Abstract
Studies in invertebrate model organisms have led to a wealth of knowledge concerning the ageing process. But which of these discoveries will apply to ageing in humans? Recently, an assessment of the degree of conservation of ageing pathways between two of the leading invertebrate model organisms, Saccharomyces cerevisiae and Caenorhabditis elegans, was completed. The results (i) quantitatively indicated that pathways were conserved between evolutionarily disparate invertebrate species and (ii) emphasized the importance of the TOR kinase pathway in ageing. With recent findings that deletion of the mTOR substrate S6K1 or exposure of mice to the mTOR inhibitor rapamycin result in lifespan extension, mTOR signalling has become a major focus of ageing research. Here, we address downstream targets of mTOR signalling and their possible links to ageing. We also briefly cover other ageing genes identified by comparing worms and yeast, addressing the likelihood that their mammalian counterparts will affect longevity.
Keywords: TOR, ageing, lifespan, longevity, rapamycin
1. Introduction
Most evolutionary biologists see ageing not as a programme, but rather as a series of events resulting in the functional decline of an individual after the pressures of natural selection have diminished. This view, if correct, necessitates a rethinking of classical approaches to understanding biology. We study development in flies, for instance, on the belief that fundamental aspects of the developmental process are programmed and thus conserved across disparate lineages. If ageing is not programmed, why should modulation of the mechanisms underlying functional decline be conserved? Nevertheless evidence has emerged that this is the case, and recent findings have quantitatively demonstrated that pathways modulating ageing are conserved between two organisms that diverged evolutionarily approximately 1.5 billion years ago: Caenorhabditis elegans (worms) and Saccharomyces cerevisiae (yeast) [1].
Prior to quantitation, evidence had already accumulated that orthologous genes could affect ageing in multiple organisms. The search for conserved ageing genes led first to the insulin/IGF-1 signal (IIS) transduction pathway. Reduced IIS signalling leads to lifespan extension in worms [2–5], flies [6] and mice [7]. Numerous studies have attempted to understand the downstream effectors of IIS signalling that are important for lifespan extension. We will only discuss the pathway in detail as it relates to TOR signalling, in part because a number of reviews of IIS signalling are available. Nevertheless, there are lessons to be learned from the more mature studies of IIS signalling, foremost of which is that lifespan extension by altered signalling through this pathway is likely to involve the coordinated action of several downstream responses that together place the organism in a state more conducive to successful ageing [8–10]. Some of these responses include altered metabolism, activation of stress response pathways and enhanced autophagy. At this point, there is no reason to think that ageing-related responses downstream of TOR signalling will be any less complex. This implies that if researchers are to follow downstream altered cell signalling pathways to understand the molecular pathology that drives ageing, a holistic approach that integrates relative contributions from several different molecular events driving ageing and a linked understanding of the contribution of different responses may ultimately prove necessary. Is there any avoiding systems biology as ageing research moves forward?
(a). The TOR pathway
In the comparison between yeast and worm ageing genes described above, 11 gene orthologue pairs were identified in which reduced expression led to lifespan extension in the yeast replicative lifespan assay (see below) and in worms [1]. Six of the 11 gene pairs, including the TOR kinase itself, could be linked to regulation of TOR pathway signalling and translation. In yeast, the TOR kinase is activated by nutrients, either in the form of amino acids or carbohydrates [11] and, when activated, promotes cell growth and proliferation (figure 1). In higher organisms, TOR kinase activation occurs in response to amino acids [12], glucose via the IIS pathway, signalling through other pathways including AMP kinase and MAP kinase, and in response to stress through the regulation of TSC1–TSC2 complex. TSC impedes Rheb-dependent activation of mTORC1 [13]. Although the mechanisms by which TOR is activated differ in the details from one organism to the next, the fundamental point remains valid: TOR is responsive to nutrient levels. Consistently, studies in yeast and worms indicate that the longevity benefits of reduced TOR signalling are linked to that of dietary restriction [14–16], defined as a reduction in caloric intake without accompanying malnutrition [17]. In yeast, for example, inhibition of TOR signalling by rapamycin appears to affect the activity of hexose transporters, which lead to glucose entry and fermentation, and also the diauxic shift, a conversion from fermentation to respiration that occurs as cells deplete fermentable energy sources. These findings further underscore the intricate relationship that exists between TOR and energy consumption [11].
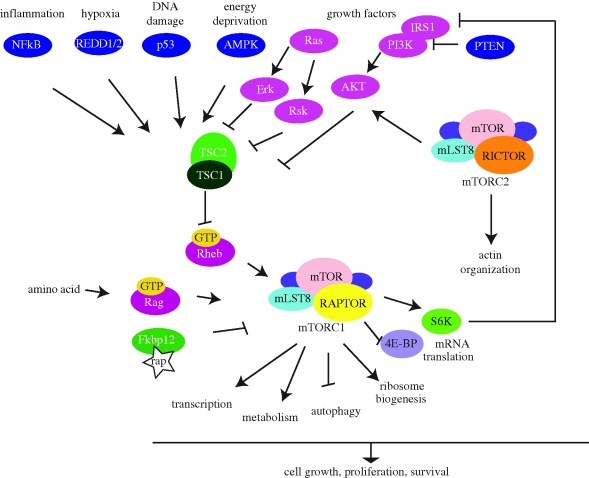
Figure 1.
mTOR signalling pathway. mTOR signalling pathways integrate environmental cues to regulate cellular growth in mammalian cells. The mTOR protein kinase is the core factor of two protein complexes, mTORC1 and mTORC2. Rapamycin-sensitive mTORC1 is responsive to nutrients and stress, and mediates diverse activities in the regulation of cellular homeostasis and growth. The GTP-bound form of the small G protein Rheb stimulates the activity of mTORC1. In turn, Rheb is regulated by the heterodimer of the tuberous sclerosis proteins TSC1 and TSC2. TSC1/2 negatively regulates mTORC1 activity by converting Rheb into an inactive GDP-bound state. Growth factors promote several kinase activities such as Akt, Erk and Rsk, which phosphorylate TSC1/2 and inhibit its activities, thereby inducing the mTORC1 signalling pathway. On the other hand, stress such as hypoxia, energy deprivation, DNA damage and inflammation activate TSC1/2 and repress mTORC1 signalling. Amino-acid activation of mTORC1 is regulated by the Rag GTPase and is independent of TSC1/2. Rapamycin-insensitive mTORC2 controls actin organization, which is essential for cell shape determination. mTORC2 is also activated by growth factors but the detailed mechanism is unclear. Arrows indicate activation, whereas bars indicate repression.
The central component of the TOR pathway, TOR kinase is encoded by two genes in yeast (TOR1 and TOR2) and one in other organisms relevant to this study (mTOR in mammals). TOR is essential for viability in organisms ranging from yeast to mammals (table 1); loss of TOR function results in embryonic lethality at the organism level and the growth inhibition of embryonic stem cells [18,19]. The kinase exists in two complexes, TORC1 and TORC2, which have different downstream targets. Among the proteins unique to TORC1 is Raptor, whereas Rictor is one protein unique to TORC2. Raptor and Rictor have both been linked to longevity, suggesting that both complexes may have age-specific functions [20]. TORC1 controls many events linked to ageing and will be the primary focus of this study. TORC2 plays a role in organization of the actin cytoskeleton, but also phosphorylates Akt, which both activates TORC1 and inhibits FOXO nuclear recruitment [21]. Recent findings in worms indicate that reduced adult expression of Rictor leads to increased fat storage, although reports differ as to the Akt dependence of this effect [22,23]. Reduced Rictor expression has also been reported to shorten lifespan of worms on a standard diet in an Akt-dependent manner, while extending lifespan on alternative nutrient-rich bacterial food sources [22].
Table 1.
