Abstract
Objective
Small integrin-binding ligand N-linked glycoproteins (SIBLINGs) are expressed in dentin and believed to control dentinogenesis. Five members of SIBLING family include bone sialoprotein (BSP), osteopontin (OPN), matrix extracellular phosphoglycoprotein (MEPE), dentin matrix protein 1 (DMP1) and dentin sialophosphoprotein (DSPP). These genes are clustered on chromosome 4q in humans and share similar biological features. DSPP and DMP1 are processed into given structural/functional fragments in rat and porcine. It still remains unclear whether these evidences occur in mouse and other SIBLING members are also processed into given fragments from their parent precursors. The aim of this study was to identify expression and processing of the five proteins in two mouse odontoblastic cell lines.
Design
Two mouse odontoblastic cells were used to study expression and processing of the five SIBLING proteins by immunohistochemistry and Western blot analyses.
Results
Immunohistochemistry study showed that all of the five SIBLING members were expressed within the cytoplasm and cellular processes in the mouse odontoblastic cell lines. Expression levels of DMP1 and DSPP were higher in differentiated mouse odontoblasts than undifferentiated mouse odontoblasts. Immunolabeling signal of DSP and MEPE was also detected within the nucleus in the two cell lines. Western blot assay indicated that all five members were processed into at least two fragments in these cells.
Conclusions
These results suggest that different processed products and expression levels of the SIBLING proteins may play distinct biological functions in tooth development and mineralization.
Keywords: SIBLING, odontoblasts, dentinogenesis, dentin, teeth
1. Introduction
Dentin is one of the major mineralized tissues of tooth and originates from odontoblasts, which synthesize collageous and non-collagenous proteins (NCPs) (1-4). Among NCPs, a family of small integrin-binding ligand N-linked glycoproteins (SIBLINGs) consists of osteopontin (OPN), matrix extracellular phosphoglycoprotein (MEPE, also known as OF45), bone sialoprotein (BSP), dentin matrix protein 1 (DMP1) and dentin sialophosphoprotein (DSPP). Genomic structures of the five SIBLING genes are located in chromosomes 4q in human and 5q in mouse (5-8). They share common gene structure features such as small non-translational first exon, start codon in exon 2 and large coding segment in the last exon (5-8). Also, these proteins contain an Arg-Gly-Asp (RGD) motif that mediates cell attachment/signaling via interaction with cell surface integrins and post-translational modification of phosphorylation and glycosylation. These SIBLING genes are highly expressed in mineralizing tissues related to tooth and bone development and mineralization (9-17).
BSP is a sulfated, phosphorylated and glycosylated protein characterized by its ability to bind to hydroxyapatite through polyglutamic acid sequences. High BSP expression is restricted to the mineralized connective tissues and seen at the onset of bone, dentin and cementum formation, indicating a role of this protein in the initial mineralization of these tissues (10, 18-19). OPN is a secreted adhesive glycoprotein expressed in both mineralized and non-mineralized tissues (20). In mineralized tissues, this gene expression is relatively abundant in bone (13) but also found in dentin (9, 21-22). Defective OPN gene in mice increases mineral content and crystallinity (23). MEPE was isolated from a human oncogenic hypophosphatemic osteomalacia (OHO) tumor cDNA library (24). Mouse and rat MEPE genes were also cloned (25-26). This protein is rich in aspartate, serine and glutamate residues and highly expressed in mineralized tissues (12, 21, 27-28). Increased expression of MEPE occurs in patients with X-linked hypophosphatemic rickets (27). Targeted disruption of MEPE gene in mice results in increased bone formation and bone mass, suggesting that MEPE inhibits mineralization (28).
DMP1 is an acidic phosphoprotein which is predominantly expressed in dentin and bone (14, 29-31). Mutations of DMP1 in human and mice cause profound defects in mineralization of both dentin and bone (32-35). DSPP is highly expressed in dentin compared to bone and other tissues (13-14, 16-17). DSPP protein is a precursor that is proteolytically processed into dentin sialoprotein (DSP) and dentin phosphoprotein (DPP) in rat teeth (36) whereas DSPP protein in porcine teeth is processed into DSP, DPP and a small segment termed dentin glycoprotein (DGP), which is located between DSP and DPP (37). Either DSP or DPP domain plays distinct biological functions during tooth development (38-44). In addition, two major fragments, 37- and 57-kDa originally from DMP1 precursor were observed in rat bone (45). Although cDNA sequences from mouse DSPP and DMP1 genes have been identified, it remains unknown if DSPP and DMP1 proteins in mouse are processed into given fragments and those proteolytical fragments are the same expressional patterns as that of rat and porcine. Furthermore, analysis of other SIBLING protein sequences shows that several proteinase cleavage sites exist in those proteins (46-47). Thus, it is not known whether OPN, MEPE and BSP proteins like DSPP and DMP1 are processed into given fragments.
Mouse is a comprehensive model for the study of tooth development and formation. Several mouse immortalized odontoblast-like cell lines have been established and used to study their biological functions responsible to intrinsic and extrinsic stimulations (17, 48-53). As the processing of SIBLING proteins in mouse odontoblasts has not been documented, in this study, we used two mouse immortalized odontoblast-like cell lines to investigate expression and processing of the five SIBLING proteins by immunohistochemistry and Western blot analyses.
2. Materials and Methods
2.1. Cell lines and cell culture
Mouse differentiated odontoblast-like (MO6-G3) and undifferentiated odontoblast-like (MD10-F2) cells were obtained from dental papilla mesenchymal cells isolated from the first mandibular molars of Swiss Webster mice at embryonic day 18 and immortalized by infection with recombinant defective retrovirus containing the temperature sensitive SV-40 large T-antigen cDNA (48). Both MO6-G3 and MD10-F2 cells express several markers of the odontoblastic genotype positive in alkaline phosphatase (ALP) staining and form Von Kossa nodules in vitro (17, 48, 53). Compared to MD10-F2, MO6-G3 cells express higher levels of the NCPs (17) and are identified as differentiated odontoblast-like cells by immunocytochemical and ultrastructural studies (53). MO6-G3 and MD10-F2 cells were grown at 33°C under 5% CO2 in alpha minimum essential medium (α-MEM) supplemented with 10% fetal calf serum, 100 units/ml penicillin/streptomycin, 50 μg/ml ascorbic acid, and 10 mM Na β-glycerophosphate (Sigma, St. Louis, MO).
2.2. Antibodies
Polyclonal rabbit anti-mouse DSP and anti-mouse DMP1 were purchased from Alpha Diagnostic International, San Antonio, TX, USA. The anti-DSP and anti-DMP1 antisera were raised in rabbit (Alpha Diagnostic International) against recombinant fragments representing the NH2-terminal portion (amino acid residues Ile18-Lys371) of mouse DSPP (54) and amino acid residues 1-508 of mouse DMP1, respectively (55). High titer polyclonal antisera as measured by enzyme-linked immunosorbent assay (ELISA) were obtained and then further purified by affinity column. Polyclonal rabbit anti-mouse MEPE (kindly provided by Dr. Peter Rowe, The Kidney Institute, University of Kansas Medical Center, MS, USA), polyclonal rabbit anti-mouse BSP (a gift from Dr. Larry Fisher, National Institute of Dental and Craniofacial Research, NIH, Bethesda, MD, USA), monoclonal mouse anti-mouse OPN, (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) were used as primary antibodies. Negative control of mouse IgG I was purchased from Dakocytomation (Carpinteria, CA, USA).
