Abstract
Asymmetrical delivery of stimuli to moving cells for perturbing spatially-heterogeneous intracellular signaling is an experimental challenge not adequately met by existing technologies. Here, we report a robust microfluidic platform allowing localized treatment of the front and/or back of moving cells which crawl through narrow channels that they completely occlude. The enabling technical element for this study is a novel design for precise, passive balancing of flow inside the microfluidic device by contacting two fluid streams before splitting them again. The microchannels constrain cell morphology and induce qualitative and quantitative changes in neutrophil chemotaxis that mimic cells crawling through tissues.
Introduction
During chemotaxis in amoeboid cells, the signaling and cytoskeleton systems responsible for chemoattractant detection and motility are activated in a polarized fashion along the direction of migration and maintained by multiple feedback loops between the front and rear of the cells.1–3 To study these feedbacks, it would be useful to expose only the front or rear of the polarized cell to chemoattractant or drugs with a high degree of spatial precision. However, current experimental techniques are ill adapted to the study of fast moving cells that rapidly change shape and direction. Local stimulation using hydrodynamic focusing of different fluid streams4,5 is ineffective if cells do not move in a direction perpendicular to the stream lines. Perfusion through pipettes6–8 has the disadvantage of exposing cells to shear stress (necessary for sharp focusing of the inhibitor) along with the drug, and a single target cell must be followed manually, making data acquisition less precise and lengthy. Furthermore, these local perfusion methods follow cells freely moving on a 2-D substrate, which is not a good physical mimic of important chemotactic events where leukocytes crawl through narrow spaces between endothelial or tissue cells. To observe cellular chemotaxis at high resolution while allowing localized treatment of the front or rear we developed a new microfluidic device that establishes robust, convection-free, stationary linear or moving steep gradients of chemoattractant (and/or drug) in an array of microchannels. Leukocytes occlude the channels as they crawl through, allowing selective drug treatment of the front or back. The channels also constrain cell morphology, making it highly consistent over time, and between cells, which will facilitate mathematical modeling.
Materials and methods
Finite element simulation
To optimize the design of our microfluidic device such that pressure was balanced as accurately as possible to minimize convection in the transversal channels and produce a stable linear gradient, we simulated the gradient generator geometry in two dimensions using finite element method (FEM) software, COMSOL Multiphysics 3.2 (Fig. 1a). The simulation was carried out under the chemical engineering module by coupling convection and diffusion and the incompressible Navier–Stokes equations and simultaneously solving them for mass and momentum balance. For the simulations, the diffusion coefficient of the chemokine fMLP (N-formyl-methionyl-leucyl-phenylalanine) was considered to be 10−10 m2 s−1,9 the density of water 103 kg m−3, and the dynamic viscosity of water 10−3 kg m−1 s−1. Chemokine concentrations of 1 and 0 M were considered for the two streams, respectively. The convective flux boundary condition at the outlet refers to purely convective mass transfer taking place at the outlet boundary and assumes that any mass flux due to diffusion across this boundary is zero. No penetration boundary conditions were set elsewhere. For solving the Navier–Stokes equation, the inlet fluid velocity for both streams was set to 10−4 m s−1, an outflow/pressure boundary condition was imposed at the outlet, and for all other surfaces, no-slip boundary conditions were used. Denser mesh was generated at the fluid–fluid interface and in the vicinity of transversal channels to achieve sufficient accuracy at locations where higher diffusion activity was expected, while coarser elements were rendered at the bulk fluid region to facilitate convergence of the solution. The initial structure consisting of 2.5 × 104 triangular elements was solved for 1.7 × 105 degrees of freedom and a converged solution was obtained in less than a minute using a 3.2 GHz dual processor Pentium-4 computer with 4 GB RAM.
Fig. 1.
Simulation results for characterization and optimization of the gradient. (a) Two solutions, A and B, of different concentration of chemoattractant are brought together in a common channel. A contact zone Dp is used to balance the pressures of the two streams. A compensation zone Dd is used to correct for the diffusion/convection mixing between the two streams. The linear gradient is generated in the transversal capillaries in the region Dg. Concentrations are shown in color scale with red being 1 M chemokine and blue being 0 M chemokine. (b–e). Simulations of concentrations in the device for equal (b and c) and 20% difference (d and e) in flow rates at the inlets, for conditions of no contact (b and d) and 250 μm long compensation zone (c and e). (f) If the compensation zone is not present (Dp = 0), the linear profile of the gradient is significantly altered by differences in flow rates of less than 1% (0.1% difference solid line, 1% dotted line, 2% dashed line, 20% dash-dotted line). (g) The compensation zone allows the maintaining of a linear gradient in the presence of unbalanced flow rates, although the slope is slightly altered from the ideal profile (0% difference solid line, 10% dotted line, 30% dashed line, 50% dash-dotted line).