Components of the TOR pathway in various eukaryotes.
| S. cerevisiae | C. elegans | Drosophila | mammals | |
|---|---|---|---|---|
| TSC complex | TSC1 | TSC1 | ||
| Gigus | TSC2 | |||
| RHEB-1 | Rheb | RHEB | ||
| TORC1 | Tor1/Tor2 | LET-363 | TOR | mTOR |
| Kog1 | DAF-15 | Raptor | RAPTOR | |
| Lst8 | C10H11.8 | CG3004 | mLST8 | |
| Tco89 | ||||
| PRAS40 | ||||
| TORC2 | Tor2 | LET-363 | TOR | mTOR |
| Lst8 | C10H11.8 | CG3004 | mLST8 | |
| Avo1 | SINH-1 | SIN1 | mSIN1 | |
| Avo2 | ||||
| Avo3 | RICT-1 | Rictor | RICTOR | |
| Bit61 | ||||
| downstream | Sch9 | RSKS-1 | S6K | S6K1/S6K2 |
| Eap1 | d4E-BP | 4EBP1-3 | ||
| Ypk2 | ||||
| IFE-2 | ||||
| SGK-1 | SGK1 | |||
| AKT-1, AKT-2 | AKT1 | AKT1 | ||
| RUVB-1 | ||||
| PHA-4 | FOXOA2 |
Dietary restriction is invoked by multiple means in different model organisms. In yeast replicative and chronological ageing, the most common method involves reduced glucose levels in the medium [24,25]. However, reduced amino-acid levels also lead to replicative lifespan extension [26]. Flies enjoy similar longevity benefits accompanying reduced yeast (amino acids) and/or sugar availability [27,28], although recent studies suggest a complex relationship between these two food sources, with the appropriate balance being important [29]. Many dietary restriction methods lead to lifespan extension in worms [30–35]. These findings call for a better understanding of the mechanisms by which dietary restriction and particularly reduced amino acids activate TOR signalling. Recent advances have been made in both yeast and mammals concerning the mechanisms by which amino acids lead to TOR activation. The fundamental mechanism, recruitment of TOR to the lysosomal (vacuolar in yeast) membrane, is conserved between yeast and mammals [36,37]. In yeast, the RAG GTPase Gtr1 interacts with and activates TOR in the presence of high amino-acid levels [38]. In mammals, mTORC1 approaches the lysosomal membrane via a similar Rag GTPases-Regulator complex [39]. An important follow-up regarding ageing will be to determine whether altered activity of these TOR recruitment factors will affect lifespan.
The direct connection leading from IIS signalling to Akt activation to TOR activation creates a union between two longevity pathways… and raises an important question. Do reduced IIS signalling and reduced TOR signalling lead to lifespan extension through the same or overlapping mechanisms? Judging from epistasis experiments in worms and flies, the pathways are not exactly the same, but may nevertheless have much in common. daf-2 and age-1 hypomorph mutants lead to lifespan extension in a manner dependent on the downstream FOXO transcription factor DAF-16 [3,4]. Lifespan extension by loss of daf-15 (CeRAPTOR) is also daf-16 dependent [20], while reduced let-363 (CeTOR) signalling increases lifespan independently of daf-16. In support of a possible functional overlap between these two pathways, it has also been shown that inhibition of let-363 (CeTOR) does not further extend the lifespan of daf-2 hypomorph mutant worms [40,41], which is consistent with the finding that daf-15 (CeRAPTOR) is itself downregulated by daf-16 [20], the activity of which should be increased in these long-lived daf-2 hypomorphs. Both TOR signalling [20] and IIS signalling [5] influence dauer formation (a diapause-like alternative larval stage in response to crowding or starvation) in worms. Finally, the possibility that these two pathways may derive from a common ancestor evolutionarily is hinted at by the fact that yeast Sch9 is a functional homologue of both S6K and Akt, as evidenced by partial rescue of sch9 mutant phenotype by mammalian Akt [42]. Independently of the degree of overlap, the fundamental question remains to what extent downstream components of both pathways contribute to modulation of longevity.
What is the molecular pathology that drives ageing in yeast, worms, flies, mice and, most importantly, humans? A major approach to answer this question has been to attempt to determine the downstream components of TOR signalling important for modulation of ageing. TOR controls several cellular processes including (but not limited to) translation initiation and elongation [43,44], autophagy [45–49], mitochondrial respiration [50–52], induction of stress response pathways [53], and the hypoxic response [54,55]. In the following sections, we discuss links between each of these pathways and longevity.
(b). Altered translational control
The two best known substrates of mTOR, S6 kinase and 4E-BP1, control translational initiation and elongation, and are both coupled to modulation of ageing in multiple organisms [56–58]. mTOR phosphorylation of S6 kinase stimulates its activity, in turn driving ribosome biogenesis and translation initiation to greater or lesser extents in yeast, worms, flies and mice. Among the downstream targets of S6 kinase are the ribosomal protein, Rps6, the initiation factor eIF4B, the elongation factor 2 kinase (eEF2K), and a range of other substrates the significance of which remains to be determined [44]. Along with TOR, S6 kinase is one of the most conserved modulators of ageing; reduced expression leads to lifespan extension in each of the three major invertebrate organisms [59]. In mice, knockout of one of two S6 kinases, S6K1, leads to enhanced longevity, with greater extension observed in females [60]. This gender specificity mirrors that observed in long-lived mice with reduced IIS signalling [7,61], as well as in rapamycin-treated mice [62]. S6K1 deletion also provides protection against high fat diet-induced diabetes [63], suggesting that the longevity benefits may be linked to altered metabolism. In the mouse longevity study, a genome-wide transcript array study in major systemic metabolic tissues including white adipose tissue, muscle and liver identified an altered transcriptional signature associated with enhanced AMP kinase activity [60]. Consistently, AMP kinase mutant, aak-2, worms were resistant to lifespan extension by rsks-1 (worm S6 kinase) RNAi [60]. While hinting at mechanism, these findings also point to the complexity of the signalling problem surrounding TOR. Reduced S6 kinase function leads to activation of AMP kinase and this, in turn, further stimulates TOR activation. How these pathways connect and which ultimate downstream targets are associated with ageing is a fundamental problem in ageing research. Another interesting question is whether long-lived S6K1−/− mice have reduced global or mRNA-specific translation and, if so, in what tissues. In myoblasts from double knockout of S6K1 and S6K2 mice, translation levels were found to be unaltered [64]; however, other key metabolic tissues remain to be tested.
TOR-mediated phosphorylation of 4E-BPs disrupts an inhibitory interaction between 4E-BP and the initiation factor, eIF4E, promoting increased translation initiation. Consistent with an important function for 4E-BPs in ageing, enhanced levels of the inhibitor (or reduced expression of eIF4E) lead to lifespan extension in both worms and flies [41,65]. Epistasis analysis in flies suggests that activation of d4E-BP/Thor may be tightly linked to the longevity benefits of dietary restriction [65]. Enhanced 4EBP1 activity in Drosophila is protective in cardiovascular and neurodegenerative disease models [66,67]. Therefore, altered activity of two key TOR substrates, S6K1 and 4E-BP, can lead to enhanced longevity and protection against age-related disease, raising the question of whether they act by the same mechanism and, if not, which of these events are more tightly coupled to lifespan extension by rapamycin.