2.3. Immunohistochemistry
MO6-G3 and MD10-F2 cells were cultured on glass slides, rinsed twice with 1 x cold phosphate buffer saline (PBS) and fixed for 10 min on ice with methanol/acetone (1:1). After washing once with 1x PBS, cells were treated with 10% normal goat serum (Sigma) for 60 min at room temperature and followed by washing three times for 2 min with PBS. The cells were then incubated with a dilution of 1:100 of primary antibodies specific for BSP, DMP1, DSP, MEPE and OPN in PBS containing 1% BSA and 10% goat serum. Negative control of mouse IgG I was purchased from Dakocytomation (Carpinteria). The cells were incubated at 4°C for overnight and then washed 3 x for 5 min with PBS containing 0.1% goat serum, followed by a 1 : 1,000 dilution of the secondary antibodies (goat-anti rabbit or goat-anti mouse) with Alexa Fluo® 488 (Molecular Probes, Eugene, OR, USA) for 60 min at room temperature. Excess secondary antibody was removed by washing the cells three times with PBS. For nucleus staining, the cells were incubated with a 1: 5,000 dilution of Hoechst (Sigma) for 5 min at room temperature. After washing the cells with PBS, the sections were mounted using Vectashield mounting medium (Vector Laboratory Inc., Burlingame, CA, USA). Images of Alexa Fluo® 488 staining of the various proteins in cultures were obtained at the Core Optical Imaging Facility at UTHSCSA under the same parameters in a Nikon inverted microscope and quantitated by means of NIS-GIEMENTS software. For each experiment, all slides were simultaneously processed for a specific antibody, so that homogeneity in the staining procedure was ensured between the samples. After the capture of these images at the same magnification, the threshold was set and maintained for each slide in the experiment. The optical density was calculated by use of the morphometric analysis within the software package.
2.4. Western blot analysis
Western blotting assay was performed as described earlier (17). Briefly, MO6-G3 and MD10-F2 cells were washed with 1x cold PBS and lysed with RIPA buffer (1x PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 10 mg/ml phenylmethylsulfonyl fluoride, 30 μl/ml aprotinin, 100 mM sodium orthovanadate; Santa Cruz Biotechnology, Inc.). Whole cell lysates (50 μg/well) were resolved by 7 % SDS-polyacrylamide gel electrophoreses (SDS-PAGE) and transferred to Trans-Blot membranes (Bio-Rad Laboratory, Inc. Hercules, CA, USA). For the detection of mouse SIBLING proteins, the membranes were blocked with 5% non-fat milk in TBST buffer (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1% Tween-20) for 60 min at room temperature. After washing, the membranes were incubated with primary antibodies against DSP, DMP1, MEPE, BSP and OPN with appropriate dilution (1:500-800) for overnight at 4°C. The secondary antibodies (horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG) were used a dilution of 1:10,000 at room temperature for 60 min. Immunoreactivity was determined using the ECL chemiluminescence reagent (Amersham Biosciences, Piscataway, NJ, USA). As a control, goat anti-mouse β-actin antibody was used (Santa Cruz Biotechnology, Inc.).
3. Results
3.1. BSP
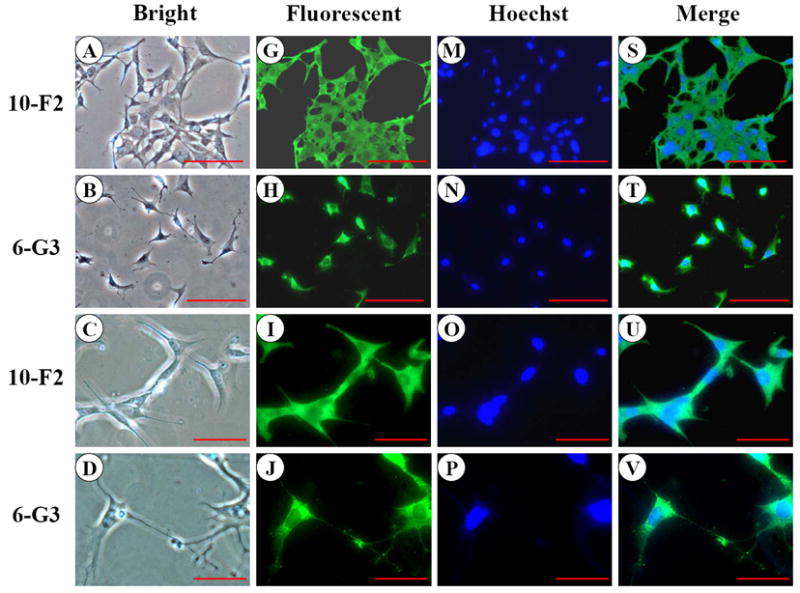
To assess BSP expression in mouse odontoblastic cells, we performed immunohistochemistry. The results in Figure 1 G-L showed that BSP expression was well distributed in the cytoplasm and cellular processes in both mouse odontoblastic cells. High magnification analysis demonstrated that BSP signal appeared more intense in the perinuclear basis (Fig. 1 K-L). To identify expressional patterns of BSP protein, we performed Western blot analysis. Data showed that approximately 39 and 27 kDa fragments of BSP protein were detected in the two odontoblastic cells (Fig.1Y).
Fig. 1. BSP expression in MD10-F2 and MO6-G3 cells.
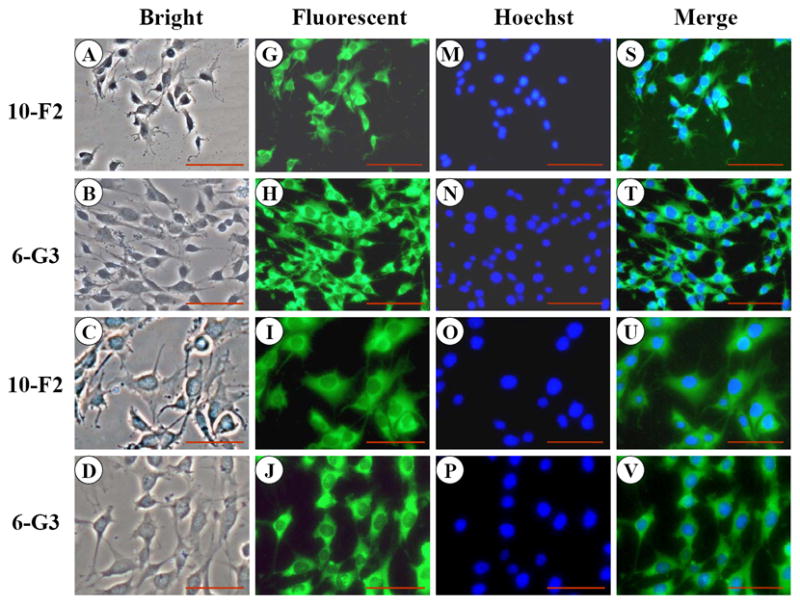
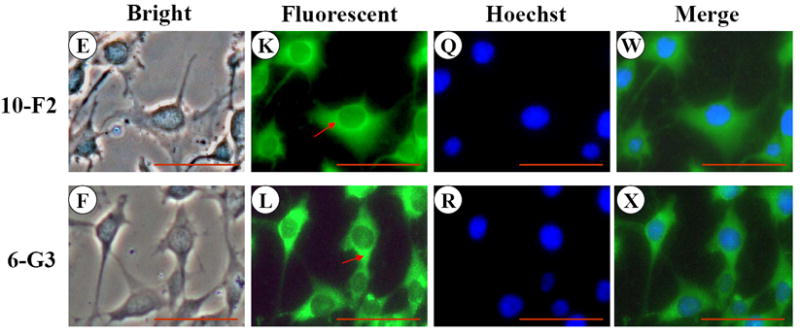
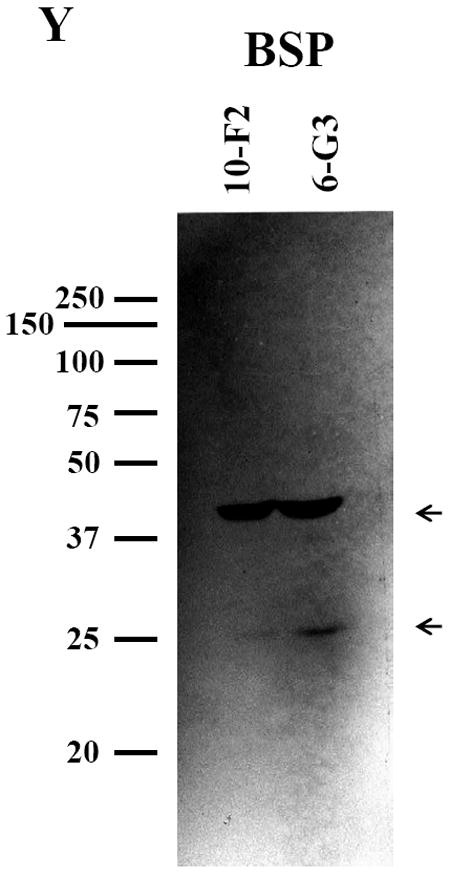
A-F, MD10-F2 and MO6-G3 cells were photographed under a light microscope using a Nikon camera. G-L, The BSP expression in these two cells was analyzed by immunostaining with a specific anti-BSP antibody. M-R, The cells were stained with Hoechst for the nucleus. S-X, Images were merged. High magnification allows seeing the BSP localization to most prominent cytoplasm area. A unipolar staining of BSP is present with a triangular basis on the perinuclear area (arrows). Bar = 100 μm. Y, Western blot analysis of BSP expression in MD10-F2 and MO6-G3 cells. Two bands of BSP protein were detected as 39-kDa and 27-kDa on a 7% SDS-PAGE gel using anti-BSP antibody. 10-F2 and 6-G3 indicate MD10-F2 and MO6-G3 cells, respectively.