We independently varied the length of the contact zone (Dp), compensation zone (Dd), and flow rates in the two inlets in order to optimize the design of the device and characterize the robustness of the linear gradient in the transversal channels. Concentration profiles were calculated along the transversal microchannel, and concentration profiles for asymmetric inlet flow rates (Cactual) were compared to the concentration profiles when the two streams have exactly equal flow rates (Cideal). Qualitative results of the simulations are presented in Fig. 1b–e. In the absence of any contact zone (Dp = 0), the pressure developed on the side of higher stream velocity forces the fluid to convectively flow through the transverse passages preventing the establishment of a diffusion gradient within the narrow channel (Fig. 1b,d). The gradient in the microchannels is linear only when the two flow rates are precisely equal, an unlikely situation experimentally (Fig. 1b). When a contact zone is used, the pressures at the ends of the microchannels are equalized and the gradient is linear for a wider range of flow rates at the two inlets (Fig. 1c,e).
To quantitatively evaluate the effect of device design and flow rate differences on spatial concentration profiles, we calculated the root mean square error (RMSE) for each case using the following equation:
| (1) |
where N (N = 200) is the number of locations along the 150 μm long transversal micro-channel where the actual (Cactual) and ideal (Cideal) concentrations are compared. Simulations were performed for differences in flow rate from 0 to 50%, contact zone length (Dp) before stream separation values from 0 to 1000 μm and length of the compensation zone (Dd) from 300 to 1800 μm. The concentration curve along microcapillaries (Fig. 1f) as well as the RMSE curve (Fig. 1g) show that when there is no interfacial flow (Dp = 0 μm), the root mean square error of the concentration profile approaches 60%, even for the slightest difference in flow rates. For interfacial lengths greater than 250 μm, the magnitude of RMSE did not vary much with the length of the contact zone (Fig. 1c,e,g). Increasing compensation zone length Dd significantly lowers the effect of flow variation and stabilizes the gradient. The optimum was obtained by setting the contact zone length of Dp = 250 μm and the compensation zone length Dd = 1800 μm. It should be noted that this observation is valid only for the specified width of the main channel and the simulation parameters may vary for other dimensions of the main channel.
Microfabrication
The microfluidic device was composed of two layers of polydimethlysiloxane (PDMS—a valve control layer 3–4 mm thick and a fluidic network layer 150 μm thick) bonded to a glass coverslip to enable high resolution imaging. Molds for each layer were fabricated by patterning photopolymer (SU8, Microchem, Newton, MA, USA) using standard photolithography techniques. The fluidic network mold combines three layers of different thicknesses, a thin layer (3 or 10 μm, depending on the design) corresponding to the transversal channels, an intermediate layer (7 μm) corresponding to the valving structures, and a thicker layer (30 μm) corresponding to the inlet and outlet channels (Fig. 2). The mold for the valve control layer uses one thickness (30 μm) corresponding to the actuation chambers and channels for the valves. Both layers were cast on their respective molds in PDMS prepared following manufacturer recommendations. The thinner fluidic network layer was prepared by spinning freshly prepared PDMS on the mold at 600 rpm followed by standard baking overnight. After curing, the valve control layer was bonded on top of the thinner fluidic network layer by exposure to oxygen plasma (25 s, 50 W, 2% oxygen, 98% nitrogen) in a plasma asher (March Inc., Concord, CA, USA) and heating at 80 °C for 5 min on a hot plate. The two layers were then bonded in a similar fashion on a thin coverslip (60 × 24 × 0.15 mm; Fisher Scientific, Pittsburgh, PA, USA). The 7 μm distance between the valve and the glass ensured that valves remained open without any additional precautions during standard bonding.