Mutation of other translation initiation factors is associated with enhanced longevity in yeast and worms [1,16,41], as well as reduced expression of ribosomal protein genes (RPGs) [41,68,69]. The latter effect has been best studied via the yeast replicative ageing assay, where the lifespan of every viable RPG knockout was determined. Of note, most ribosomal protein genes are duplicated in yeast, meaning that knockout of one gene, e.g. RPL31A, typically leads to reduction but not ablation of that protein, since RPL31B encodes a second copy. Strikingly, all of the long-lived RPG deletions encoded components of the large subunit of the ribosome, precluding the simplest model that reduced translation was coupled to lifespan extension, since many strains lacking small subunit components have reduced translation: e.g. long-lived yeast rpl31aΔ, rpl20bΔ, and rpl21aΔ strains have been shown by depressed overall polysome profiles to have decreased overall translation [68]. Instead, the mechanism, at least in part, entails activation of translation of GCN4, a transcription factor the own expression of which is regulated at the translational level. GCN4 targets include amino-acid biosynthetic and stress response factors, including components of the unfolded protein response. Translation is activated during starvation conditions including dietary restriction, at a time when most mRNAs experience reduced translation. The mechanism involves small regulatory upstream open reading frames in the 5′ UTR of GCN4. Initiation at these uORFs, particularly the one most proximal to the start site of GCN4, inhibits GCN4 translation since a strong termination signal promotes dissociation of the ribosome, and the short distance between the uORF and the GCN4 ORF precludes re-initiation. Amino-acid or carbohydrate limitation both lead to reduced translation initiation, allowing the small ribosomal subunit to scan past the uORFs before associating with the large subunit and initiation factors, leading paradoxically to more GCN4 translation. Specifically in the case of carbohydrates, glucose limitation (0.05% glucose as opposed to 2% glucose) induces GCN4 translation by activation of GCN2 protein kinase [70]. Reduced 60S subunit levels, brought about by deletion of a range of large subunit components, phenocopy starvation since 40S subunits have a harder time finding their 60S counterparts, leading to scanning past the uORFs more frequently prior to initiation. The importance of this pathway for longevity is illustrated by the findings that deletion of GCN4 blocks much of the lifespan extension associated with reduced 60S subunit biogenesis (although some extension still occurs), pointing to the existence of other mechanisms. Similarly, deletion of GCN4 reduced lifespan extension by dietary restriction, tor1Δ and sch9Δ (yeast S6 kinase). Another report indicated that deletion of one small subunit component could also extend yeast replicative lifespan [69], again pointing to additional mechanisms. Finally, while reduced expression of worm RPGs also leads to enhanced longevity, the involvement of the GCN4 orthologue, atf-5, is currently unknown and, since reduced expression of both 40S and 60S subunits can promote longevity, mechanistic differences are likely.
Combining the data described above, a strong case for altered translation associated with reduced TOR activity in the modulation of longevity can be made; however, the details remain to be fully elucidated. Furthermore, equally strong cases can be made for other downstream regulatory pathways (see below).
(c). A role for autophagy
Autophagy is a well-studied process by which cells degrade damaged molecules through their recruitment to the lysosome [71]. This pathway is also used to recycle cellular components (both organelles and individual proteins). This latter activity implies that autophagy should be responsive to cellular nutrient status; TOR signalling represents a primary signalling pathway that controls its activity [45–49]. Under high nutrient levels, where energy is easy to come by, high TOR signalling suppresses autophagy and the converse is also true. Nutrient deprivation and concomitantly reduced TOR signalling lead to elevated autophagy. TOR-mediated suppression of autophagy probably occurs through inhibition of a protein complex containing Atg13 that is required for induction of the autophagic response. Indeed, TORC1 does directly phosphorylate Atg13 on at least eight serine residues [72,73].
Autophagy has been closely linked to ageing [74]. In C. elegans, impaired autophagy by knockdown (mutant) of worm bec-1, homologue of yeast VPS30/mammalian beclin1 [75], as well as atg-7 and atg-12 [76], blocks lifespan extension by a daf-2 mutant. Similarly, lifespan extension via reduced TOR signalling (let-363 aka CeTOR RNAi) has been shown to depend on bec-1, and extension by dietary restriction (eat-2 mutants) has been shown to depend on both bec-1 and vps-34 [77]. In yeast chronological ageing, autophagy is important for normal survival as well as lifespan extension by rapamycin [78,79]. Finally, in Drosophila inhibition of autophagy abrogates the rapaymcin-dependent lifespan extension [80].
While these findings have not been extended to other organisms, they strongly suggest that induced autophagy may be an important downstream consequence of reduced TOR signalling with respect to longevity. But TOR may also be downstream of autophagy. Enhanced Atg1 expression in Drosophila inhibits TOR signalling, presumably as a regulatory mechanism to further enhance induction of autophagy [81]. Feedback mechanisms such as these are pervasive in TOR signalling [82–84]. Presumably, this regulation allows pinpoint control over the responses to nutrient signalling. In the context of research on ageing, however, sorting out the downstream signalling components linked to lifespan extension is significantly complicated by these high levels of connectivity.
(d). Enhanced mitochondrial function
Levels of mTOR activity directly modulate cellular metabolism, in part by shifting the balance between modes of energy production and usage. In tumour cells, activation of the PI3K/mTOR pathway leads to increased glycolysis [85–87]. Glycolysis is globally used by many human tumour cells and stem cells. Glycolytic metabolism is not used to maximize energy production; however, it does generate more biomass than mitochondrial oxidation, an attribute favourable to tumour cell growth [85–87]. One molecule of glucose is completely oxidized in the mitochondrial tricarboxylic acid (TCA) cycle to generate 36 ATPs with minimal production of lactate. On the other hand, glycolysis only produces 2 ATP per molecule of glucose with large amounts of lactate, which regenerates NAD+ from NADH. The NAD+/NADH ratio influences the rate of biosynthetic pathways that use intermediates derived from glucose metabolism, so glycolysis generates more material for nucleotides, lipid and protein biosynthesis.
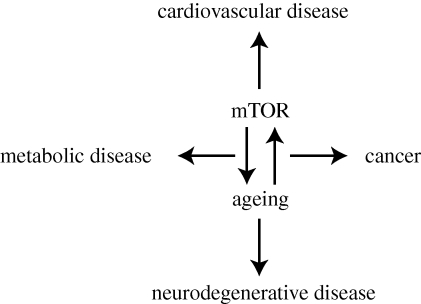
Repression of FoxO3a activity by PI3K pathway activation or by RNAi knockdown of FoxO3a transcriptionally down-regulates TSC1. As a consequence, suppression of TSC1 increases mTORC1 activity, enhancing glycolysis and survival after growth factor withdrawal in murine haematopoietic cells [82]. Consistent with this notion, inhibition of the mTOR pathway or glycolysis can suppress growth and/or induce death in cancer [88–91]. Inactivation of the TOR pathway increases mitochondrial number and induces mitochondrial gene expression via translational regulation, leading to enhanced cellular respiration. Similarly, enhanced d4E-BP1 activity in Drosophila melanogaster or S6K1 deletion in mouse results in increased mitochondrial biogenesis and in both cases results in lifespan extension [60,65]. In the case of Drosophila, dietary restriction leads to d4E-BP-dependent increased translation (but not transcription) of several components of the mitochondrial electron transport chain [65]. In this context, the increased lifespan of the flies upon dietary restriction has been shown to be dependent on both d4E-BP and on some of the upregulated electron transport chain components [65]. The positive correlation of mitochondrial biogenesis and longevity has been shown in several organisms [92,93]. Increased mitochondrial biogenesis could be the keystone of TOR-dependent lifespan extension (figure 2).
Figure 2.
Deregulation of mTOR signalling in human diseases. Recently, inhibition of mammalian TOR (mTOR) by rapamycin was shown to extend longevity even when administered in older mice. Application of rapamycin in transgenic animal models of age-related disease such as diabetes, neurodegenerative diseases and cancer show promising outcomes. Combination therapy using rapamycin and PI3K pathway inhibitors is also being tested in clinical trials to treat breast cancer, leukaemia, neurofibroblastoma, non-small cell lung cancer and others forms of cancer, with results still pending. Currently there are several clinical trials of rapamycin in treatment of coronary artery and heart disease, type I diabetes, age-related macular degeneration, kidney disease and autoimmune diseases (ClinicalTrials.gov). Thus, mTOR appears to be an attractive molecular target for pharmacological interventions to treat ageing and age-related diseases.