3.2. OPN
Like BSP, OPN expression was predominantly detected in the cytoplasm and cellular processes in MD10-F2 and MO6-G3 cells (Fig.2 G-L). Expressional patterns of OPN in the mouse odontoblasts were analyzed by Western blot assay using anti-OPN antibody. Figure 2Y showed that three major fragments of OPN polypeptides were seen in a 7% SDS-PAGE gel.
Fig. 2. OPN expression in MD10-F2 and MO6-G3 cells.
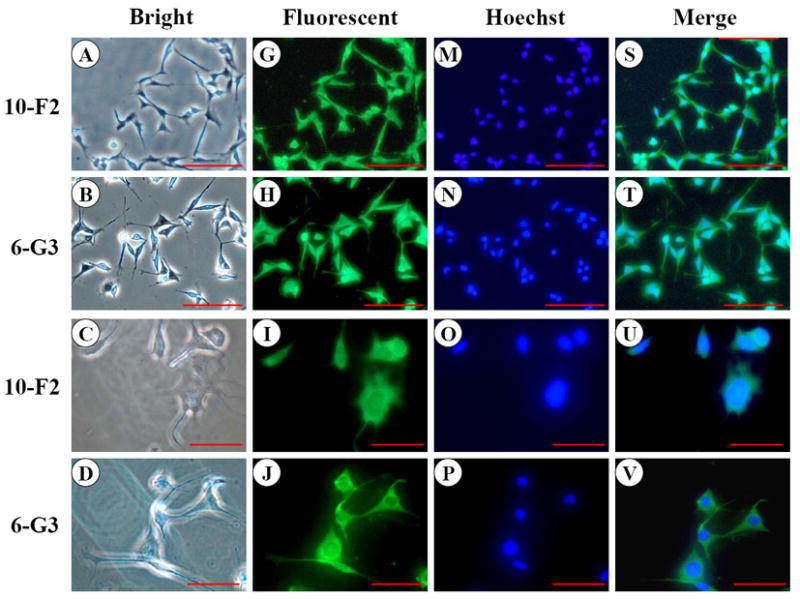
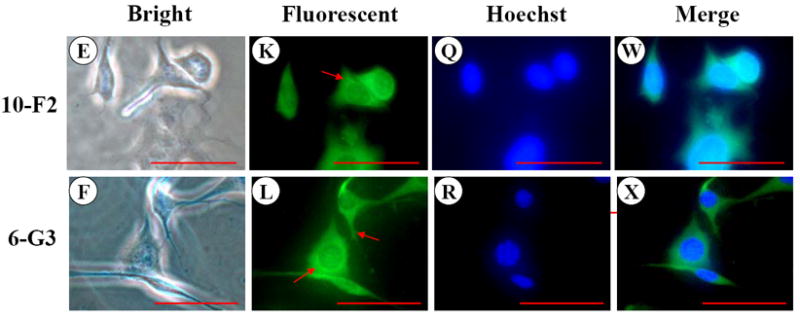
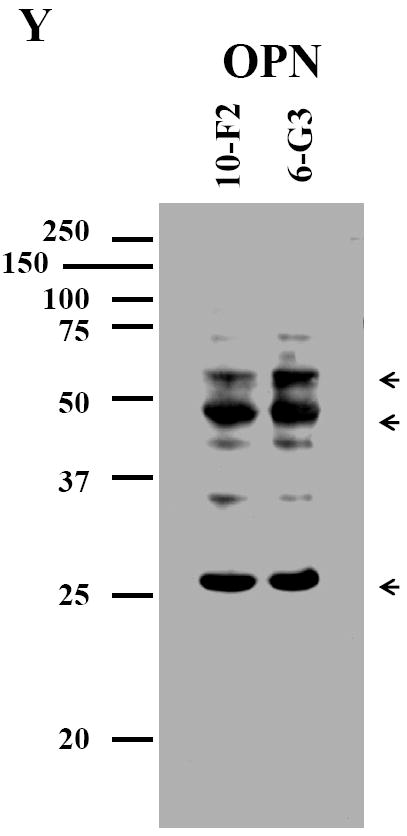
A-F, Cells were photographed under a light microscope. G-L, The OPN expression in the two cells was analyzed by immunostaining with a primary anti-OPN antibody. M-R, Cells were stained with Hoechst for the nucleus. S-X, Images were merged from G-L and M-R. OPN signal was detected in the cytoplasm and cellular processes in both the two cells (arrows). Bar = 100 μm. Y, Western blot analysis of OPN expression in MD10-F2 and MO6-G3 cells. Three major bands of OPN protein were detected by protein blot analysis as 55-kDa, 45-kDa and 27-kDa polypeptides in MD10-F2 and MO6-G3 cells on a 7% SDS-PAGE gel. 10-F2 and 6-G3 denote MD10-F2 and MO6-G3 cells, respectively.
3.3. DMP1
DMP1 signal was distributed in the cytoplasm and cellular processes in MD10-F2 and MO6-G3 cells by immunohistochemical analysis (Fig.3 G-L). Compared to MD10-F2 cells, DMP1 expression level in MO6-G3 cells was more profound (Fig.3 K-L). The same processed fragments of DMP1 protein in the two mouse odontoblastic cells were seen at molecular weight of about 53, 51 and 35 kDa, respectively in a 7 % SDS-PAGE gel using immunoblotting assay (Fig. 3Y). The processed patterns of mouse DMP1 polypeptides were similar to that of full-length mouse recombinant DMP1 protein cleaved by BMP1/tolloid-like proteinases in vitro (56).
Fig. 3. DMP1 expression in MD10-F2 and MO6-G3 cells.