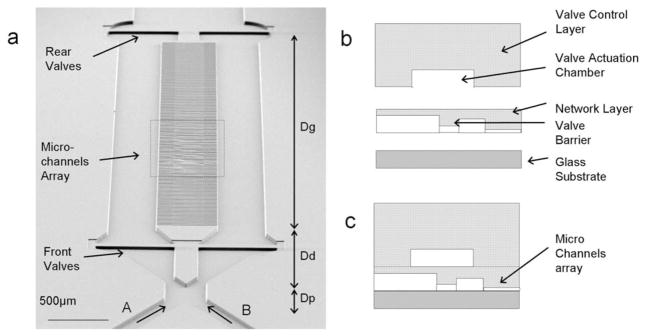
Fig. 2.
Microfluidic device design and manufacturing. (a) Scanning electron microscopy of the microfluidic device. The inlets, at the bottom of the figure, converge into a common channel and then are split again before the transversal microchannel array. A region Dp is used to balance the pressures of the two streams. A region Dd is used to correct for the transversal diffusion between the two streams. A linear gradient is generated in the transversal microchannels in the region Dg. Two valves in each of the arm can be used to direct the flow through the transversal microchannels during cell loading. (b) A valve control (3 mm thick) and a network layer (150 μm) were cast in PDMS using a microfabricated mold. The control layer contains a 30 μm tall actuation chamber for the valves. The network layer contains features of three different heights, corresponding to the main channels (30 μm), valves (7 μm) and microcapillaries (either 10 or 3 μm depending on the device). (c) The control layer is aligned on top of the network layer and both layers are bonded on top of a thin glass coverslip (150 μm).
Device setup
Microfluidic connections between the device and outside reservoirs or syringes for actuating the valves were accomplished using Tygon tubing (Small Parts, Miami Lakes, FL, USA). Before experimentation, the device was functionalized by chemisorption of 70 μg ml−1 solution of human fibronectin (BD Clontech) in PBS for 1 h at room temperature, then it was passivated with casein (Sigma, St. Louis, MO, USA) for 45 min and finally with human serum albumin (Sigma) for 15 min to reduce non-specific adhesion of the cells to the device surfaces. After priming, the two inlets were connected to two reservoirs containing the control and the chemoattractant solutions, and gravity flow was established by the height difference (~3 cm) between the inlet and outlet.
HL60 cell differentiation and loading
HL60 cells were cultured at 37 °C with 5% CO2 in RPMI1640 (Mediatech, Herndon, VA, USA) with 10% FCS, penicillin/streptomycin, and 25 mM HEPES. Cells were differentiated by exposure to 1.3% DMSO for a week.10 Prior to experimentation, differentiated cells were pelleted and resuspended in mHBSS (Invitrogen, Carlsbad, CA, USA) with 1.8% HSA. All experiments were performed in mHBSS with 1.8% HSA.
The cells were loaded in the device, after priming and after closing one front valve and one rear valve on opposite streams, by the application of pressure via syringes. With two valves in opposite streams and at opposite ends of the device closed, the flow is directed from one main channel to the other through the transversal microchannels. Cells are too large to easily flow through the transversal microchannels and are trapped at their inlets. The initial volume of air in each of the syringes was 0.5 mL and this was reduced to 0.2 mL to close the valves, and subsequently increased to 0.8 mL by moving the syringe pistons, to completely open the valves and facilitate the removal of un-attached cells. The valves were designed such that their actuation was necessary only during cell loading and not afterwards. After cell loading, the chemoattractant gradient was established by connecting the two inlets to the reservoirs with control solution and chemoattractant, respectively. The chemoattractant reservoir contained 100 nM fMLP (MW 438) and a fluorescent tracer of comparable molecular size (tetra-methyl-rhodamine (MW 430), Alexa 488 (MW 885), or Alexa 647 (MW 589)) to enable imaging of the chemoattractant gradient. Once the chemoattractant gradient was established, cells migrated up the channels in a matter of minutes.
To image the actin distribution in live migrating cells, an HL60 cell line stably expressing actin-mRFP was created by retroviral infection and subsequent selection with 500 μg ml−1 G418 (Sigma) using the retroviral vector previously described.11 Cells were differentiated in the same way as wild type cells.