In contrast to the in vivo studies described above, cell culture studies of the links between mTOR activity and mitochondrial function have pointed to a different relationship. mTOR has been shown to physically interact with the transcription factor yin-yang 1 (YY1) [52], which regulates mitochondrial gene expression, as well as the mitochondrial outer-membrane proteins Bcl-xl and VDAC1 [95]. Inhibition of mTORC1 activities by RNAi knockdown of genes coding components of the mTORC1 complex or rapamycin results in the reduction of oxygen consumption and decreased mitochondrial respiration in vitro [94]. The difference in outcomes between in vivo and in vitro studies points to the need to further elucidate the relationship between TOR signalling and respiration in mammals.
In yeast, a more clarified picture has emerged. Reduced TOR function leads to enhanced respiratory capacity in the absence of enhanced mitochondrial biogenesis, and this is required for chronological lifespan extension [50,95]. Instead of enhanced biogenesis, the result of reduced TOR signalling is more OXPHOS complexes per mitochondrion [96]. Moreover, ATP production is not elevated. Rather, the result is enhanced uncoupling activity and reduced inner membrane potential. These findings lead the authors to speculate that these changes, which happen during cell proliferation and early cell cycle stages, precondition cells to a state promoting prolonged survival during the growth arrested period. Deletion of yeast S6 kinase (sch9), which also promotes enhanced chronological lifespan [97], is also linked to these changes in respiration [96,98]. Unlike chronological ageing, extension of yeast replicative lifespan by sch9Δ is not impaired by blocking accompanying enhancement of respiration [98].
(e). Induction of stress response pathways
In yeast, reduced TOR signalling or deletion of sch9 leads to nuclear relocalization and/or activation of a set of stress-responsive transcription factors including the Msn2/4 complex and Gis1 [53]. The kinase Rim15 is also retained in the cytoplasm in cells with high TOR activity [99,100]. The mechanism of TOR-dependent cytoplasmic retention of Msn2/4 and Rim 15 may involve inhibitory binding of the transcription factor complex by the 14-3-3 proteins Bmh1 and Bmh2 [99,101]. Rim15, when translocated to the nucleus, promotes activation of the Msn2/4 and Gis1 transcription factors [102,103]. Among the targets of Msn2/4 and Gis1 are components of oxidative defence pathways including superoxide dismutase. This pathway is required for chronological lifespan extension by tor1Δ or sch9Δ [15,101,104], although increased superoxide dismutase levels only account for part of the effect [97,104,105]. Thus, other targets of these transcription factors are likely to be important. Extension of replicative lifespan by reduced TOR signalling has been linked to enhanced Msn2/4-dependent transcription of PNC1 [106], which in turn promotes activation of the protein deacetylase SIR2 [107,108]. The links between TOR signalling and SIR2 are unclear however, since it has been reported that tor1Δ and sch9Δ robustly extend lifespan in strains lacking SIR2, as long as extrachromosomal rDNA circles are maintained at low levels [109]. In worms, let-363 (CeTOR) mutants have been shown to have increased thermotolerance [41].
Another mechanism has recently been proposed for yeast chronological lifespan extension by dietary restriction and for many long-lived mutants [110]. In rich medium, yeast prefer fermentative growth, only relying on low levels of respiration. During fermentative growth, yeast produce and secrete high levels of ethanol and organic acids such as acetic acid. Dietary restriction involves starting yeast cultures under conditions of lower glucose concentration, which results in a robust increase in survival during chronological ageing [111–113]. A recent study reported that the long lifespan associated with dietary restriction in chronological ageing derives from less fermentative growth, resulting in less secretion of organic acids and maintenance of a higher extracellular pH [110]. High extracellular acetic acid levels and low pH together formed a toxic combination that accelerated yeast mortality in the post-mitotic environment and appeared to be largely responsible for many measured differences in yeast chronological lifespan, as reversing these effects reversed the lifespan differences. Relevant to this study, both tor1Δ and sch9Δ are reported to result in enhanced respiratory activity, possibly limiting acetic acid production by shunting fermentative products into respiratory metabolism [50,98]. Furthermore, the sch9Δ was found to be resistant to toxicity associated with high levels of acetic acid in a manner requiring RIM15 [110]. How this reduction of yeast chronological lifespan by extracellular acetic acid is related to other proposed mechanisms for chronologic ageing, or ageing in other model organisms, remains to be determined.
(f). The hypoxic response
Under hypoxic conditions, many organisms exhibit a conserved transcriptional response mediated by transcription factor Hypoxia Inducible Factor 1 (HIF-1) [114]. Oxygen-dependent prolyl hydroxylases normally target one subunit of HIF-1, HIF-1α, for recognition by the E3 ubiquitin ligase von Hippel–Lindau protein (VHL), leading to HIF-1α's subsequent ubiquitination and degradation [115–117]. Under hypoxic conditions, HIF-1α degradation by this oxygen-dependent mechanism is decreased, and HIF-1α accumulates to transcribe its target genes, after dimerization with HIF-1β/ARNT [118]. This hypoxic response has been linked to ageing in C. elegans [119–121], although the effect of HIF-1 on lifespan appears to be context specific [122]. Both increased and reduced HIF-1 activity can lead to lifespan extension, but the mechanisms appear to be different [119–121]. These findings emphasize the importance of the hypoxic response in longevity and call for detailed studies to place hif-1 in the context of other known longevity pathways.
One transcriptional target of HIF1 in flies and mammals is Regulated in Development and DNA Damage Responses 1 (REDD1) [54,55,123]. REDD1 activates the TSC1/TSC2 tuberous sclerosis complex by releasing TSC2 from inhibitory 14-3-3 proteins, allowing TSC2 to interact with TSC1, resulting in TORC1 inhibition [54,55,124]. While HIF-1 regulates TOR, the converse is also true. TOR controls either HIF-1 translation [125] or nuclear localization [126] in mammalian cells and Drosophila, respectively. Additionally, a protein tyrosine phosphatase, Ptp61F, has been shown in Drosophila cells to be required for hypoxic suppression of translation through TOR signalling [127]. These studies point to a complex relationship between nutrient levels, TOR signalling, HIF1 and ageing, raising the possibility that lifespan extension by reduced TOR signalling may be linked to altered HIF1 activity.
2. Conclusions
The good news is that convincing evidence has been provided by several laboratories linking reduced TOR signalling to lifespan extension in yeast, worms, flies and mice (table 2). A central nexus linking nutrient levels to cell growth, proliferation and survival, it is perhaps not surprising that TOR modulates ageing. The bad news is that, being a central nexus, TOR regulates a range of downstream pathways, many of which have been linked to ageing. Why does reduced TOR signalling result in lifespan extension and, by working downstream, will it be possible to determine the age-related pathologies that are mitigated? Integrated approaches are likely to be required to answer these questions. These will probably include (i) genetic and biochemical approaches to understand TOR signalling in the context of other longevity pathways, (ii) studies to determine in which tissues reduced TOR signalling promotes longevity, and (iii) systems biology to integrate information from large datasets.
Table 2.
Some core components (excluding downstream targets) of the TOR pathway implicated in longevity.
| S. cerevisiae | C. elegans | Drosophila | mammals | |
|---|---|---|---|---|
| TSC complex | overexpression of TSC1 | |||
| overexpression of Gigus | ||||
| RNAi knockdown of rheb-1 | ||||
| TORC1 | TOR1 deletion, rapamycin or MSX treatment | null mutant or RNAi knockdown of let-363 | overexpression of TOR dominant negative mutation or rapamycin treatment | rapamycin treatment |
| TORC2 | daf-15 hypomorph mutant |
Given the findings last year that the TOR inhibitor rapamycin extends mouse lifespan, even when administered late in life [62], it seems clear that the good outweighs the bad. This study not only linked TOR to mammalian ageing, but demonstrated that pharmacological interventions can influence longevity. Whether rapamycin will ever be used to delay ageing as a means to target age-related disease is a matter of debate, awaiting further studies. It seems reasonable to assert, however, that if there is one drug that impacts ageing, then there are likely to be others (and candidates affecting other targets already exist), increasing the likelihood that one will be efficacious in humans. By better understanding TOR signalling in the context of ageing, it may also be possible to identify more specific targets to extend lifespan and healthspan. Exciting discoveries in ageing research are surely ahead.