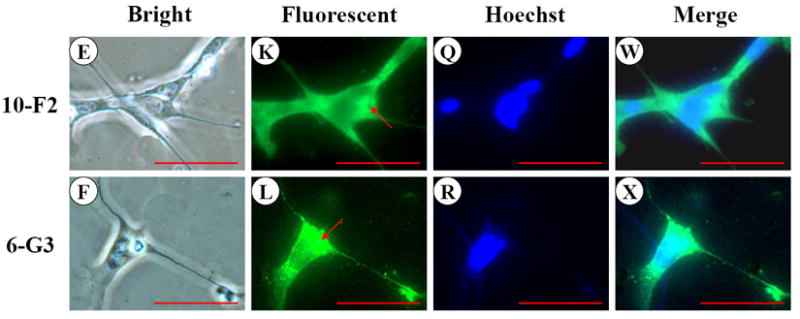
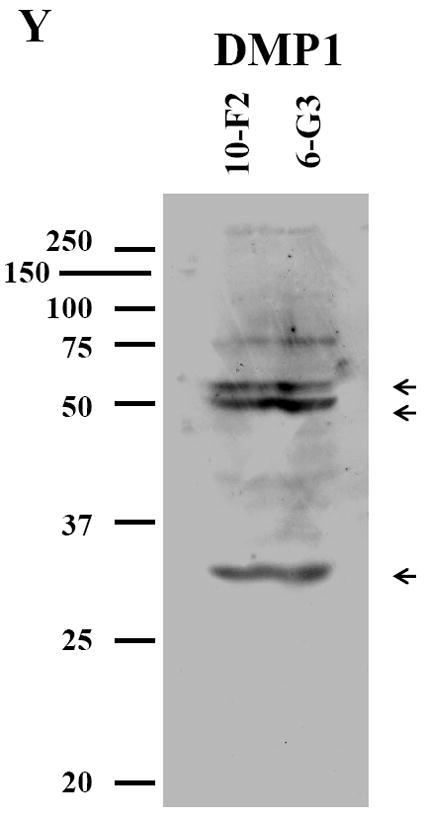
A-F, MD10-F2 and MO6-G3 cells were photographed under a light microscope using a Nikon camera. G-L, The DMP1 expression in two odontoblastic cells was analyzed by immunostaining with a primary anti-DMP1 antibody. M-R, Cells were incubated with Hoechst dye for the nucleus staining. S-X, Images are composites of G-L and M-R. DMP1 was expressed in the cellular processes and cytoplasm in the two odontoblastic cells. Arrows represent high DMP1 expression in specific areas in the cells by high magnification. Bar = 100 μm. Y, Western blot analysis of DMP1 expression in MD10-F2 and MO6-G3 cells. Three major fragments of DMP1 were seen as 53-kDa, 51-kDa and 35-kDa, respectively in a SDS-PAGE gel using anti-DMP1 antibody. 10-F2 and 6-G3 represent MD10-F2 and MO6-G3 cells, respectively.
3.4. MEPE
To examine MEPE expression in the mouse odontoblastic cells, we performed immunohistochemistry assay and found MEPE expression within both the cytoplasm and nucleus in MD10-F2 and MO6-G3 cells (Fig. 4G-L). Expressional patterns of MEPE were analyzed by Western blot assay. The results showed that three major fragments of MEPE protein were seen in the mouse odontoblastic cells (Fig.4Y).
Fig. 4. MEPE expression in MD10-F2 and MO6-G3 cells.
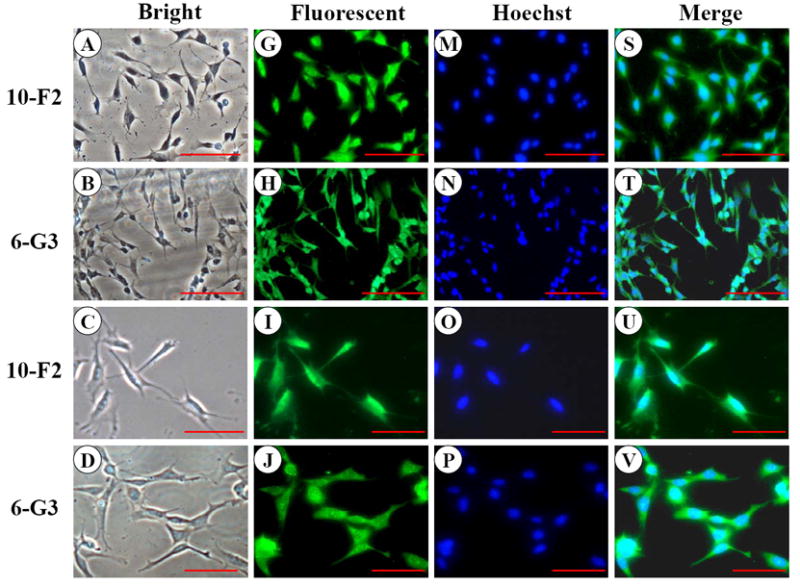
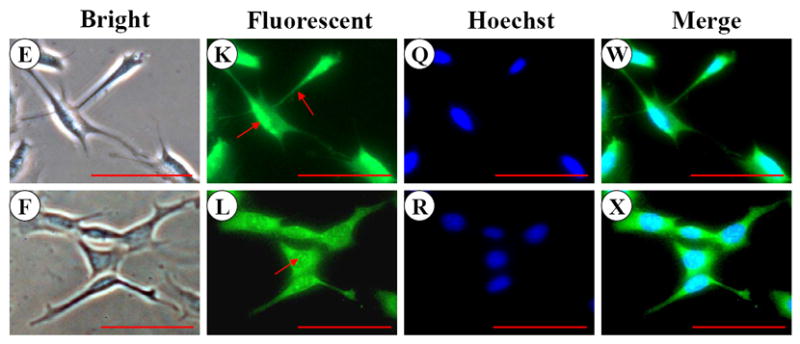
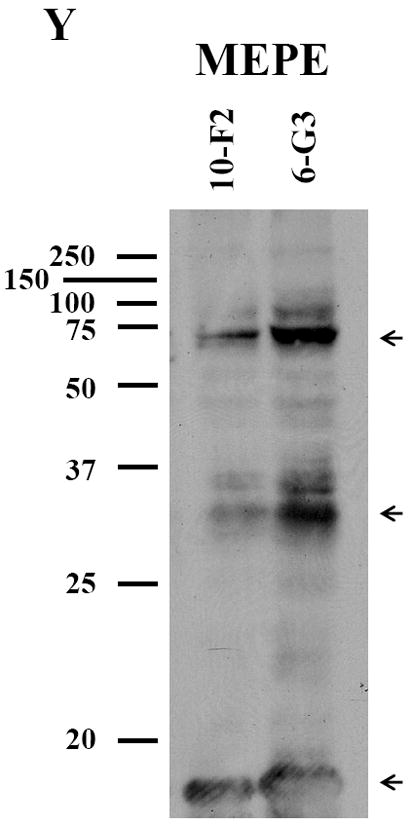
A-F, Cells were photographed under a light microscope. G-L, The MEPE expression in the two cells was analyzed by immunostaining with a primary anti-MEPE antibody. M-R, Cells were stained with Hoechst for the nucleus. S-X, Images were merged from G-L and M-R. MEPE signal was detected in the cytoplasm and nucleus in both mouse odontoblastic cells. Arrows show MEPE signaling within the nucleus. Bar = 100 μm. Y, Western blot analysis of MEPE expression in MD10-F2 and MO6-G3 cells. Three major bands of MEPE protein were detected by protein blot analysis as 70-, 30- and 10-kDa polypeptides in MD10-F2 and MO6-G3 cells on a 7% SDS-PAGE gel. 10-F2 and 6-G3 denote MD10-F2 and MO6-G3 cells, respectively.
3.5. DSP
Like MEPE, DSP signal was present within the cytoplasm and nucleus in the two odontoblastic cells by immunocytotistochemistry (Fig. 5G-L). However, DSP expression level in MO6-G3 cells was more intense than that of MD10-F2 cells (Fig.5G-L). To determine expressional patterns of DSP protein analyzed by Western blot assay, we were able to detect multiple fragments of DSP polypeptides in MD10-F2 and MO6-G3 cells using anti-DSP antibody (Fig.5Y). Four major bands were seen at molecular weight of about 250, 170, 70, 15 kDa, respectively.
Fig. 5. DSP expression in MD10-F2 and MO6-G3 cells.