Fixation and immunostaining
After we had ascertained that cells were migrating into the transversal channels, at the time of choice, glutaraldehyde and formaldehyde were added to both the control and chemoattractant reservoirs to final concentrations of 0.1% and 2%, respectively. We empirically determined the time needed for fixatives to reach the cells by adding a fluorescent tracer simultaneously. The time lag was ~15 min with 10 cm tubing connecting the reservoirs to the inlets. We incubated cells with fixative for a further 15 min at room temperature. Once the cells were fixed, they were firmly wedged into the channels and it was possible to exchange solutions in the devices by forcing fluid through with a syringe. The cells were washed for 15 min in antibody dilution buffer (TBS, 0.1% tritonX, 2% BSA) and given a second wash. Then, they were incubated for 45 min at room temperature with 1 μg ml−1 rhodamine-phalloidin (Sigma) to visualize F-actin and FITC labeled monoclonal anti-tubulin antibody (1 : 400 dilution, DM1α, Sigma) in antibody dilution buffer. The cells were then washed 3 times for 5 min each in antibody dilution buffer and were examined by confocal microscopy.
Microscopy
All imaging was done using on a Nikon TE-2000 inverted microscope (Nikon, Yokohama, Japan). Both a 100× 1.3 NA oil immersion objective and a 40 × 0.75 NA dry lens objective were used for imaging. Fluorophores were excited either with epifluorescence illumination from a mercury lamp and the appropriate filter sets, or a 3 W tunable laser. For confocal microscopy, a Perkin Elmer spinning disk confocal (Wellesley, MA, USA) was interfaced to the microscope. Images were captured on an Orca ER CCD camera (Hamamatsu, Hamamatsu, Japan) and acquired on a PC using Metamorph software (Molecular Devices, Sunnyvale, CA, USA). Images of the cells were acquired at either 488 nm wavelength for FITC or 568 nm wavelength for rhodamine. For display, images were low-pass filtered and scaled such that background fluorescence was minimal. In the confocal images, the rhodamine-phalloidin staining was gamma-enhanced to show the actin cortex at the uropod which had a much weaker staining than at the leading edge. In some cases, projection images were created by projecting the maximal intensity of fluorescence of all optical sections onto one plane.
Localized application of inhibitor
Localized application of inhibitor was possible because the cells fully occluded the microcapillaries. We used two different approaches to treat the leading edge of migrating cells with the PI3-kinase inhibitor wortmannin. First, we established a chemoattractant gradient and when cells started migrating, we substituted the original chemoattractant solution for one containing chemoattractant, 4 μM wortmannin, and a different fluorophore than the original solution (e.g. Alexa 647, FITC). Cell migration was imaged and the onset of chemical treatment could be visualized using the intensity of the fluorescent tracer. This technique could also be used to apply inhibitor to the rear of the cell. In the second method, we established the chemoattractant gradient directly with a solution containing inhibitor. Before the cells fully penetrated into the channels and occluded them, the concentration of inhibitor they were exposed to was calculated, considering the flow conditions inside the device, to be less than one tenth of the maximum concentration. However, once the cells occluded the microchannels, the concentration of inhibitor increased with time due to diffusion from the chemoattractant stream until it equalized. Because of the length of the channels, concentration equalization still required a significant amount of time. For a small peptide such as fMLP, the diffusion time is of the order of 2 min (t ~ l2/D ~ 150 s where D ~ 10−10 m2 s−1 is the diffusion constant of fMLP9 and l ~ 125 μm the length of the channels).
Cell velocity and cell volume measurements
Cell velocities were estimated by tracking the position of the leading edge using kymographs. To create the kymographs we used the specific tool implemented in Metamorph. Briefly, one column of individual pixels along a chosen line (e.g. the dotted line in Fig. 5d) is stored from each and every frame of a time sequence. All pixels are then presented side by side in a figure where each column of pixels represents one time point, and each row one location along the channel. The positions of the front, back, and different features inside the cells could then be easily tracked on the kymograph. We measured both the maximal velocity of cells by searching for the line of shallowest angle and the average velocity of cells by measuring the distance traveled over the duration of acquisition (10 or 20 min). Cells migrating in 3 μm high microchannels contacted both the top and bottom substrates, therefore cell volume could be estimated by measuring cell area and knowing the channel height. Cell area was determined by tracing the contour of each cell manually for time points 90 s apart.
Fig. 5.