Footnotes
One contribution of 15 to a Discussion Meeting Issue ‘The new science of ageing’.
References
- 1.Smith E. D., et al. 2008Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 18, 564–570 10.1101/gr.074724.107 (doi:10.1101/gr.074724.107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman D. B., Johnson T. E.1988A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics 118, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R.1993A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464 10.1038/366461a0 (doi:10.1038/366461a0) [DOI] [PubMed] [Google Scholar]
- 4.Dorman J. B., Albinder B., Shroyer T., Kenyon C.1995The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics 141, 1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura K. D., Tissenbaum H. A., Liu Y., Ruvkun G.1997daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277, 942–946 10.1126/science.277.5328.942 (doi:10.1126/science.277.5328.942) [DOI] [PubMed] [Google Scholar]
- 6.Garofalo R. S.2002Genetic analysis of insulin signaling in Drosophila. Trends Endocrinol. Metab. 13, 156–162 10.1016/S1043-2760(01)00548-3 (doi:10.1016/S1043-2760(01)00548-3) [DOI] [PubMed] [Google Scholar]
- 7.Bartke A.2008Impact of reduced insulin-like growth factor-1/insulin signaling on aging in mammals: novel findings. Aging Cell 7, 285–290 10.1111/j.1474-9726.2008.00387.x (doi:10.1111/j.1474-9726.2008.00387.x) [DOI] [PubMed] [Google Scholar]
- 8.Kenyon C. J.The genetics of ageing. Nature 464, 504–512 10.1038/nature08980 (doi:10.1038/nature08980) [DOI] [PubMed] [Google Scholar]
- 9.Parrella E., Longo V. D.Insulin/IGF-I and related signaling pathways regulate aging in nondividing cells: from yeast to the mammalian brain. Scient. World J. 10, 161–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giannakou M. E., Partridge L.2007Role of insulin-like signalling in Drosophila lifespan. Trends Biochem. Sci. 32, 180–188 10.1016/j.tibs.2007.02.007 (doi:10.1016/j.tibs.2007.02.007) [DOI] [PubMed] [Google Scholar]
- 11.Hardwick J. S., Kuruvilla F. G., Tong J. K., Shamji A. F., Schreiber S. L.1999Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl Acad. Sci. USA 96, 14 866–14 870 10.1073/pnas.96.26.14866 (doi:10.1073/pnas.96.26.14866) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nobukuni T., et al. 2005Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl Acad. Sci. USA 102, 14 238–14 243 10.1073/pnas.0506925102 (doi:10.1073/pnas.0506925102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polak P., Hall M. N.2009mTOR and the control of whole body metabolism. Curr. Opin. Cell Biol. 21, 209–218 10.1016/j.ceb.2009.01.024 (doi:10.1016/j.ceb.2009.01.024) [DOI] [PubMed] [Google Scholar]
- 14.Honjoh S., Yamamoto T., Uno M., Nishida E.2009Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature 457, 726–730 10.1038/nature07583 (doi:10.1038/nature07583) [DOI] [PubMed] [Google Scholar]
- 15.Wei M., Fabrizio P., Hu J., Ge H., Cheng C., Li L., Longo V. D.2008Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 4, e13. 10.1371/journal.pgen.0040013 (doi:10.1371/journal.pgen.0040013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ching T. T., Paal A. B., Mehta A., Zhong L., Hsu A. L.2010drr-2 encodes an eIF4H that acts downstream of TOR in diet-restriction-induced longevity of C. elegans. Aging Cell 9, 545–557 10.1111/j.1474-9726.2010.00580.x (doi:10.1111/j.1474-9726.2010.00580.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piper M. D., Bartke A.2008Diet and aging. Cell Metab. 8, 99–104 10.1016/j.cmet.2008.06.012 (doi:10.1016/j.cmet.2008.06.012) [DOI] [PubMed] [Google Scholar]
- 18.Murakami M., Ichisaka T., Maeda M., Oshiro N., Hara K., Edenhofer F., Kiyama H., Yonezawa K., Yamanaka S.2004mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol. Cell. Biol. 24, 6710–6718 10.1128/MCB.24.15.6710-6718.2004 (doi:10.1128/MCB.24.15.6710-6718.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gangloff Y. G., et al. 2004Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol. Cell. Biol. 24, 9508–9516 10.1128/MCB.24.21.9508-9516.2004 (doi:10.1128/MCB.24.21.9508-9516.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia K., Chen D., Riddle D. L.2004The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 131, 3897–3906 10.1242/dev.01255 (doi:10.1242/dev.01255) [DOI] [PubMed] [Google Scholar]
- 21.Bhaskar P. T., Hay N.2007The two TORCs and Akt. Dev. Cell. 12, 487–502 10.1016/j.devcel.2007.03.020 (doi:10.1016/j.devcel.2007.03.020) [DOI] [PubMed] [Google Scholar]
- 22.Soukas A. A., Kane E. A., Carr C. E., Melo J. A., Ruvkun G.2009Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 23, 496–511 10.1101/gad.1775409 (doi:10.1101/gad.1775409) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones K. T., Greer E. R., Pearce D., Ashrafi K.2009Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 7, e60. 10.1371/journal.pbio.1000060 (doi:10.1371/journal.pbio.1000060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin S. J., Defossez P. A., Guarente L.2000Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289, 2126–2128 10.1126/science.289.5487.2126 (doi:10.1126/science.289.5487.2126) [DOI] [PubMed] [Google Scholar]
- 25.Fabrizio P., Battistella L., Vardavas R., Gattazzo C., Liou L. L., Diaspro A., Dossen J. W., Graua E. B., Longo V. D.2004Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J. Cell Biol. 166, 1055–1067 10.1083/jcb.200404002 (doi:10.1083/jcb.200404002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang J. C., Jaruga E., Repnevskaya M. V., Jazwinski S. M.2000An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J. 14, 2135–2137 [DOI] [PubMed] [Google Scholar]
- 27.Grandison R. C., Piper M. D., Partridge L.2009Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 462, 1061–1064 10.1038/nature08619 (doi:10.1038/nature08619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chapman T., Partridge L.1996Female fitness in Drosophila melanogaster: an interaction between the effect of nutrition and of encounter rate with males. Proc. R. Soc. Lond. B 263, 755–759 10.1098/rspb.1996.0113 (doi:10.1098/rspb.1996.0113) [DOI] [PubMed] [Google Scholar]
- 29.Skorupa D. A., Dervisefendic A., Zwiener J., Pletcher S. D.2008Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell 7, 478–490 10.1111/j.1474-9726.2008.00400.x (doi:10.1111/j.1474-9726.2008.00400.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakowski B., Hekimi S.1998The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 95, 13 091–13 096 10.1073/pnas.95.22.13091 (doi:10.1073/pnas.95.22.13091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hosono R., Nishimoto S., Kuno S.1989Alterations of life span in the nematode Caenorhabditis elegans under monoxenic culture conditions. Exp. Gerontol. 24, 251–264 10.1016/0531-5565(89)90016-8 (doi:10.1016/0531-5565(89)90016-8) [DOI] [PubMed] [Google Scholar]
- 32.Greer E. L., Dowlatshahi D., Banko M. R., Villen J., Hoang K., Blanchard D., Gygi S. P., Brunet A.2007An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 17, 1646–1656 10.1016/j.cub.2007.08.047 (doi:10.1016/j.cub.2007.08.047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houthoofd K., Braeckman B. P., Lenaerts I., Brys K., De Vreese A., Van Eygen S., Vanfletern J. R.2002Axenic growth up-regulates mass-specific metabolic rate, stress resistance, and extends life span in Caenorhabditis elegans. Exp. Gerontol. 37, 1371–1378 10.1016/S0531-5565(02)00173-0 (doi:10.1016/S0531-5565(02)00173-0) [DOI] [PubMed] [Google Scholar]
