Abstract
Ageing in humans is typified by the decline of physiological functions in various organs and tissues leading to an increased probability of death. Some individuals delay, escape or survive much of this age-related decline and live past age 100. Studies comparing centenarians to average-aged individuals have found polymorphisms in genes that are associated with long life, including APOE and FOXOA3, which have been replicated many times. However, the associations found in humans account for small percentages of the variance in lifespan and many other gene associations have not been replicated in additional populations. Therefore, ageing is probably a highly polygenic trait. In humans, it is important to also consider differences in age-related decline that occur within and among tissues. Longitudinal data of age-related traits can be used in association studies to test for polymorphisms that predict how an individual will change over time. Transcriptional and genetic association studies of different tissues have revealed common and unique pathways involved in human ageing. Genomic convergence is a method that combines multiple types of functional genomic information such as transcriptional profiling, expression quantitative trait mapping and gene association. The genomic convergence approach has been used to implicate the gene MMP20 in human kidney ageing. New human genetics technologies are continually in development and may lead to additional breakthroughs in human ageing in the near future.
Keywords: human ageing, centenarians, tissue ageing, longitudinal studies, genomic convergence
1. Introduction
Ageing is characterized by a decline of multiple physiological functions leading to an increasing probability of death. Some ageing-related changes affect appearance, such as wrinkled skin and grey hair, whereas others affect organ function, such as decreased kidney filtration rate and decreased muscular strength. Ageing is a major risk factor in most human diseases, including heart disease and cancer. Average lifespan has increased at a steady pace of almost three months per year in both males and females since 1840 [1]. Japanese women have a life expectancy of 85 years, the highest in the developed world [1]. Initially, this increase in lifespan was probably attributed to decreases in juvenile mortality through treatment of infectious diseases [2]. In the latter half of the twentieth century, improvements in survival after age 65, due in part to improved treatments for ageing-related diseases, propelled the rise in lifespan [2]. For Japanese women, the chance of surviving from age 65 to 100 soared from less than 1 in 1000 in 1950 to 1 in 20 in 2002 [1]. Centenarians now constitute the fastest-growing segment of the US population, increasing in number from 3700 in 1940 to roughly 61 000 in 2006 [3]. Finding differences in genes between centenarians and average-aged individuals may point to molecular pathways important in the ageing process.
Ageing trajectories vary among individuals. Some individuals live to age 100, but most do not. Twin studies have shown that lifespan tends to be more correlated in monozygotic twin pairs than same-sex dizygotic twin pairs, with these differences yielding heritability estimates ranging from 0.23 to 0.33 [4,5]. Yet, little is known about specific genes that affect the rate of ageing or human lifespan.
2. Centenarian studies
One way to find potential ageing genes is to search for genetic differences between centenarians and average-aged individuals. The average lifespan in the US is 77 years and only one in 10 000 individuals survive to age 100 [6]. Siblings of centenarians have an eight to 17-fold greater relative risk of surviving to age 100 than those without centenarian siblings [7]. Alleles enriched in centenarians probably affect genes that are important for longevity. A summary of relevant gene association studies in long-lived individuals is shown in table 1. A genome-wide linkage scan of 400 short tandem repeats using 137 long-lived sibling groups of European descent from the Boston area identified a locus on chromosome 4 linked to exceptional longevity [8]. Genotyping 2000 single nucleotide polymorphisms (SNPs) within this 12 Mb locus revealed an association between microsomal transfer protein (MTP) and human lifespan [9]. The cases were 653 individuals aged 98 years and older from the same Boston population and the controls were Caucasians less than 50 years old from several sources in the US [9]. This finding could not be replicated in French or German populations [10].
Table 1.
Candidate gene association studies in long-lived individuals (LLI).
| gene | initial population | number LLIa | polymorphism | p-value | replication population? | references |
|---|---|---|---|---|---|---|
| APOE | French Caucasian | 325 | 2 SNP haplotype | 0.001 | many | [15,12,82] |
| MTP | US Caucasian | 653 | 2 SNP haplotype | 0.0005 | none | [9,10] |
| APOC3 | Ashkenazi | 213 | SNP rs2542052 | 0.0001 | none | [6] |
| IGFIR | Ashkenazi | 384 | 2 rare SNPs | 0.02 | none | [27] |
| FOXO3A | Japanese | 213 | SNP rs2802292 | 0.00009 | many | [26,28–31] |
| hTERT | Ashkenazi | 74 | 4 SNP haplotype | 0.007 | none | [19] |
aSome studies included nonagenarians and centenarians.