Cell migration characterization and localized inhibitor treatment of cells. (a) Kymograph of neutrophil displacement in a 10 × 3 μm microchannel shows uniform velocity over a long period of time. (b) Velocity is very irregular with marked phases of forward movement and retraction in the 10 × 10 μm microchannels (white arrows). (c) Box plot comparison of the average migration velocity through capillaries of different sizes. Differentiated HL60 cells migrate at higher average speed in microchannels with a 10 × 3 μm cross-section, which they completely obstruct, than in larger 10 × 10 μm cross-section microchannels, which they do not completely block. The boundary of the box closest to zero indicates the 25th percentile, a line within the box marks the median, and the boundary of the box farthest from zero indicates the 75th percentile. Whiskers above and below the box indicate the 90th and 10th percentiles. In addition, the mean and outlying points are graphed. (d) DIC image of the morphology of a migrating cell prior to and after exposure to wortmannin. A reservoir containing chemoattractant and wortmannin was substituted for the chemoattractant reservoir. The morphology of the cell changes with drug treatment and the cell stalls. The dye front is shown in red. Scale bar = 5 μm. (e) Kymograph of treatment with 4 μM wortmannin acquired along the dotted line in (d). The timing of drug treatment is shown in red. Prior to drug treatment, the cell moves at a constant velocity of 4 μm min−1. Upon drug treatment, the velocity decreases, the cell stops, and finally retracts. (f) Neutrophil migration is impaired following local exposure to wortmannin and fMLP in an evolving gradient. Once the chemoattractant gradient is established, cells penetrate into the capillaries and occlude them (t = 5:30 min). Over time, the concentration of wortmannin increases and the cells stop migrating (t = 40:00 min). Alexa 488 is used to visualize drug exposure. Cells are marked by an asterisk. Scale bar = 10 μm.
Results and discussion
Perfect axial symmetry inside the device was accomplished by the passive correction of flow rates at the inlets, using a surprising design enabled by the laminar flow at microscale. The two inlet streams are brought in contact for a short distance (Dp) and then evenly split to the sides of an array of parallel microchannels (Fig. 1 and 2). The contact zone between streams and the geometrical axial symmetry of the device downstream of it allow for pre-balancing of downstream pressures and prevent convective flow in the transversal direction through the microchannels. The linear compensation zone (Dd) corrects for the changes in concentration of the two streams due to diffusion occurring in the contact region, such that the ends of the transversal microchannels are exposed to concentrations within one tenth change of those of the two initial solutions. A perfectly linear, stable gradient is formed along the microchannels by diffusion between the two streams. In the absence of the contact zone, input flow rate imbalances as small as 1% between the two streams would lead to convective flow and prevent the establishment of a gradient (Fig. 1d). In practice, flow imbalances always exist and are often larger than 1% due to the combined effect of variations in the macro-to-micro coupling, different physical characteristics of the two solutions, capillary effects, or variations in the inlet tubing size.12 Our design overcomes these issues and can compensate for imbalances in flow rates of up to 50% without affecting the linearity of the gradient or the convection-free conditions inside the microcapillaries. An additional design element for the device are the microstructured membrane valves13 to facilitate gentle cell loading, precisely at the entrance of the transversal microchannels, by temporarily directing flow through the microchannels (Fig. 2). The advantages of this approach are that samples with small number of cells can be used, and the cells are immediately available for migration in a newly established chemotactic gradient, with similar starting conditions for all cells. Moreover, the regular spacing of the microchannels and the predictable location and trajectory of the cells could potentially help in tracking all cells migrating in microchannels in the same device by using a motorized stage and time-lapse imaging.
We observed the entry and migration of DMSO differentiated HL-60 cells (a common neutrophil model) inside channels of different lengths (150 or 400 μm) and cross-sections (10 × 10, 10 × 3, and 5 × 3 μm, Fig. 3). For capillaries with 10 × 3 μm cross-sections and smaller, the cells had a leading edge that fully filled the channel (Fig. 3a–e) rather than a thin flat one, and an often oversized uropod (the bulge at the trailing edge). For channels with a 10 × 3 μm cross-section or smaller, the entry of cells completely occluded the channels, as evidenced by a progressive increase in fluorophore intensity at the leading edge of the cells when compared to the same location in unobstructed channels (arrows, Fig. 3a). After a sufficient equilibration time, the ends of the migrating cells were exposed only to the solutions from each side channel, and altered less than 10% compared to the inlet solutions, in a steep concentration change across the cell (Fig. 3b,c). When the cells reached the end of the channels and exited, the steep gradient disappeared and a linear gradient was re-established (Fig. 3c). Cells did not occlude channels with a 10 × 10 μm cross section, and their morphology was similar to that observed on glass coverslips with a thin ruffling leading edge at the front and a small uropod at the rear (Fig. 3f).
Fig. 3.