- 34.Kaeberlein T. L., Smith E. D., Tsuchiya M., Welton K. L., Thomas J. H., Fields S., Kennedy B. K., Kaeberlein M.2006Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell 5, 487–494 10.1111/j.1474-9726.2006.00238.x (doi:10.1111/j.1474-9726.2006.00238.x) [DOI] [PubMed] [Google Scholar]
- 35.Lee G. D., Wilson M. A., Zhu M., Wolkow C. A., de Cabo R., Ingram D. K., Zou S.2006Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell 5, 515–524 10.1111/j.1474-9726.2006.00241.x (doi:10.1111/j.1474-9726.2006.00241.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim E., Guan K. L.2009RAG GTPases in nutrient-mediated TOR signaling pathway. Cell Cycle 8, 1014–1018 [DOI] [PubMed] [Google Scholar]
- 37.Wang X., Proud C. G.2009Nutrient control of TORC1, a cell-cycle regulator. Trends Cell Biol. 19, 260–267 10.1016/j.tcb.2009.03.005 (doi:10.1016/j.tcb.2009.03.005) [DOI] [PubMed] [Google Scholar]
- 38.Binda M., Peli-Gulli M. P., Bonfils G., Panchaud N., Urban J., Sturgill T. W., Loewith R., De Virgilo C.2009The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell. 35, 563–573 10.1016/j.molcel.2009.06.033 (doi:10.1016/j.molcel.2009.06.033) [DOI] [PubMed] [Google Scholar]
- 39.Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., Sabatini D. M.Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vellai T., Takacs-Vellai K., Zhang Y., Kovacs A. L., Orosz L., Muller F.2003Genetics: influence of TOR kinase on lifespan in C. elegans. Nature 426, 620. 10.1038/426620a (doi:10.1038/426620a) [DOI] [PubMed] [Google Scholar]
- 41.Hansen M., Taubert S., Crawford D., Libina N., Lee S. J., Kenyon C.2007Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6, 95–110 10.1111/j.1474-9726.2006.00267.x (doi:10.1111/j.1474-9726.2006.00267.x) [DOI] [PubMed] [Google Scholar]
- 42.Geyskens I., Kumara S. H. M. C., Donaton M. C. V., Bergsma J. C. T., Thevelein J. M., Wera S.2000Expression of mammalian PKB partially complements deletion of the yeast protein kinase Sch9. In Molecular mechanisms of signal transduction (ed. Bos J. L.), pp. 117–126 Amsterdam, The Netherlands: IOS Press [Google Scholar]
- 43.Sonenberg N., Hinnebusch A. G.2007New modes of translational control in development, behavior, and disease. Mol. Cell 28, 721–729 10.1016/j.molcel.2007.11.018 (doi:10.1016/j.molcel.2007.11.018) [DOI] [PubMed] [Google Scholar]
- 44.Ma X. M., Blenis J.2009Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. 10, 307–318 10.1038/nrm2672 (doi:10.1038/nrm2672) [DOI] [PubMed] [Google Scholar]
- 45.Hay N., Sonenberg N.2004Upstream and downstream of mTOR. Genes Dev. 18, 1926–1945 10.1101/gad.1212704 (doi:10.1101/gad.1212704) [DOI] [PubMed] [Google Scholar]
- 46.Noda T., Ohsumi Y.1998Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273, 3963–3966 10.1074/jbc.273.7.3963 (doi:10.1074/jbc.273.7.3963) [DOI] [PubMed] [Google Scholar]
- 47.Scott R. C., Schuldiner O., Neufeld T. P.2004Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell 7, 167–178 10.1016/j.devcel.2004.07.009 (doi:10.1016/j.devcel.2004.07.009) [DOI] [PubMed] [Google Scholar]
- 48.Rusten T. E., Lindmo K., Juhasz G., Sass M., Seglen P. O., Brech A., Stenmark H.2004Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev. Cell 7, 179–192 10.1016/j.devcel.2004.07.005 (doi:10.1016/j.devcel.2004.07.005) [DOI] [PubMed] [Google Scholar]
- 49.Blommaart E. F., Luiken J. J., Blommaart P. J., van Woerkom G. M., Meijer A. J.1995Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 270, 2320–2326 [DOI] [PubMed] [Google Scholar]
- 50.Bonawitz N. D., Chatenay-Lapointe M., Pan Y., Shadel G. S.2007Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 5, 265–277 10.1016/j.cmet.2007.02.009 (doi:10.1016/j.cmet.2007.02.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schieke S. M., Phillips D., McCoy J. P., Jr, Aponte A. M., Shen R. F., Balaban R. S., Finkel T.2006The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem. 281, 27 643–27 652 10.1074/jbc.M603536200 (doi:10.1074/jbc.M603536200) [DOI] [PubMed] [Google Scholar]
- 52.Cunningham J. T., Rodgers J. T., Arlow D. H., Vazquez F., Mootha V. K., Puigserver P.2007mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 450, 736–740 10.1038/nature06322 (doi:10.1038/nature06322) [DOI] [PubMed] [Google Scholar]
- 53.Beck T., Hall M. N.1999The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402, 689–692 10.1038/45287 (doi:10.1038/45287) [DOI] [PubMed] [Google Scholar]
- 54.Brugarolas J., Lei K., Hurley R. L., Manning B. D., Reiling J. H., Hafen E., Witters L. A., Ellisen L. W., Kaelin W. G., Jr2004Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 18, 2893–2904 10.1101/gad.1256804 (doi:10.1101/gad.1256804) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reiling J. H., Hafen E.2004The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 18, 2879–2892 10.1101/gad.322704 (doi:10.1101/gad.322704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laplante M., Sabatini D. M.2009mTOR signaling at a glance. J. Cell Sci. 122, 3589–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin D. E., Powers T., Hall M. N.2006Regulation of ribosome biogenesis: where is TOR? Cell Metab. 4, 259–260 10.1016/j.cmet.2006.09.002 (doi:10.1016/j.cmet.2006.09.002) [DOI] [PubMed] [Google Scholar]
- 58.Choo A. Y., Blenis J.2009Not all substrates are treated equally: implications for mTOR, rapamycin-resistance and cancer therapy. Cell Cycle 8, 567–572 [DOI] [PubMed] [Google Scholar]
- 59.Stanfel M. N., Shamieh L. S., Kaeberlein M., Kennedy B. K.2009The TOR pathway comes of age. Biochim. Biophys. Acta 1790, 1067–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Selman C., et al. 2009Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326, 140–144 10.1126/science.1177221 (doi:10.1126/science.1177221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anderson R. M., Shanmuganayagam D., Weindruch R.2009Caloric restriction and aging: studies in mice and monkeys. Toxicol. Pathol. 37, 47–51 10.1177/0192623308329476 (doi:10.1177/0192623308329476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harrison D. E., et al. 2009Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460, 392–395 10.1038/nature.08221 (doi:10.1038/nature.08221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Um S. H., et al. 2004Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431, 200–205 10.1038/nature02866 (doi:10.1038/nature02866) [DOI] [PubMed] [Google Scholar]
- 64.Mieulet V., Roceri M., Espeillac C., Sotiropoulos A., Ohanna M., Oorschot V., Klumperman J., Sandri M., Pende M.2007S6 kinase inactivation impairs growth and translational target phosphorylation in muscle cells maintaining proper regulation of protein turnover. Am. J. Physiol. 293, C712–C722 10.1152/ajpcell.00499.2006 (doi:10.1152/ajpcell.00499.2006) [DOI] [PubMed] [Google Scholar]