Most of the work comparing the genotypes of long-lived individuals to average-aged individuals has taken place in candidate genes rather than by genome-wide analysis. For example, the ɛ4 allele of apolipoprotein E (APOE) is well known to increase risk of Alzheimer's disease and cardiovascular disease [11,12]. The ɛ4 allele (112Arg) of APOE differs from the most common ɛ3 allele (112Cys) at a single amino-acid [13]. While the exact function of the ɛ4 allele remains to be fully determined, evidence has been presented suggesting an interaction with the amyloid β protein. One characteristic of Alzheimer's disease is the formation of amyloid beta plaques. In mice, the ɛ4 form of APOE exhibits an impaired ability to promote amyloid β proteolysis compared to the ɛ2 and ɛ3 isoforms [14]. The ɛ4 allele is found in significantly lower proportions of nonagenarians and centenarians, which suggests that individuals with this allele do not live as long as those without it [11,12,15]. By contrast, the ɛ2 allele is enriched in long-lived individuals and may offer a protective effect for Alzheimer's disease and cardiovascular disease [12,15].
A study of 35 additional genes related to cardiovascular disease found that an allele in apolipoprotein C3 (APOC3) is enriched in centenarians and their offspring compared to average-aged controls in an Ashkenazi Jewish population [6]. APOC3 is a very low density lipoprotein (VLDL) involved in fat metabolism and may delay the catabolism of triglyceride-rich particles. The enriched allele in centenarians was associated with low levels of APOC3 and a favourable lipid profile [6].
Shortening of the telomeres at the ends of chromosomes has been associated with age-related disease and mortality [16–18]. A recent study identified a common haplotype of four SNPs in the human telomerase reverse transcriptase gene (hTERT) that is enriched in centenarians and associated with longer telomere length [19]. It was also shown that centenarians and their offspring maintain longer telomeres compared with controls and that longer telomeres are associated with protection from age-related diseases, better cognitive function and lipid profiles of healthy ageing [19].
In addition to testing genes known to be associated with age-related diseases and phenotypes for association with longevity, genes known to promote longevity in model organisms have been examined in human populations. Mutations in insulin or insulin-like signalling pathway genes have been shown to extend lifespan in Caenorhabditis elegans [20], Drosophila melanogaster [21,22] and mice [23,24]. The insulin-signalling pathway negatively regulates the forkhead (FOXO) transcription factor [25]. When insulin or insulin-like growth factor signalling is low, FOXO is activated and lifespan extension occurs [26]. An overrepresentation of rare insulin-like growth factor I receptor (IGFIR) mutations has been observed in centenarians [27]. These mutations are associated with reduced activity of IGFIR as measured in transformed lymphocytes [27].
The forkhead box O3A (FOXO3A) transcription factor contains alleles associated with longevity in multiple Asian and European populations [26,28–31]. In all cases, the alleles associated with longevity are intronic and unlinked to known coding SNPs, so the functional SNPs may affect gene expression rather than protein activity, but this has not yet been shown. In a male population of Japanese descent, the odds ratio for homozygous minor versus homozygous major alleles for SNP rs2802292 in FOXO3A between the long-lived individuals (greater than or equal to 95 years) and controls was 2.75 [26]. In two replication studies including both sexes, the odds ratios of the corresponding alleles were 1.26 for a German population and 1.36 for a Chinese population [29,30]. These enriched alleles in long-lived individuals may promote better health and contribute towards extended lifespan by increasing expression or activity of FOXO3A.
The only two genes associated with human longevity that have been replicated in multiple populations are FOXO3A and APOE [11,12,15,26,28–31]. The effect sizes of these two genes for longevity are small with odds ratios of 1.26 and 1.45 for survival to age 100 in replicate studies for FOXO3A and APOE, respectively [10,29]. These genes account for only a small portion of the genetic contribution to longevity measured through family heritability studies [4,5]. Therefore, much of the heritability of lifespan remains to be explained.