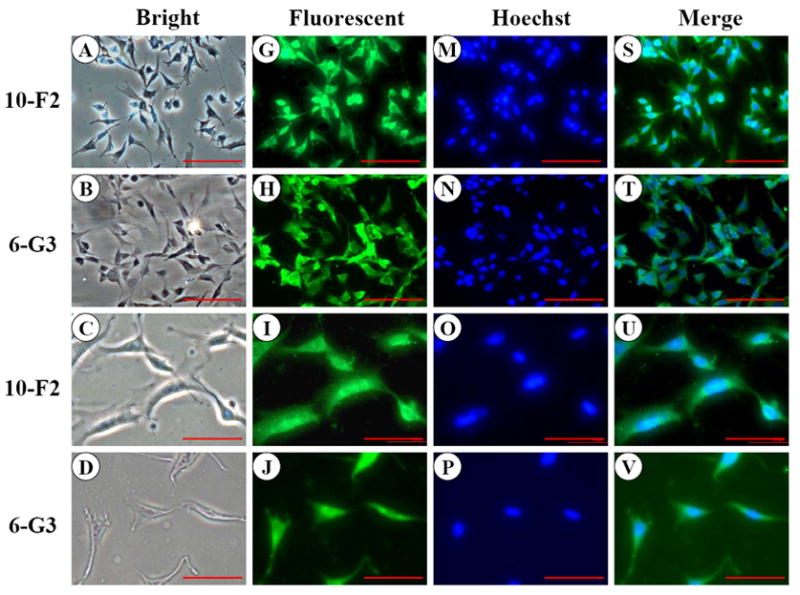
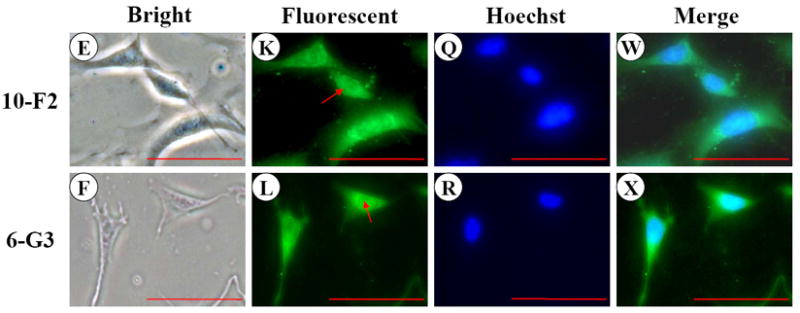
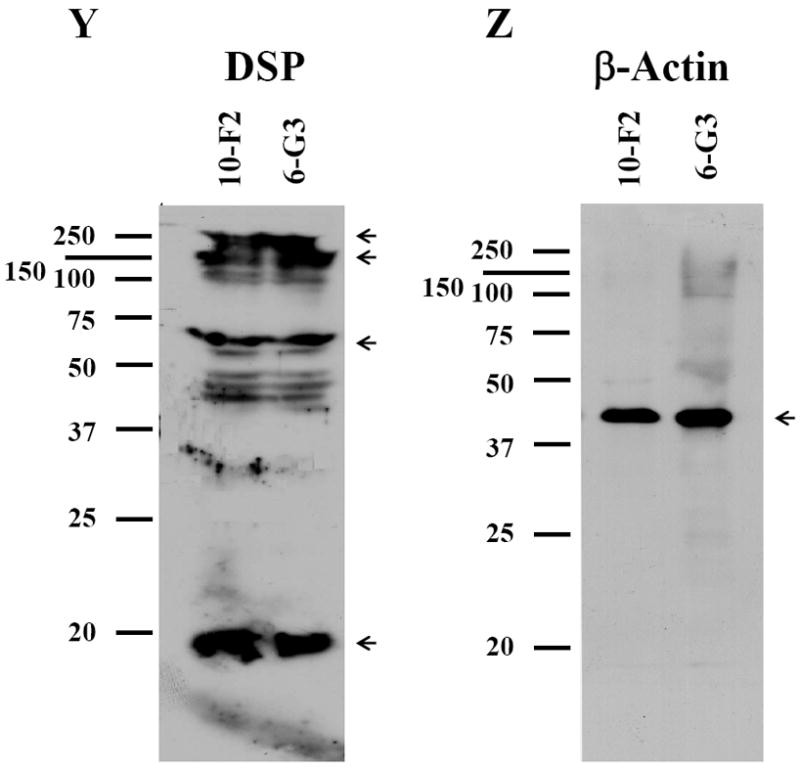
A-F, Cells were photographed under a light microscope. G-L, The DSP expression in the two cells was analyzed by immunostaining with a primary anti-DSP antibody. M-R, Cells were stained with Hoechst for the nucleus. S-X are composites of G-L and M-R. DSP signal was seen in both the cytoplasm and nucleus in the two mouse odontoblastic cells. Arrows indicate DSP signaling within the nucleus. Bar = 100 μm. Y, Western blot analysis of DSP expression in MD10-F2 and MO6-G3 cells. Multiple fragments of DSP polypeptides were detected in MD10-F2 and MO6-G3 cells using anti-DSP antibody. Major bands are 260-, 170-, 70- and 15-kDa as indicated by arrows. 10F2 and 6G3 represent MD10-F2 and MO6-G3 cells, respectively. Z, β-actin was used as control for Western blot assay.
3.6. Control study
Negative control for SIBLING proteins examined above demonstrated no positive staining by immunohistochemistry (data not shown). β-actin as internal control was used to detect whether proteins from the two odontoblastic cells were degraded during protein isolation process (Fig.5Z).
4. Discussion
SIBLING family is highly expressed in mineralized tissues and shares similar biological features as well as plays significant roles during dentinogenesis (5-8). It is well known that DSPP and DMP1 proteins are proteolytically processed into given fragments in tooth and bone tissues (36-37, 45). These processed fragments from DSPP and DMP1 precursors have their unique biological roles during formation and mineralization of teeth and bones (38-44, 57-58). To determine whether the other members of SIBLING family exhibit the same phenomena as the DSPP and DMP1, we assessed expression and processing of five members of the SIBLING family; BSP, OPN, MEPE, DMP1 and DSPP in two mouse odontoblastic cells. In this study, our results for the first time showed that the five SIBLING proteins were expressed and processed into given fragments in the two mouse odontoblastic cells. In addition, sizes and patterns of processed fragments from mouse DSPP and DMP1 precursors were different from that of rat and porcine described earlier (36-37, 45, 59).
For BSP, immunohistochemistry study indicated that its signal was dominantly seen in cytoplasm and cellular processes in MD10-F2 and MO6-G3 cells (Fig. 1 G-L) in agreement with previous reports in vitro and in vivo (10, 18). Expressional patterns of BSP protein analyzed by Western blot assay demonstrated that two bands were detected at positions, ~39- and 27-kDa, respectively in the two mouse odontoblastic cells (Fig. 1Y). Based on our observation, this suggests that intact BSP protein was processed within the mouse odontoblastic cells before being secreted. However, we have not known what enzymes catalyze BSP and which positions of BSP are cleaved by the enzymes as well as their biological roles.
Assessing OPN expression and processing, we found that in addition to OPN expression in the two mouse odontoblastic cells, two major fragments of OPN were detected at molecular masses of ~45- and 27-kDa by Western blot analysis (Fig. 2Y). Analysis of OPN protein sequences indicates that this protein contains a thrombin cleavage site (20). Boukpessi et al. reported that two fragments of OPN protein exist in human teeth using Western blot assay (21). However, sizes of the two OPN fragments with ~60- and 46-kDa in human teeth are different from that we observed. The differences may be due to species-species variations.
Although MEPE is an extracellular matrix protein, it is interesting that its expression was detectable in both the cytoplasm and nucleus (Fig. 4G-L). However, its role in the nucleus remains obscure. To examine expressional patterns of MEPE, we observed three major fragments that were recognized by anti-MEPE antibody in the mouse odontoblastic cells (Fig. 4Y). Guo et al. found that MEPE protein is cleaved by cathepsin B into several fragments in vitro (47). For its biological roles, knock-out of MEPE gene in mice increased bone formation and bone mass, indicating that MEPE inhibits mineralization process (28). However, recent studies have found that given domains of MEPE protein have different biological functions. Rowe et al. reported that a small peptide released from COOH-terminus of MEPE was able to inhibit mineralization processes in vitro (60) whereas another fragment from N-terminal MEPE with RGD motif accelerated mineralization (61). In this study, we identified the three major fragments of MEPE polypeptides in the mouse odontoblastic cells. Which fragment is derived from which portion of MEPE protein needs to be further investigated.