Cell migration in microcapillaries. (a) A linear chemoattractant gradient is present in the empty channels and directs the migration of the HL60 cells (middle channel). After a cell enters one microchannel, a steep concentration difference is established between the front and back of the cell (top and bottom channels). Arrows indicate that the fluorescence intensity is different depending on whether the channel is obstructed or not. (b) Migration of a HL60 cells towards chemoattractant and establishment of a steep gradient across the cells. Alexa 647 was added to the stream with chemoattractant (shown in blue) and FITC labelled dextran (MW 100 000) to the stream with buffer (shown in green). The sides of the channels are imaged in brightfield. (c) Migration of a neutrophil towards chemoattractant. Alexa 647 was added to the stream with chemoattractant (shown in blue). The cell was incubated with a cytoplasmic marker (in red). At t = 0 min, the cell fully occludes the channel and there is no fluorescence to the rear of the cell. At t = 10 min, the cell exits the channel and a linear gradient of chemoattractant is re-established. Scale bar = 10 μm. (d) DIC image of a neutrophil migrating through a 10 μm wide and 3 μm tall microchannel. The cell is polarized with a recognizable leading edge and uropod. Scale bar = 10 μm. (e) DIC image of a neutrophil migrating in a 3 × 5 μm capillary. The cell was very elongated and the nucleus (marked as N) very compressed. Scale bar = 4 μm. (f) DIC image of a neutrophil migrating through a 10 × 10 μm microchannel. The cell is polarized with a recognizable leading edge and uropod and does not occlude the microchannel. Scale bar = 10 μm.
One important feature of our device is that it allows high resolution imaging with most forms of optical microscopy. Differentiated HL-60 cells stably expressing actin-mRFP were imaged during migration through 10 × 3 μm channels. In these cells, actin was predominantly localized at the leading edge in front of the nucleus (Fig. 4a). Microspikes reminiscent of those observed in ruffles in two-dimensional migration were observed and actin localized strongly to the sides of the leading edge (Fig. 4a, red arrow). Cells migrating in the channels were fixed and immunostained for F-actin with rhodamine-phalloidin by flowing staining reagents through the device, and showed a similar distribution as actin-mRFP in live cells (Fig. 4b,c). Immunostaining of microtubules showed that they were mainly localized to the rear of the cell, reached past the nucleus, but stopped at the leading edge (Fig. 4b,c). Contrary to two-dimensional migration, the leading edge was not a thin lamellipodium14 but a rectangular prism, filling the whole channel top to bottom (Fig. 4c). One implication of this channel-filling morphology is that cell volume can be estimated by measuring cell area over time knowing the height of the channel. Differentiated HL60 cells migrating in response to 100 nM fMLP had an average volume of 850 ± 141 μm3 (N = 25) and this varied by ±3.15% on average for individual cells, suggesting tight control of cell volume during migration.
Fig. 4.
Cytoskeletal distribution in live and fixed cells. (a) Actin distribution in a live cell stably expressing actin-mRFP. Actin localizes predominantly to the leading edge but also to the sides of the leading edge in contact with the microchannel walls (white arrows). Microspikes similar to those observed in lamellipodia can be seen (red arrow). Scale bar = 5 μm. (b) Localization of actin and tubulin in fixed migrating cells. Actin (in red) localizes predominantly to the leading edge of the cell, but is also present in the form of a thin cortex at the rear. Microtubules (in green) are localized to the rear, reach past the nucleus but stop at the leading edge. This image is a maximum projection of a confocal stack. The yellow color indicates co-localization of actin and microtubules. Scale bar = 10 μm. (c) Top and side view of the middle cell in b. Actin (in red) is predominantly localized at the leading edge and on the sides in contact with the microchannel wall similar to the distribution in live cells (white arrows). A thin actin cortex is present at the cell rear. Microtubules (in green) are mainly localized behind the nucleus but extend from the leading edge to the uropod, and from the top to the bottom of the cell. The cell contacts both the bottom and top walls of the microchannel. No thin lamellipodium can be seen at the leading edge. Actin is slightly enriched close to the glass coverslip. Height = 3 μm, scale bar = 5 μm.