3. Longitudinal studies of human ageing
In addition to examining differences in lifespan in centenarian studies, important human ageing molecular pathways may be found by examining physiological ageing. Physiological age serves as an indicator of an individual's general health status and may also serve as an indicator of remaining healthy lifespan [32]. More important than simply reaching age 100 is the health of the individual. For example, some centenarians appear frail and look like they have lived 100 years whereas others appear more fit and have a physical appearance more like that of a 70 year old. Because the rate of ageing varies immensely among individuals, humans become increasingly different from each other with age. Thus, chronological age fails to provide an accurate indicator of the ageing process. Measuring the physiological age of the whole individual is difficult because tissues age at different rates. Therefore, it is easier to measure physiological age in individual organs or tissues in hopes of understanding the molecular basis of ageing.
Measuring how a tissue changes with chronological age in a population can identify biomarkers that can be used as an index of physiological age [33]. The biomarker can then be used to determine if an individual is physiologically younger or older than his or her chronological age. Determining genes and pathways that associate with the measures of physiological age can reveal molecular processes important for the ageing of particular tissues. Studies in different tissues can be compared to find specific and common regulators of ageing.
Several longitudinal studies have been developed to collect data on many diverse phenotypes. The rationale for these studies is that the ageing process might be better understood by studying the same individuals throughout their lifespan, collecting serial assessments rather than by comparing individuals of different ages from different environments [34]. Along with multiple phenotypic measurements per individual, the cohorts have been genotyped for hundreds of thousands of SNPs across the genome.
The Baltimore Longitudinal Study of Ageing (BLSA) is the longest running scientific study of human ageing in the US [35]. Healthy volunteers aged 18 and older were enrolled in the study starting in 1958. These subjects visit the National Institute on Aging at regular intervals for two days of medical, physiological and psychological testing. Examples of measurements taken include electrocardiograms, memory tests, glomerular filtration rates and various serum metabolite levels [36–39]. Currently, the study population has 1450 active participants, aged 18–97 years (http://www.grc.nia.nih.gov/branches/blsa/blsa.htm).
Another longitudinal study of ageing is the InCHIANTI study, which consists of residents of two small towns in Tuscany, Italy [40]. The study includes 1320 participants (age range 20–102 years), who were randomly selected from the population registry of Greve in Chianti (population 11 709) and Bagno a Ripoli (population 4704) starting in 1998 [40]. Over 90 per cent of the population that were over the age of 65 participated in this study, and thus the cohort is a good representation of normal ageing. Since the baseline measurements were taken in 1998, three sets of follow-up measurements have been collected. The focus of the InCHIANTI study is the decline in mobility that occurs with age. Many physiological measurements are available for researchers including clinical histories, serum metabolite levels, glomerular filtration rates and muscle strength levels (http://www.inchiantistudy.net).
Within the past two years, both the BLSA and InCHIANTI cohorts have been genotyped for approximately 550 000 SNPs, making genome-wide association studies for various age-related traits possible [41,42]. Association studies using longitudinal data to investigate age-related traits are listed in table 2. One study investigated genetic variants that may affect normal serum iron concentrations [43]. Iron homeostasis is crucial because too little iron causes anaemia and too much iron is toxic. Iron can react with oxygen species to form free radicals, leading to protein damage that accumulates with age [44]. One significant association between serum iron concentration and an exonic SNP in transmembrane protease serine 6 (TMPRSS6) was found [43]. TMPRSS6 encodes an enzyme that promotes iron absorption and recycling by inhibiting hepcidin antimicrobial peptide transcription [43]. The allele that is associated with lower serum iron concentrations may help prevent oxidative damage, but this has not yet been tested.
Table 2.
Gene associations with age-related traits found using longitudinal study data.