- 65.Zid B. M., Rogers A. N., Katewa S. D., Vargas M. A., Kolipinski M. C., Lu T. A., Benzer S., Kapatri P.20094E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 139, 149–160 10.1016/j.cell.2009.07.034 (doi:10.1016/j.cell.2009.07.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tain L. S., Mortiboys H., Tao R. N., Ziviani E., Bandmann O., Whitworth A. J.2009Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat. Neurosci. 12, 1129–1135 10.1038/nn.2372 (doi:10.1038/nn.2372) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wessells R., et al. 2009d4eBP acts downstream of both dTOR and dFoxo to modulate cardiac functional aging in Drosophila. Aging Cell 8, 542–552 10.1111/j.1474-9726.2009.00504.x (doi:10.1111/j.1474-9726.2009.00504.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steffen K. K., et al. 2008Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133, 292–302 10.1016/j.cell.2008.02.037 (doi:10.1016/j.cell.2008.02.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chiocchetti A., et al. 2007Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp. Gerontol. 42, 275–286 10.1016/j.exger.2006.11.002 (doi:10.1016/j.exger.2006.11.002) [DOI] [PubMed] [Google Scholar]
- 70.Yang R., Wek S. A., Wek R. C.2000Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol. Cell. Biol. 20, 2706–2717 10.1128/MCB.20.8.2706-2717.2000 (doi:10.1128/MCB.20.8.2706-2717.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang R. C., Levine B.Autophagy in cellular growth control. FEBS Lett. 584, 1417–1426 10.1016/j.febslet.2010.01.009 (doi:10.1016/j.febslet.2010.01.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kamada Y., Yoshino K., Kondo C., Kawamata T., Oshiro N., Yonezawa K., Ohsumi Y.2010Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell. Biol. 30, 1049–1058 10.1128/MCB.01344-09 (doi:10.1128/MCB.01344-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kamada Y.Prime-numbered Atg proteins act at the primary step in autophagy: unphosphorylatable Atg13 can induce autophagy without TOR inactivation. Autophagy 6, 415–416 10.4161/auto.6.3.11390 (doi:10.4161/auto.6.3.11390) [DOI] [PubMed] [Google Scholar]
- 74.Cuervo A. M.2008Autophagy and aging: keeping that old broom working. Trends Genet. 24, 604–612 10.1016/j.tig.2008.10.002 (doi:10.1016/j.tig.2008.10.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Melendez A., Talloczy Z., Seaman M., Eskelinen E. L., Hall D. H., Levine B.2003Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387–1391 10.1126/science.1087782 (doi:10.1126/science.1087782) [DOI] [PubMed] [Google Scholar]
- 76.Hars E. S., Qi H., Ryazanov A. G., Jin S., Cai L., Hu C., Liu L.2007Autophagy regulates ageing in C. elegans. Autophagy 3, 93–95 [DOI] [PubMed] [Google Scholar]
- 77.Hansen M., Chandra A., Mitic L. L., Onken B., Driscoll M., Kenyon C.2008A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 4, e24. 10.1371/journal.pgen.0040024 (doi:10.1371/journal.pgen.0040024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alvers A. L., Wood M. S., Hu D., Kaywell A. C., Dunn W. A., Jr, Aris J. P.2009Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy 5, 847–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alvers A. L., Fishwick L. K., Wood M. S., Hu D., Chung H. S., Dunn W. A., Jr, Aris J. P.2009Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell 8, 353–369 10.1111/j.1474-9726.2009.00469.x (doi:10.1111/j.1474-9726.2009.00469.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bjedov I., Toivonen J. M., Kerr F., Slack C., Jacobson J., Foley A., Partridge L.2010Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 11, 35–46 10.1016/j.cmet.2009.11.010 (doi:10.1016/j.cmet.2009.11.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scott R. C., Juhasz G., Neufeld T. P.2007Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 17, 1–11 10.1016/j.cub.2006.10.053 (doi:10.1016/j.cub.2006.10.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kockel L., Kerr K. S., Melnick M., Bruckner K., Hebrok M., Perrimon N.2010Dynamic switch of negative feedback regulation in Drosophila Akt-TOR signaling. PLoS Genet. 6, e1000990. 10.1371/journal.pgen.1000990 (doi:10.1371/journal.pgen.1000990) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee J. H., et al. 2010Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science 327, 1223–1228 10.1126/science.1182228 (doi:10.1126/science.1182228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lempiainen H., Uotila A., Urban J., Dohnal I., Ammerer G., Loewith R., Shore D.2009Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol. Cell 33, 704–716 10.1016/j.molcel.2009.01.034 (doi:10.1016/j.molcel.2009.01.034) [DOI] [PubMed] [Google Scholar]
- 85.DeBerardinis R. J., Lum J. J., Hatzivassiliou G., Thompson C. B.2008The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7, 11–20 10.1016/j.cmet.2007.10.002 (doi:10.1016/j.cmet.2007.10.002) [DOI] [PubMed] [Google Scholar]
- 86.Vander Heiden M. G., Cantley L. C., Thompson C. B.2009Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 10.1126/science.1160809 (doi:10.1126/science.1160809) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hsu P. P., Sabatini D. M.2008Cancer cell metabolism: Warburg and beyond. Cell 134, 703–707 10.1016/j.cell.2008.08.021 (doi:10.1016/j.cell.2008.08.021) [DOI] [PubMed] [Google Scholar]
- 88.Thompson J. E., Thompson C. B.2004Putting the rap on Akt. J. Clin. Oncol. 22, 4217–4226 10.1200/JCO.2004.01.103 (doi:10.1200/JCO.2004.01.103) [DOI] [PubMed] [Google Scholar]
- 89.Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., Fleming M. D., Schreiber S. L., Cantley L. C.2008The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 10.1038/nature06734 (doi:10.1038/nature06734) [DOI] [PubMed] [Google Scholar]
- 90.Fantin V. R., St-Pierre J., Leder P.2006Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9, 425–434 10.1016/j.ccr.2006.04.023 (doi:10.1016/j.ccr.2006.04.023) [DOI] [PubMed] [Google Scholar]
- 91.Khatri S., Yepiskoposyan H., Gallo C. A., Tandon P., Plas D. R.2010FOXO3a regulates glycolysis via transcriptional control of tumor suppressor TSC1. J. Biol. Chem. 285, 15 960–15 965 10.1074/jbc.M110.121871 (doi:10.1074/jbc.M110.121871) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Finley L. W., Haigis M. C.2009The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Ageing Res. Rev. 8, 173–188 10.1016/j.arr.2009.03.003 (doi:10.1016/j.arr.2009.03.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hur J. H., Cho J., Walker D. W.2010Aging: Dial M for mitochondria. Aging 2, 69–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ramanathan A., Schreiber S. L.2009Direct control of mitochondrial function by mTOR. Proc. Natl Acad. Sci. USA 106, 22 229–22 232 10.1073/pnas.0912074106 (doi:10.1073/pnas.0912074106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bonawitz N. D., Shadel G. S.2007Rethinking the mitochondrial theory of aging: the role of mitochondrial gene expression in lifespan determination. Cell Cycle 6, 1574–1578 10.4161/cc.6.13.4457 (doi:10.4161/cc.6.13.4457) [DOI] [PubMed] [Google Scholar]
- 96.Pan Y., Shadel G. S.2009Extension of chronological life span by reduced TOR signaling requires down-regulation of Sch9p and involves increased mitochondrial OXPHOS complex density. Aging 1, 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fabrizio P., Pozza F., Pletcher S. D., Gendron C. M., Longo V. D.2001Regulation of longevity and stress resistance by Sch9 in yeast. Science 292, 288–290 10.1126/science.1059497 (doi:10.1126/science.1059497) [DOI] [PubMed] [Google Scholar]
