha-L/beta-2 (ITGAL/ITGB2) that together form a receptor for ICAM-1, and are involved in a variety of immune phenomena, including leukocyte-endothelial cell interaction (36). Moreover, E-selectin (SELE) and P-selectin (SELP) were down-regulated further contributing to compromised leukocyte recruitment (26). Indeed P/E-selectin-deficient mice showed increased bacteria colonization of the oral cavity (30). Recognition of P. gingivalis MAMPs in calvarial tissue may also be compromised and there was down regulation of TLR1, TLR2, TLR4 (along with CD14 and LBP), TLR6 and the downstream signaling molecule MyD88. In some strains of mice knockout of TLR4 contributes to alveolar bone loss following infection with P. gingivalis (6).
The significant increase in osteoclast numbers and full thickness calvarial bone resorption defects observed in response to P. gingivalis infection were similar to the effects that we observed in the calvaria of mice following local injections of IL-1 (1), as well as with other periodontal pathogens C. rectus and F. nucleatum (42).
In conclusion, the present study reports findings from a comprehensive gene expression profile of inflamed soft tissue and calvarial bone that accompanied a localized, acute infection of mice with P. gingivalis. Importantly, many of the most highly affected genes in tissues are related to biological processes not historically emphasized within the literature related to periodontal disease initiation and progression (24, 37). It is likely that host responses and their contribution to protection or to tissue destruction are highly context dependent, and vary according to parameters such as dose of infecting organism, duration of infection, and genetic background. The substantive impact of P. gingivalis in down-regulating host inflammatory/innate immune responses in this model supports the need for additional studies to explore the role of selected genes in the periodontal infections, including the effect of P. gingivalis on host response patterns as a component of the polymicrobial infection in periodontal disease.
Supplementary Material
Acknowledgements
We thank Drs. Arnold Stromberg, Cidambaram Srinivasan, Christopher Saunders, and Matt Hersh (Department of Statistics, University of Kentucky, Lexington) for initial data management and microarray data analysis. We also thank Dr. Quey-Chen and Ms. Donna Wall (University of Kentucky, Lexington, Microarray Facility) for microarray technical assistance. This study was supported by National Institute of Dental and Craniofacial Research DE015720 (L.K), and NIAMS AR43510 (B.F.B).
Footnotes
Disclosures
The authors have no financial conflict of interest.
References
- 1.Boyce BF, Aufdemorte TB, Garrett IR, Yates AJP, Mundy GR. Effects of interleukin-1 on bone turnover in normal mice. Endocrinology. 1989;125:1142–1150. doi: 10.1210/endo-125-3-1142. [DOI] [PubMed] [Google Scholar]
- 2.Baker PJ, DuFour L, Dixon M, D. C. Roopenian DC. Adhesion molecule deficiencies increase Porphyromonas gingivalis-induced alveolar bone loss in mice. Infect Immun. 2000;68:3103–3107. doi: 10.1128/iai.68.6.3103-3107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang H, Brown CW, Matzuk MM. Genetic analysis of the mammalian transforming growth factor-beta superfamily. Endocr Rev. 2002;23:787–823. doi: 10.1210/er.2002-0003. [DOI] [PubMed] [Google Scholar]
- 4.Chou HH, Yumoto H, Davey M, Takahashi Y, Miyamoto T, Gibson FC, 3rd, Genco CA. Porphyromonas gingivalis fimbria-dependent activation of inflammatory genes in human aortic endothelial cells. Infect Immun. 2005;73:5367–5378. doi: 10.1128/IAI.73.9.5367-5378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collazo CM, Yap GS, Hieny H, Caspar P, Feng CG, Taylor GA, Sher A. The function of gamma interferon-inducible GTP-binding protein IGTP in host resistance to Toxoplasma gondii is Stat1 dependent and requires expression in both hematopoietic and nonhematopoietic cellular compartments. Infect Immun. 2002;70:6933–6939. doi: 10.1128/IAI.70.12.6933-6939.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costalonga M, Batas L, Reich BJ. Effects of Toll-like receptor 4 on Porphyromonas gingivalis-induced bone loss in mice. J Periodontal Res. 2009;44:537–542. doi: 10.1111/j.1600-0765.2008.01152.x. [DOI] [PubMed] [Google Scholar]