In the present study, three major fragments of DMP1 were detected in the two mouse odontoblastic cells (Fig. 3Y). The expressional patterns of the three DMP1 fragments were similar to that of a full length recombinant mouse DMP1 cleaved by BMP1/tolloid-like proteinases (56). However, the processed patterns of mouse DMP1 polyepeptides were different from that of rat and porcine (45, 62). Two DMP1 fragments, ~37- and 57-kDa were detected in rat bone (45) whereas two bands of porcine DMP1 were seen at molecular masses of ~90- and 60-kDa in porcine pulp-derived cells (62). It is known that rat DSPP is processed into DSP and DPP (36) whereas porcine DSPP is processed into DSP, DGP and DPP (37). Like DSPP, whether differences of the DMP1 processed products among mouse, rat and porcine are due to species-species variation needs to be further investigated. For DMP1 biological roles, DMP1 null mice showed teeth with failure of maturation of pre-dentin into dentin, enlarged pulp chambers, increased width of pre-dentin zone with reduced dentin wall and hypomineralization (35). Recent studies have found that these two fragments from rat DMP1 play different roles during mineralization processes (57-58). A fragment from the DMP1 COOH terminus (~57 kDa) isolated from rat bone enhanced the hydroxyapatite nucleation whereas the other fragment from DMP1 NH2-terminal domain (~37 kDa) inhibited nucleation of hydroxyapatite. Which fragment (37 kDa or 57 kDa) of DMP1 plays its dominant role during different stages of tooth and bone development and mineralization is currently unknown. We also observed that DMP1 was only expressed in the cytoplasm, but not in the nucleus in the two mouse odontoblastic cells (Fig.3G-L). Narayanan et al. (63) found that calcium is able to trigger DMP1 protein translocation from the nucleus into the cytoplasm in mouse pre-osteoblastic (MC3T3-E1) cells. However, the phenomenon occurred in MC3T3-E1 cells rather than in mouse fibroblast (NIH3T3) cells, suggesting cell-type specific. In addition, DMP1 expression in MO6-G3 cells was higher than MD10-F2 cells (Fig.3G-L). This indicates that its expression is relevant to odontoblastic differentiation and mineralization (14, 29-31).
Like DMP1, higher levels of DSP expression were detected in MO6-G3 cells and correspond to odontoblastic differentiation (13-14, 17). It is interesting to note that DSP protein signal was also present within the nucleus in the two mouse odontoblastic cells (Fig.5G-L). However, its role within the nucleus remains unknown. Furthermore, multiple processed fragments of mouse DSP polypeptides were detected by Western blot analysis (Fig.5Y). Qin et al. characterized 13 peptides of various regions of DSP peptides from rat teeth and identified major fragments ending at amino acid residues 409 and 421 (36). Yamakoshi et al. found that DSPP is processed into DSP, DPP and DGP in porcine teeth (37). DGP molecular weight on SDS-PAGE is 19 kDa and it contains an 81 amino acid segment located between the DSP and DPP domain. DGP has not been identified in rat and mouse teeth so far. DSP and DPP domains play different biological roles during tooth development and formation. Heterogeneous mutations of DSP domain in human are associated with dentinogenesis imperfecta type II (DGI-II) and dentin dysplasia type II (DDII) (38-39, 41-42) whereas DPP domain mutations cause dentinogenesis imperfecta type III (DGI-III) (40). DSPP null mice showed teeth similar to human DGI-III with discolored teeth, enlarged pulp chambers, a wider pre-dentin zone with thin dentin wall and hypomineralization (43). In addition to dentinogenesis, DSP and DPP also have distinct functions in enamel formation. Paine et al. reported that overexpression of DSP in transgenic mice resulted in increased rate of enamel formation, but DPP overexpression created “pitted” and “chalky” enamel of non-uniform thickness that is more prone to wear as well as resulted in abnormal primatic enamel structures (44). Most of studies of DSPP-derived proteins have been performed in rat and porcine tissues (36-37). In the present study, we found the four major DSP fragments existed in the two mouse odontoblastic cells and expressional patterns of mouse DSP were different from that of rat and porcine (36-37). The differences among mouse, rat and porcine may be due to species-species variations. As a full-length DSPP protein has not been detected within cells so far, it is speculated that DSPP is catalyzed by given enzymes within cells before being secreted. Moreover, based on our observations, it indicates that BSP, OPN and MEPE proteins were processed into given fragments within the mouse odontoblastic cells.
In conclusions, our first approach was to detect expression of the SIBLING family; BSP, OPN, MEPE, DMP1, and DSP in the two mouse odontoblastic cells by immunohistochemistry assay. Besides their expression in cytoplasm, signal of MEPE and DSP was also seen in the nucleus. The second approach was to investigate processing of the five SIBLING proteins in the two mouse odontoblastic cells by Western blot analysis. The results showed that these five proteins were processed into given fragments within the two odontoblastic cells before being secreted.
Acknowledgments
This work was supported by NIDCR, National Institutes of Health, Grant PO1 DE113221 (M.M.) and RO3 DE014484 (S.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thesleff I. Epithelial-mesenchymal signalling regulating tooth morphogenesis. J Cell Sci. 2003;116:1647–8. doi: 10.1242/jcs.00410. [DOI] [PubMed] [Google Scholar]
- 2.Maas R, Bei M. The genetic control of early tooth development. Crit Rev Oral Biol Med. 1997;8:4–39. doi: 10.1177/10454411970080010101. [DOI] [PubMed] [Google Scholar]
- 3.Linde A, Goldberg M. Dentiogenesis. Crit Rev Oral Biol Med. 1993;4:679–728. doi: 10.1177/10454411930040050301. [DOI] [PubMed] [Google Scholar]
- 4.Butler WT. Dentin matrix proteins. Eur J Oral Sci. 1998;106:204–210. doi: 10.1111/j.1600-0722.1998.tb02177.x. [DOI] [PubMed] [Google Scholar]
- 5.Fisher LW, Fedarko NS. Six genes expressed in bones and teeth encode the current members of the SIBLING family of proteins. Connect Tissue Res. 2003;1:33–40. [PubMed] [Google Scholar]
- 6.Qin C, Baba O, Butler WT. Post-translational modifications of sibling proteins and their roles in osteogenesis and dentinogenesis. Crit Rev Oral Biol Med. 2004;15:126–36. doi: 10.1177/154411130401500302. [DOI] [PubMed] [Google Scholar]
- 7.MacDougall M, Dong J, Acevedo AC. Molecular basis of human dentin diseases. Am J Med Genet A. 2006;140:2536–46. doi: 10.1002/ajmg.a.31359. [DOI] [PubMed] [Google Scholar]
- 8.Huq NL, Cross KJ, Ung M, Reynolds EC. A review of protein structure and gene organization for proteins associated with mineralized tissue and calcium phosphate stabilization encoded on human chromosome 4. Arch Oral Biol. 2005;50:599–609. doi: 10.1016/j.archoralbio.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 9.Bosshardt DD, Degen T, Lang NP. Sequence of protein expression of bone sialoprotein and osteopontin at the developing interface between repair cementum and dentin in human deciduous teeth. Cell & Tissue Res. 2005;320:399–407. doi: 10.1007/s00441-005-1106-8. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Sasaguri K, Sodek J, Aufdemorte TB, Jiang H, Thomas HF. Enamel epithelium expresses bone sialoprotein (BSP) Eur J Oral Sci. 1998;106:331–6. doi: 10.1111/j.1600-0722.1998.tb02194.x. [DOI] [PubMed] [Google Scholar]