- 98.Lavoie H., Whiteway M.2008Increased respiration in the sch9{Delta} mutant is required for increasing chronological life span but not replicative life span. Eukaryot. Cell 7, 1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wanke V., Pedruzzi I., Cameroni E., Dubouloz F., De Virgilio C.2005Regulation of G0 entry by the Pho80-Pho85 cyclin-CDK complex. EMBO J. 24, 4271–4278 10.1038/sj.emboj.7600889 (doi:10.1038/sj.emboj.7600889) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pedruzzi I., Dubouloz F., Cameroni E., Wanke V., Roosen J., Winderickx J., De Virgilio C.2003TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol. Cell 12, 1607–1613 10.1016/S1097-2765(03)00485-4 (doi:10.1016/S1097-2765(03)00485-4) [DOI] [PubMed] [Google Scholar]
- 101.Wang C., Skinner C., Easlon E., Lin S. J.2009Deleting the 14–3-3 protein Bmh1 extends life span in Saccharomyces cerevisiae by increasing stress response. Genetics 183, 1373–1384 10.1534/genetics.109.107797 (doi:10.1534/genetics.109.107797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Swinnen E., Wanke V., Roosen J., Smets B., Dubouloz F., Pedruzzi I., Cameroni E., De Virgilio C., Winderickx J.2006Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae. Cell Div. 1, 3. 10.1186/1747-1028-1-3 (doi:10.1186/1747-1028-1-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cameroni E., Hulo N., Roosen J., Winderickx J., De Virgilio C.2004The novel yeast PAS kinase Rim 15 orchestrates G0-associated antioxidant defense mechanisms. Cell Cycle 3, 462–468 [PubMed] [Google Scholar]
- 104.Powers R. W., III, Kaeberlein M., Caldwell S. D., Kennedy B. K., Fields S.2006Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 20, 174–184 10.1101/gad.1381406 (doi:10.1101/gad.1381406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fabrizio P., Gattazzo C., Battistella L., Wei M., Cheng C., McGrew K., Longo V. D.2005Sir2 blocks extreme life-span extension. Cell 123, 655–667 10.1016/j.cell.2005.08.042 (doi:10.1016/j.cell.2005.08.042) [DOI] [PubMed] [Google Scholar]
- 106.Medvedik O., Lamming D. W., Kim K. D., Sinclair D. A.2007MSN2 and MSN4 link calorie restriction and TOR to Sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 5, 230–241 10.1371/journal.pbio.0050261 (doi:10.1371/journal.pbio.0050261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Sinclair D. A.2003Nicotinamide and PNC1 govern lifespan extension by caloric restriction in Saccharomyces cerevisiae. Nature 423, 181–185 10.1038/nature01578 (doi:10.1038/nature01578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gallo C. M., Smith D. L. J., Smith J. S.2004Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol. Cell Biol. 24, 1301–1312 10.1128/MCB.24.3.1301-1312.2004 (doi:10.1128/MCB.24.3.1301-1312.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kaeberlein M., et al. 2005Regulation of yeast replicative life-span by TOR and Sch9 in response to nutrients. Science 310, 1193–1196 10.1126/science.115535 (doi:10.1126/science.115535) [DOI] [PubMed] [Google Scholar]
- 110.Burtner C. R., Murakami C. J., Kennedy B. K., Kaeberlein M.2009A molecular mechanism of chronological aging in yeast. Cell Cycle 8, 1256–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fabrizio P., Longo V. D.2003The chronological life span of Saccharomyces cerevisiae. Aging Cell 2, 73–81 10.1046/j.1474-9728.2003.00033.x (doi:10.1046/j.1474-9728.2003.00033.x) [DOI] [PubMed] [Google Scholar]
- 112.Murakami C. J., Burtner C. R., Kennedy B. K., Kaeberlein M.2008A method for high throughput quantitative analysis of yeast chronological life span. J. Gerontol. A Biol. Sci. 63, 113–121 [DOI] [PubMed] [Google Scholar]
- 113.Smith D. L., Jr, McClure J. M., Matecic M., Smith J. S.2007Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell 6, 649–662 [DOI] [PubMed] [Google Scholar]
- 114.Wang G. L., Semenza G. L.1993General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc. Natl Acad. Sci. USA 90, 4304–4308 10.1073/pnas.90.9.4304 (doi:10.1073/pnas.90.9.4304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Maxwell P. H., et al. 1999The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399, 271–275 [DOI] [PubMed] [Google Scholar]
- 116.Jaakkola P., et al. 2001Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 10.1126/science.1059796 (doi:10.1126/science.1059796) [DOI] [PubMed] [Google Scholar]
- 117.Kamura T., Sato S., Iwai K., Czyzyk-Krzeska M., Conaway R. C., Conaway J. W.2000Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc. Natl Acad. Sci. USA 97, 10 430–10 435 10.1073/pnas.190332597 (doi:10.1073/pnas.190332597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang G. L., Jiang B. H., Rue E. A., Semenza G. L.1995Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl Acad. Sci. USA 92, 5510–5514 10.1073/pnas.92.12.5510 (doi:10.1073/pnas.92.12.5510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen D., Thomas E. L., Kapahi P.2009HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet. 5, e1000486. 10.1371/journal.pgen.1000486 (doi:10.1371/journal.pgen.1000486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Y., Shao Z., Zhai Z., Shen C., Powell-Coffman J. A.2009The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans. PLoS One 4, e6348. 10.1371/journal.pone.0006348 (doi:10.1371/journal.pone.0006348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mehta R., Steinkraus K. A., Sutphin G. L., Ramos F. J., Shamieh L. S., Huh A., Davis C., Chandler-Brown D., Kaeberlein M.2009Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science 324, 1196–1198 10.1126/science.1173507 (doi:10.1126/science.1173507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kaeberlein M., Kapahi P.2009The hypoxic response and aging. Cell Cycle 8, 2324. [DOI] [PubMed] [Google Scholar]
- 123.Jin H. O., An S., Lee H. C., Woo S. H., Seo S. K., Choe T. B., et al. 2007Hypoxic condition- and high cell density-induced expression of Redd1 is regulated by activation of hypoxia-inducible factor-1alpha and Sp1 through the phosphatidylinositol 3-kinase/Akt signaling pathway. Cell Signal. 19, 1393–1403 10.1016/j.cellsig.2006.12.014 (doi:10.1016/j.cellsig.2006.12.014) [DOI] [PubMed] [Google Scholar]
- 124.DeYoung M. P., Horak P., Sofer A., Sgroi D., Ellisen L. W.2008Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14–3-3 shuttling. Genes Dev. 22, 239–251 10.1101/gad.1617608 (doi:10.1101/gad.1617608) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Thomas G. V., Tran C., Mellinghoff I. K., Welsbie D. S., Chan E., Fueger B., Czernin J., Sawyers C. L.2006Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nat. Med. 12, 122–127 10.1038/nm1337 (doi:10.1038/nm1337) [DOI] [PubMed] [Google Scholar]
- 126.Dekanty A., Lavista-Llanos S., Irisarri M., Oldham S., Wappner P.2005The insulin-PI3K/TOR pathway induces a HIF-dependent transcriptional response in Drosophila by promoting nuclear localization of HIF-alpha/Sima. J. Cell Sci. 118, 5431–5441 [DOI] [PubMed] [Google Scholar]
- 127.Lee S. J., Feldman R., O'Farrell P. H.2008An RNA interference screen identifies a novel regulator of target of rapamycin that mediates hypoxia suppression of translation in Drosophila S2 cells. Mol. Biol Cell 19, 4051–4061 10.1091/mbc.E08-03-0265 (doi:10.1091/mbc.E08-03-0265) [DOI] [PMC free article] [PubMed] [Google Scholar]