- 7.Demmer RT, Behle JH, Wolf DL, Handfield M, Kebschull M, Celenti R, Pavlidis P, Papapanou PN. Transcriptomes in healthy and diseased gingival tissues. J Periodontol. 2008;79:2112–2124. doi: 10.1902/jop.2008.080139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 9.Draghici S, Khatri P, Eklund AC, Szallasi Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006;22:101–109. doi: 10.1016/j.tig.2005.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feezor RJ, Oberholzer C, Baker HV, Novick D, Rubinstein M, Moldawer LL, et al. Molecular characterization of the acute inflammatory response to infections with gram-negative versus gram-positive bacteria. Infect Immun. 2003;71:5803–5813. doi: 10.1128/IAI.71.10.5803-5813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gemmell E, Drysdale KE, Seymour GJ. Gene expression in splenic CD4 and CD8 cells from BALB/c mice immunized with Porphyromonas gingivalis. J Periodontol. 2006;77:622–633. doi: 10.1902/jop.2006.050211. [DOI] [PubMed] [Google Scholar]
- 13.Gemmell E, Winning TA, Bird PS, Seymour GJ. Cytokine profiles of lesional and splenic T cells in Porphyromonas gingivalis infection in a murine model. J Periodontol. 1998;69:1131–1138. doi: 10.1902/jop.1998.69.10.1131. [DOI] [PubMed] [Google Scholar]
- 14.Han X, Amar S. Identification of genes differentially expressed in cultured human periodontal ligament fibroblasts vs. human gingival fibroblasts by DNA microarray analysis. J Dent Res. 2002;81:399–405. doi: 10.1177/154405910208100609. [DOI] [PubMed] [Google Scholar]
- 15.Handfield M, Mans JJ, Zheng G, Lopez MC, Mao S, Progulske-Fox A, Narasimhan G, Baker HV, Lamont RJ. Distinct transcriptional profiles characterize oral epithelium-microbiota interactions. Cell Microbiol. 2005;7:811–823. doi: 10.1111/j.1462-5822.2005.00513.x. [DOI] [PubMed] [Google Scholar]
- 16.Handfield M, Baker HV, Lamont RJ. Beyond good and evil in the oral cavity: insights into host-microbe relationships derived from transcriptional profiling of gingival cells. J Dent Res. 2008;87:203–223. doi: 10.1177/154405910808700302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasigawa Y, Tribble GD, Baker HV, Mans JJ, Handfield M, Lamont RJ. Role of Porphyromonas gingivalis SerB in gingival epithelial cell cytoskeletal remodeling and cytokine production. Infect Immun. 2008;76:2420–2427. doi: 10.1128/IAI.00156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis. Periodontol 2000. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 19.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasamatsu A, Uzawa K, Shimada K, Shiiba M, Otsuka Y, Seki N, Abiko Y, Tanzawa H. Elevation of galectin-9 as an inflammatory response in the periodontal ligament cells exposed to Porphylomonas gingivalis lipopolysaccharide in vitro and in vivo. Int J Biochem Cell Biol. 2005;37:397–408. doi: 10.1016/j.biocel.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Khatri P, Draghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21:3857–3895. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kesavalu L, Chandrasekar B, Ebersole JL. In vivo induction of proinflammatory cytokines in mouse tissue by Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 2002;17:177–180. doi: 10.1034/j.1399-302x.2002.170307.x. [DOI] [PubMed] [Google Scholar]
- 23.Kesavalu L, Ebersole JL, Machen RL, Holt SC. Porphyromonas gingivalis virulence in mice: induction of immunity to bacterial components. Infect Immun. 1992;60:1455–1464. doi: 10.1128/iai.60.4.1455-1464.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinane DF, Podmore M, Murray MC, Hodge PJ, Ebersole JL. Etiopathogenesis of periodontitis in children and adolescents. Periodontol 2000. 2001;26:54–91. doi: 10.1034/j.1600-0757.2001.2260104.x. [DOI] [PubMed] [Google Scholar]
- 25.Lamont RJ, Jenkinson HF. Life below the gum line: Pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ley K, Kansas GS. Selectins in T-cell recruitment to non-lymphoid tissues and sites of inflammation. Nat Rev Immunol. 2004;4:325–335. doi: 10.1038/nri1351. [DOI] [PubMed] [Google Scholar]