- 11.Igarashi M, Kamiya N, Ito K, Takagi M. In situ localization and in vitro expression of osteoblast/osteocyte factor 45 mRNA during bone cell differentiation. Histochem J. 2002;34:255–63. doi: 10.1023/a:1021745614872. [DOI] [PubMed] [Google Scholar]
- 12.Liu H, Li W, Shi S, Habelitz S, Gao C, Denbesten P. MEPE is downregulated as dental pulp stem cells differentiate. Arch Oral Biol. 2005;50:923–8. doi: 10.1016/j.archoralbio.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Moses KD, Butler WT, Qin C. Immunohistochemical study of small integrin-binding ligand, N-linked glycoproteins in reactionary dentin of rat molars at different ages. Eur J Oral Sci. 2006;114:216–22. doi: 10.1111/j.1600-0722.2006.00353.x. [DOI] [PubMed] [Google Scholar]
- 14.D’Souza RN, Cavender A, Sunavala G, Alvarez J, Ohshima T, Kulkarni AB, et al. Gene expression patterns of murine dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (DSPP) suggest distinct developmental functions in vivo. J Bone Miner Res. 1997;12:2040–9. doi: 10.1359/jbmr.1997.12.12.2040. [DOI] [PubMed] [Google Scholar]
- 15.Papagerakis P, Berdel A, Mesbah M, Peuchmaur M, Malaval L, Nydegger J, et al. Investigation of ostoeocalcin, osteonectin, and dentin sialophosphrotein in developing human teeth. Bone. 2002;30:377–385. doi: 10.1016/s8756-3282(01)00683-4. [DOI] [PubMed] [Google Scholar]
- 16.Qin C, Brunn JC, Cadena E, Ridall A, Butler WT. Dentin sialoprotein in bone and dentin sialophosphoprotein gene expressed by osteoblasts. Connect Tissue Res. 2003;44:179–83. [PubMed] [Google Scholar]
- 17.Chen S, Rani S, Wu Y, Unterbrink A, Gu TT, Gluhak-Heinrich J, et al. Differential regulation of dentin sialophosphoprotein expression by Runx2 during odontoblast cytodifferentiation. J Biol Chem. 2005;280:29717–27. doi: 10.1074/jbc.M502929200. [DOI] [PubMed] [Google Scholar]
- 18.Sasaguri K, Ganss B, Sodek J, Chen JK. Expression of bone sialoprotein in mineralized tissues of tooth and bone and in buccal-pouch carcinomas of Syrian golden hamsters. Arch Oral Biol. 2000;45:551–62. doi: 10.1016/s0003-9969(00)00022-4. [DOI] [PubMed] [Google Scholar]
- 19.Sodek J. Bone sialoprotein. Crit Rev Oral Biol Med. 1999;10:79–98. doi: 10.1177/10454411990100010401. [DOI] [PubMed] [Google Scholar]
- 20.Denhardt DT, Noda M, O’Regan AW, Pavlin D, Berman JS. Osteopontin as a means to cope with environmental insults: regulation of inflammation, tissue remodeling, and cell survival. J Clin Invest. 2001;107:1055–61. doi: 10.1172/JCI12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boukpessi T, Septier D, Bagga S, Garabedian M, Goldberg M, Chaussain-Miller C. Dentin alteration of deciduous teeth in human hypophosphatemic rickets. Calcif Tissue Int. 2006;79:294–300. doi: 10.1007/s00223-006-0182-4. [DOI] [PubMed] [Google Scholar]
- 22.Bellows CG, Reimers SM, Heersche JN. Expression of mRNAs for type-I collagen, bone sialoprotein, osteocalcin, and osteopontin at different stages of osteoblastic differentiation and their regulation by 1, 25 dihydroxyvitamin D3. Cell & Tissue Res. 1999;297:249–59. doi: 10.1007/s004410051353. [DOI] [PubMed] [Google Scholar]
- 23.Boskey AL, Spevak L, Paschalis E, Doly SB, McKee MD. Osteopontin deficiency increases mineral content and mineral crystallinity in mouse bone. Calcif Tissue Int. 2002;71:145–154. doi: 10.1007/s00223-001-1121-z. [DOI] [PubMed] [Google Scholar]
- 24.Rowe PS, de Zoysa PA, Dong R, Wang HR, White KE, Econs MJ, et al. MEPE, a new gene expressed in bone marrow and tumors causing osteomalacia. Genomics. 2000;67:54–68. doi: 10.1006/geno.2000.6235. [DOI] [PubMed] [Google Scholar]
- 25.Petersen DN, Tkalcevic GT, Mansolf AL, Rivera-Gonzalez R, Brown TA. Identification of osteoblast/osteocyte factor 45 (OF45), a bone-specific cDNA encoding an RGD-containing protein that is highly expressed in osteoblasts and osteocytes. J Biol Chem. 2000;275:36172–80. doi: 10.1074/jbc.M003622200. [DOI] [PubMed] [Google Scholar]
- 26.Argiro L, Desbarats M, Glorieux FH, Ecarot B. Mepe, the gene encoding a tumor-secreted protein in oncogenic hypophosphatemic osteomalacia, is expressed in bone. Genomics. 2001;74:342–51. doi: 10.1006/geno.2001.6553. [DOI] [PubMed] [Google Scholar]
- 27.Bresler D, Bruder J, Mohnike K, Fraser WD, Rowe PS. Serum MEPE-ASARM-peptides are elevated in X-linked rickets (HYP): implications for phosphaturia and rickets. J Endocrinol. 2004;183:R1–9. doi: 10.1677/joe.1.05989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gowen LC, Petersen DN, Mansolf AL, Qi H, Stock JL, Tkalcevic GT, et al. Targeted disruption of the osteoblast/osteocyte factor 45 gene (OF45) results in increased bone formation and bone mass. J Biol Chem. 2003;278:1998–2007. doi: 10.1074/jbc.M203250200. [DOI] [PubMed] [Google Scholar]
- 29.George A, Gui J, Jenkins NA, Gilbert DJ, Copeland NG, Veis A. In situ localization and chromosomal mapping of the Ag1 (DMP1) gene. J Histochem Cytochem. 1994;42:1527–1531. doi: 10.1177/42.12.7983353. [DOI] [PubMed] [Google Scholar]
- 30.Toyosawa S, Shintani S, Fujiwara T, Ooshima T, Sato A, Ijuhin N, et al. Dentin matrix protein 1 is predominantly expressed in chicken and rat osteocytes but not in osteoblasts. J Bone Miner Res. 2001;16:2017–26. doi: 10.1359/jbmr.2001.16.11.2017. [DOI] [PubMed] [Google Scholar]
- 31.Kamiya N, Takagi M. Differential expression of dentin matrix protein 1, type I collagen and osteocalcin genes in rat developing mandibular bone. Histochem J. 2001;33:545–52. doi: 10.1023/a:1014955925339. [DOI] [PubMed] [Google Scholar]
- 32.Lorenz-Depiereux B, Bastepe M, Benet-Pages A, Amyere M, Wagenstaller J, Muller-Barth U, et al. DMP1 mutations in autosomal recessive hypophosphatemia implicate a bone matrix protein in the regulation of phosphate homeostasis. Nat Genet. 2006;38:1248–50. doi: 10.1038/ng1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng JQ, Ward LM, Liu S, Lu Y, Xie Y, Yuan B, et al. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat Genet. 2006;38:1310–5. doi: 10.1038/ng1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye L, Mishina Y, Chen D, Huang H, Dallas SL, Dallas MR, et al. Dmp1-deficient mice display severe defects in cartilage formation responsible for a chondrodysplasia-like phenotype. J Biol Chem. 2005;280:6197–203. doi: 10.1074/jbc.M412911200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye L, MacDougall M, Zhang S, Xie Y, Zhang J, Li Z, et al. Deletion of dentin matrix protein-1 leads to a partial failure of maturation of predentin into dentin, hypomineralization, and expanded cavities of pulp and root canal during postnatal tooth development. J Biol Chem. 2004;279:19141–8. doi: 10.1074/jbc.M400490200. [DOI] [PubMed] [Google Scholar]