We observed qualitative and quantitative differences between cells migrating in channels with different cross sections. HL-60 cells migrated inside 10 × 3 μm crossection channels with very uniform velocities and cell morphology was also remarkably constant over the entire time of observation, as is evident from the constant slopes for leading edge displacement in the kymograph in Fig. 5a and in supplementary video 1, ESI.† The morphological characteristics of migration are more suggestive for continuous creation and extinction of interactions with the microchannel walls,15 rather than the three-step model (attraction, contraction, and detachment) for haptokinetic cell migration,16 and may be adhesion independent.17 Migration in the deeper channels (10 × 10 μm cross-section) resembled the more irregular velocity and morphology of cells chemotaxing on two-dimensional surfaces (white arrows, Fig. 5b and supplementary video 2, ESI†) and was significantly slower compared to shallower channels (average velocity 2.1 μm min−1 vs. 3.2 μm min−1). The significant difference (p = 0.02) was due to more persistent and uniform movement rather than a difference in maximum velocities (Fig. 5c).
Occlusion of the channels by migrating cells made it easy to deliver drugs locally to the front or back. We tested two protocols for delivering stimuli or inhibitors to only the leading edge or only the uropod of migrating cells, using as our test drug the PI3-kinase inhibitor, wortmannin, that is known to inhibit chemotactic signaling.18 First, we substituted the chemoattractant fluid stream for one containing chemoattractant and wortmannin. Second, we ran the chemotaxis assays with the drug added to the chemoattractant fluid stream. In the first case, cells migrated normally until wortmannin concentration reached a sufficient concentration by diffusion in the microcapillary, whereupon the cells slowed and eventually stopped (Fig. 5d). Analysis of the kymograph along with the evolution of intensity of drug treatment shows that inhibition occurs 2–3 minutes after contact of wortmannin with the leading edge (Fig. 5e). In the second case, cells entered the channels and, after they occluded the channel, wortmannin concentration rose progressively by diffusion from the lateral channels, until it was sufficiently high to inhibit movement (Fig. 5f). Cells were able to migrate after entering the microchannels because, for the low concentrations present at the entrance of the microchannels, they are more sensitive to the chemoattractant fMLP than to the inhibitor wortmannin. The different locations where cells stop are a consequence of migration rate and sensitivity to the inhibitor, with cells that are migrating faster and more resistant to inhibition advancing further in the microchannels. While the first method may be preferred when detailed observations at single cell level are targeted, the second method may be more appropriate for population studies or end point assays.
The new ability for polar stimulation of moving cells, with the added capabilities of constraining cells in 3D channels, in a well controlled environment over time and in the absence of shear stress, are all important features of the new device that distinguish it from other existing devices for the study of motile cells (Table 1). None of the other existing technologies allows one to expose multiple cells to distinct environment at the front and back of the cells, during cell migration, without the need to individually track each of the cells, and without the need to re-adjust the delivery system with cell displacement. The ability to visualize cells at high resolution, during migration in a 3D-like environment could provide a simpler and better controlled alternative to more sophisticated in vivo cell tracking systems. The high permeability for oxygen of the PDMS material that constitutes three of the four walls of the microchannles may also help creating in vivo-like conditions around the moving cells. With the exception of polar stimulation of motile cells, other features of the device may not be unique, and be shared with other existing technologies, but it is the first time all the capabilities are integrated in one single device.
Table 1.
Comparison between features of the present method and existing in vitro methods for studying motile cells
| Present method | Boyden chamber19 | Dunn chamber20 | Micro-pipette | Microfluidic devices |
||
|---|---|---|---|---|---|---|
| flow21 | no flow22 | |||||
| Polar stimulation of moving cells | YES | — | — | — | — | — |
| Constrained migration (3D) | YES | YES | — | — | — | — |
| Stable gradient | YES | — | — | — | YES | — |
| Absence of shear stress | YES | YES | YES | — | — | YES |
| High resolution imaging | YES | — | YES | YES | YES | YES |
| Small number of cells | YES | — | YES | YES | YES | YES |
Conclusions
We have demonstrated the use of a microfluidic device for the study of chemotaxis that enables precise application of localized stimuli to the leading edge or the trailing edge of motile cells migrating in microchannels along with simultaneous imaging at high resolution using optical microscopy. By constraining cells in three dimensions, our device removes the variability in cell morphology seen with unconstrained cells and mimics the mechanical challenge leukocytes face when migrating between cells or when they leave blood vessels during the inflammatory response.
Supplementary Material
Acknowledgments
The authors acknowledge funding from National Institutes of Health under grants P41 EB002503 (M.T.), and GM48027 (T.M.). GC wishes to acknowledge the support of the Royal Society. The authors wish to thank the Nikon Imaging Center at Harvard Medical School, as well as its director Dr J. Waters.