- 27.Li B. Bone morphogenetic protein-Smad pathway as drug targets for osteoporosis and cancer therapy. Endocr Metab Immune Disord Drug Targets. 2008;8:208–219. doi: 10.2174/187153008785700127. [DOI] [PubMed] [Google Scholar]
- 28.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lian JB, Stein GS, Javed A, van Wijnen AJ, Stein JL, Montecino M, Hassan MQ, Gaur T, Lengner CJ, Young DW. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev. Endocr Metab Disord. 2006;7:1–16. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- 30.Niederman R, Westernoff T, Lee C, Mark LL, Kawashima N, Ullman-Culler M, Dewhirst FE, Paster BJ, Wagner DD, Mayadas T, Hynes RO, Stashenko P. Infection-mediated early-onset periodontal disease in P/E-selectin-deficient mice. J Clin Periodontol. 2001;28:569–575. doi: 10.1034/j.1600-051x.2001.028006569.x. [DOI] [PubMed] [Google Scholar]
- 31.Ohno T, Okahashi N, Kawai S, Kato T, Inaba H, Shibata Y, Morisaki I, Abiko Y, Amano A. Proinflammatory gene expression in mouse ST2 cell line in response to infection by Porphyromonas gingivalis. Microbes Infect. 2006;8:1025–1034. doi: 10.1016/j.micinf.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 32.Orozco A, Gemmell E, Bickel M, Seymour GJ. Interleukin 18 and periodontal disease. J Dent Res. 2007;86:586–593. doi: 10.1177/154405910708600702. [DOI] [PubMed] [Google Scholar]
- 33.Oshikawa M, Sugano N, Koshi R, Ikeda K, Ito K. Differential gene induction in macrophage-like human cells by two types of Porphyromonas gingivalis: a microarray study. J Oral Sci. 2004;46:9–14. doi: 10.2334/josnusd.46.9. [DOI] [PubMed] [Google Scholar]
- 34.Piana C, Wirth M, Gerbes S, Viernstein H, Gabor F, Toegel S. Validation of reference genes for qPCR studies on caco-2 cell differentiation. Eur J Pharm Biopharm. 2008;69:1187–1192. doi: 10.1016/j.ejpb.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Sasaki H, Okamatsu Y, Kawai T, Kent R, Taubman M, Stashenko P. The interleukin-10 knockout mouse is highly susceptible to Porphyromonas gingivalis-induced alveolar bone loss. J Periodontal Res. 2004;39:432–441. doi: 10.1111/j.1600-0765.2004.00760.x. [DOI] [PubMed] [Google Scholar]
- 36.Smith A, Stanley P, Jones K, Svensson L, McDowall A, Hogg N. The role of the integrin LFA-1 in T-lymphocyte migration. Immunol Rev. 2007;218:135–146. doi: 10.1111/j.1600-065X.2007.00537.x. [DOI] [PubMed] [Google Scholar]
- 37.Tatakis DN, Kumar PS. Etiology and pathogenesis of periodontal diseases. Dent Clin North Am. 2005;49:491–516. doi: 10.1016/j.cden.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Tsuchida K, Nakatani M, Uezumi A, Murakami T, Cui X. Signal transduction pathway through activin receptors as a therapeutic target of musculoskeletal diseases and cancer. Endocr J. 2008;55:11–21. doi: 10.1507/endocrj.kr-110. [DOI] [PubMed] [Google Scholar]
- 39.Wada T, Nakashima T, Hiroshi N, Penninger JM. RANKL-RANK signaling in osteoclastogenesis and bone disease. Trends Mol Med. 2006;12:17–25. doi: 10.1016/j.molmed.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Wang PL, Ohura K, Fujii T, Oido-Mori M, Kowashi Y, Kikuchi M, Suetsugu Y, Tanaka J. DNA microarray analysis of human gingival fibroblasts from healthy and inflammatory gingival tissues. Biochem Biophys Res Commun. 2003;305:970–973. doi: 10.1016/s0006-291x(03)00821-0. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Q, Amar S. Identification of proteins differentially expressed in human monocytes exposed to Porphyromonas gingivalis and its purified components by high-throughput immunoblotting. Infect Immun. 2006;74:1204–1214. doi: 10.1128/IAI.74.2.1204-1214.2006. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zubery Y, Dunstan CR, Story BM, Kesavalu L, Ebersole JL, Holt SC, Boyce BF. Bone resorption caused by three periodontal pathogens in vivo in mice is mediated in part by prostaglandin. Infect Immun. 1998;66:4158–4162. doi: 10.1128/iai.66.9.4158-4162.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.