- 36.Qin C, Cook RG, Orkiszewski RS, Butler WT. Identification and characterization of the carboxylterminal region of rat dentin sialoprotein. J Biol Chem. 2001;276:904–9. doi: 10.1074/jbc.M006271200. [DOI] [PubMed] [Google Scholar]
- 37.Yamakoshi Y, Hu JC, Fukae M, Zhang H, Simmer JP. Dentin glycoprotein: the protein in the middle of the dentin sialophosphoprotein chimera. J Biol Chem. 2005;280:17472–9. doi: 10.1074/jbc.M413220200. [DOI] [PubMed] [Google Scholar]
- 38.Xiao S, Yu C, Chou X, Yuan W, Wang Y, Bu L, et al. Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat Genet. 2001;27:201–4. doi: 10.1038/84848. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Zhao J, Li C, Gao S, Qiu C, Liu P, et al. DSPP mutation in dentinogenesis imperfecta Shields type II. Nat Genet. 2001;27:151–2. doi: 10.1038/84765. [DOI] [PubMed] [Google Scholar]
- 40.Dong J, Gu T, Jeffords L, MacDougall M. Dentin phosphoprotein compound mutation in dentin sialophosphoprotein causes dentinogenesis imperfecta type III. Am J Med Genet A. 2005;132:305–309. doi: 10.1002/ajmg.a.30460. [DOI] [PubMed] [Google Scholar]
- 41.Rajpar MH, Koch MJ, Davies RM, Mellody KT, Kielty CM, Dixon MJ. Mutation of the signal peptide region of the bicistronic gene DSPP affects translocation to the endoplasmic reticulum and results in defective dentine biomineralization. Hum Mol Genet. 2002;11:2559–65. doi: 10.1093/hmg/11.21.2559. [DOI] [PubMed] [Google Scholar]
- 42.Kim JW, Simmer JP. Hereditary dentin defects. J Dent Res. 2007;86:392–9. doi: 10.1177/154405910708600502. [DOI] [PubMed] [Google Scholar]
- 43.Sreenath T, Thyagarajan T, Hall B, Longenecker G, D’Souza R, Hong S, et al. Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta type III. J Biol Chem. 2003;278:24874–80. doi: 10.1074/jbc.M303908200. [DOI] [PubMed] [Google Scholar]
- 44.Paine ML, Luo W, Wang HJ, Bringas P, Jr, Ngan AY, Miklus VG, et al. Dentin sialoprotein and dentin phosphoprotein overexpression during amelogenesis. J Biol Chem. 2005;280:31991–8. doi: 10.1074/jbc.M502991200. [DOI] [PubMed] [Google Scholar]
- 45.Qin C, Brunn JC, Cook RG, Orkiszewski RS, Malone JP, Veis A, et al. Evidence for the proteolytic processing of dentin matrix protein 1. Identification and characterization of processed fragments and cleavage sites. J Biol Chem. 2003;278:34700–8. doi: 10.1074/jbc.M305315200. [DOI] [PubMed] [Google Scholar]
- 46.Rangaswami H, Bulbule A, Kundu GC. Osteopontin: role in cell signaling and cancer progression. Trends Cell Biol. 2006;16:79–87. doi: 10.1016/j.tcb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 47.Guo R, Rowe PS, Liu S, Simpson LG, Xiao ZS, Quarles LD. Inhibition of MEPE cleavage by Phex. Biochem Biophys Res Commun. 2002;297:38–45. doi: 10.1016/s0006-291x(02)02125-3. [DOI] [PubMed] [Google Scholar]
- 48.MacDougall M, Thiemann F, Ta H, Hsu P, Chen LS, Snead ML. Temperature sensitive Simian Virus 40 Large T antigen immortalization of murine odontoblast cell cultures: establishment of clonal odontoblast cell line. Connect Tissue Res. 1995;33:97–103. doi: 10.3109/03008209509016988. [DOI] [PubMed] [Google Scholar]
- 49.Hanks CT, Fang D, Sun Z, Edwards CA, Butler WT. Dentine-specific proteins in MDPC-23 cell line. Eur J Oral Sci. 1998;106:260–6. doi: 10.1111/j.1600-0722.1998.tb02185.x. [DOI] [PubMed] [Google Scholar]
- 50.Hanks CT, Sun ZL, Fang DN, Edwards CA, Wataha JC, Ritchie HH, et al. Cloned 3T6 cell line from CD-1 mouse fetal molar dental papillae. Connect Tissue Res. 1998;37:233–49. doi: 10.3109/03008209809002442. [DOI] [PubMed] [Google Scholar]
- 51.Hao J, Narayanan K, Ramachandran A, He G, Almushayt A, Evans C, et al. Odontoblast cells immortalized by telomerase produce mineralized dentine-like tissue both in vitro and in vivo. J Biol Chem. 2002;277:19976–81. doi: 10.1074/jbc.M112223200. [DOI] [PubMed] [Google Scholar]
- 52.Caton J, Bringas P, Jr, Zeichner-David M. Establishment and characterization of an immortomouse-derived odontoblast-like cell line to evaluate the effect of insulin-like growth factors on odontoblast differentiation. J Cellular Bioch. 2007;100:450–63. doi: 10.1002/jcb.21053. [DOI] [PubMed] [Google Scholar]
- 53.Mesgouez C, Oboeuf M, Mauro N, Colon P, MacDougall M, Machtou P, et al. Ultrastructural and immunocytochemical characterization of immortalized odontoblast MO6-G3. Int Endod J. 2006;39:453–63. doi: 10.1111/j.1365-2591.2006.01089.x. [DOI] [PubMed] [Google Scholar]
- 54.MacDougall M, Simmons D, Luan X, Nydegger J, Feng J, Gu TT. Dentin phosphoprotein and dentin sialoprotein are cleavage products expressed from a single transcript coded by a gene on human chromosome 4: Dentin phosphoprotein DNA sequence determination. J Biol Chem. 1997;272:835–842. doi: 10.1074/jbc.272.2.835. [DOI] [PubMed] [Google Scholar]
- 55.MacDougall M, Gu TT, Luan X, Simmons D, Chen J. Identification of a novel isoform of mouse dentin matrix protein 1: spatial expression in mineralized tissues. J Bone Miner Res. 1998;13:422–31. doi: 10.1359/jbmr.1998.13.3.422. [DOI] [PubMed] [Google Scholar]
- 56.Steiglitz BM, Ayala M, Narayanan K, George A, Greenspan DS. Bone morphogenetic protein 1/Tolloid-like proteinases process dentin matrix protein-1. J Biol Chem. 2004;279:980–6. doi: 10.1074/jbc.M310179200. [DOI] [PubMed] [Google Scholar]
- 57.Tartaix PH, Doulaverakis M, George A, Fisher LW, Butler WT, Qin C, et al. In vitro effects of dentin matrix protein-1 on hydroxyapatite formation provide insights into in vivo functions. J Biol Chem. 2004;279:18115–20. doi: 10.1074/jbc.M314114200. [DOI] [PubMed] [Google Scholar]
- 58.Gajjeraman S, Narayanan K, Hao J, Qin C, George A. Matrix macromolecules in hard tissues control the nucleation and hierarchical assembly of hydroxyapatite. J Biol Chem. 2007;282:1193–204. doi: 10.1074/jbc.M604732200. [DOI] [PubMed] [Google Scholar]
- 59.Yamakoshi Y, Hu JC, Iwata T, Kobayashi K, Fukae M, Simmer JP. Dentin sialophosphoprotein is processed by MMP-2 and MMP-20 in vitro and in vivo. J Biol Chem. 2006;281:38235–43. doi: 10.1074/jbc.M607767200. [DOI] [PubMed] [Google Scholar]
- 60.Rowe PS, Garrett IR, Schwarz PM, Carnes DL, Lafer EM, Mundy GR, et al. Surface plasmon resonance (SPR) confirms that MEPE binds to PHEX via the MEPE-ASARM motif: a model for impaired mineralization in X-linked rickets (HYP) Bone. 2005;36:33–46. doi: 10.1016/j.bone.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayashibara T, Hiraga T, Yi B, Nomizu M, Kumagai Y, Nishimura R, et al. A synthetic peptide fragment of human MEPE stimulates new bone formation in vitro and in vivo. J Bone Miner Res. 2004;19:455–62. doi: 10.1359/JBMR.0301263. [DOI] [PubMed] [Google Scholar]
- 62.Iwata T, Yamakoshi Y, Simmer JP, Ishikawa I, Hu JC. Establishment of porcine pulp-derived cell lines and expression of recombinant dentin sialoprotein and recombinant dentin matrix protein-1. Eur J Oral Sci. 2007;115:48–56. doi: 10.1111/j.1600-0722.2007.00426.x. [DOI] [PubMed] [Google Scholar]
- 63.Narayanan K, Ramachandran A, Hao J, He G, Park KW, Cho M. Dual functional roles of dentin matrix protein 1. Implications in biomineralization and gene transcription by activation of intracellular Ca2+ store. J Biol Chem. 2003;278:17500–8. doi: 10.1074/jbc.M212700200. [DOI] [PubMed] [Google Scholar]