Footnotes
Electronic supplementary information (ESI) available: Two movies showing typical cellular responses in tall and shallow channels during polar stimulation. See DOI: 10.1039/b710524j
Contributor Information
Daniel Irimia, Email: dirimia@hms.harvard.edu.
Guillaume Charras, Email: g.charras@ucl.ac.uk.
References
- 1.Janetopoulos C, Borleis J, Vazquez F, Iijima M, Devreotes P. Dev Cell. 2005;8:467–477. doi: 10.1016/j.devcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Xu J, Wang F, Van Keymeulen A, Herzmark P, Straight A, Kelly K, Takuwa Y, Sugimoto N, Mitchison T, Bourne HR. Cell. 2003;114:201–214. doi: 10.1016/s0092-8674(03)00555-5. [DOI] [PubMed] [Google Scholar]
- 3.Nathan C. Nat Rev Immunol. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 4.Sawano A, Takayama S, Matsuda M, Miyawaki A. Dev Cell. 2002;3:245–257. doi: 10.1016/s1534-5807(02)00224-1. [DOI] [PubMed] [Google Scholar]
- 5.Takayama S, Ostuni E, Leduc P, Naruse K, Ingber DE, Whitesides GM. Nature. 2001;411:1016. doi: 10.1038/35082637. [DOI] [PubMed] [Google Scholar]
- 6.O’connell CB, Warner AK, Wang Y. Curr Biol. 2001;11:702–707. doi: 10.1016/s0960-9822(01)00181-6. [DOI] [PubMed] [Google Scholar]
- 7.Popov S, Brown A, Poo MM. Science. 1993;259:244–246. doi: 10.1126/science.7678471. [DOI] [PubMed] [Google Scholar]
- 8.Charras GT, Yarrow JC, Horton MA, Mahadevan L, Mitchison TJ. Nature. 2005;435:365–369. doi: 10.1038/nature03550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moghe PV, Nelson RD, Tranquillo RT. J Immunol Methods. 1995;180:193–211. doi: 10.1016/0022-1759(94)00314-m. [DOI] [PubMed] [Google Scholar]
- 10.Servant G, Weiner OD, Neptune ER, Sedat JW, Bourne HR. Mol Biol Cell. 1999;10:1163–1178. doi: 10.1091/mbc.10.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charras GT, Hu CK, Coughlin M, Mitchison TJ. J Cell Biol. 2006;175:477–490. doi: 10.1083/jcb.200602085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paliwal S, Iglesias PA, Campbell K, Hilioti Z, Groisman A, Levchenko A. Nature. 2007;446:46–51. doi: 10.1038/nature05561. [DOI] [PubMed] [Google Scholar]
- 13.Irimia D, Toner M. Lab Chip. 2006;6:345–352. doi: 10.1039/b515983k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stossel TP. Science. 1993;260:1086–1094. doi: 10.1126/science.8493552. [DOI] [PubMed] [Google Scholar]
- 15.Lee J, Ishihara A, Theriot JA, Jacobson K. Nature. 1993;362:167–171. doi: 10.1038/362167a0. [DOI] [PubMed] [Google Scholar]
- 16.Friedl P, Brocker EB. Cell Mol Life Sci. 2000;57:41–64. doi: 10.1007/s000180050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malawista SE, De Boisfleury Chevance A, Boxer LA. Cell Motil Cytoskeleton. 2000;46:183–189. doi: 10.1002/1097-0169(200007)46:3<183::AID-CM3>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 18.Ui M, Okada T, Hazeki K, Hazeki O. Trends Biochem Sci. 1995;20:303–307. doi: 10.1016/s0968-0004(00)89056-8. [DOI] [PubMed] [Google Scholar]
- 19.Boyden S. J Exp Med. 1962;115:453. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zicha D, Dunn GA, Brown AF. J Cell Sci. 1991;99:769–775. doi: 10.1242/jcs.99.4.769. [DOI] [PubMed] [Google Scholar]
- 21.Jeon NL, Baskaran H, Dertinger SKW, Whitesides GM, Van De Water L, Toner M. Nat Biotechnol. 2002;20:826–830. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- 22.Frevert CW, Boggy G, Keenan TM, Folch A. Lab Chip. 2006;6:849–856. doi: 10.1039/b515560f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.