| age-related trait | longitudinal dataset | N | gene | SNP | p-value | references |
|---|---|---|---|---|---|---|
| serum iron | BLSA, InCHIANTI | 1919 | TMPRSS6 | rs855791 | 3.9 × 10−7 | [43] |
| serum B6 | InCHIANTI | 1178 | ALPL | rs4654748 | 1.2 × 10−8 | [42] |
| serum B12 | BLSA, SardiNIA, InCHIANTI | 2927 | FUT2 | rs602662 | 2.4 × 10−12 | [42] |
| plasma arachidonic acid | InCHIANTI | 1075 | FADS1 | rs174537 | 6.0 × 10−46 | [48] |
| plasma eicosapentanoic acid | InCHIANTI | 1075 | FADS1 | rs174537 | 1.1 × 10−14 | [48] |
| weight | Framingham heart study | 2073 | FTO | rs1121980 | 2.0 × 10−4 (SNP × age) | [52] |
| BMI | Framingham heart study | 2073 | FTO | rs1121980 | 1.0 × 10−4 (SNP × age) | [52] |
| glomerular filtration rate | BLSA, InCHIANTI | 2190 | MMP20 | rs1711437 | 3.6 × 10−5 | [73] |
Another genome-wide association study included data from a third longitudinal study of ageing, SardiNIA, along with BLSA and InCHIANTI [42]. The SardiNIA cohort includes more than 6000 individuals from the island of Sardinia, a founder population [45]. Because Sardinians are more closely related than people living in other societies, they share much of the same genetic information, which makes it easier to track genetic effects through generations. In the SardiNIA cohort, 98 quantitative traits have been measured, including blood chemistry components, cardiovascular traits and personality facets (http://sardinia.nia.nih.gov). This particular genome-wide association study sought to find genetic variants that affect circulating levels of B vitamins. B vitamins have a protective effect against mortality and age-related diseases, like coronary artery disease and colon cancer [46,47]. In this study, serum vitamin B6 levels were significantly associated with SNPs in tissue-nonspecific alkaline phosphatase (ALPL) and serum vitamin B12 levels were significantly associated with SNPs in fucosyltransferase 2 (FUT2) [42]. ALPL probably influences the catabolism of vitamin B6 while FUT2 interferes with absorption of vitamin B12 [42]. The alleles associated with lower vitamin levels may indicate a risk for vitamin deficiencies and age-related diseases.
In a third genome-wide association study, plasma polyunsaturated fatty acid (PUFA) levels were found to associate with SNPs in the genes FADS1 in the InCHIANTI cohort and the GOLDN replication cohort [48]. GOLDN is the Genetics of Lipid Lowering Drugs and Diet Network study and this population consists of 1120 white men and women from the US [49]. GOLDN is not a longitudinal study, but has measurements of PUFA levels from one time point. FADS1 is a fatty acid desaturase involved in the metabolism of PUFAs. The alleles associated with higher PUFA levels may help protect against age-related diseases. High plasma PUFA concentrations have been shown to reduce the risk of mortality and cardiovascular disease [50].
Genome-wide association models that incorporate information about how an individual's phenotype changes over time have been developed [51]. Instead of simply finding genotype–phenotype associations, longitudinal methods have the potential to reveal alleles that are associated with the rate of ageing. Both the age at which physiological function begins to decline and the rate of such decline varies among individuals. In the BLSA, one third of individuals showed no decrease in glomerular filtration rate measured over a 20-year period, whereas the remainder of the population showed a distinct decline [37]. For those individuals who showed a significant decline in filtration rate, the slope of the decrease varied widely [37].
One association study examined changes in weight and body mass index (BMI), two traits that increase with age, in the Framingham Heart Study cohort [52]. The cohort included 7130 Caucasian individuals each with weight and BMI data at up to four time points. An allele of FTO (fat mass and obesity associated), a gene of unknown function, was previously found to associate with weight and BMI in two cross-sectional genome-wide association studies [53,54]. Luan et al. found that homozygotes for the minor allele of rs1121980 in FTO had the most pronounced increase in weight and BMI [52].
4. Transcriptional studies of human ageing
To elucidate molecular differences associated with ageing, an attractive approach is to use DNA microarrays to scan the entire genome for genes that change expression with age (age-regulated genes). Changes in gene expression levels may lead to important functional consequences in the process of ageing. Transcriptional profiles of many human tissues, including brain [55,56], blood [57], eye [58], kidney [59,60], muscle [61–63] and skin [64] have been generated. A major aim of these studies is the identification of novel biomarkers that can be used as indices of physiological age. Determining genes and pathways that associate with the measures of physiological age have revealed molecular processes important for the ageing of particular tissues. Studies in different tissues have been compared to find specific and common regulators of ageing. Ageing DNA microarray studies have been reviewed extensively [65,66], but two studies will be highlighted here because they have led to a new type of analysis known as genomic convergence.
In their analysis of kidney ageing, Rodwell et al. showed that age-regulated markers could predict physiological age [60]. They measured levels of gene expression in kidney samples from 72 patients between 27 and 92 years of age, and they identified a set of 447 age-regulated genes. Next, they tested whether the overall level of expression of the 447 genes was a signature of physiological age for different individuals. The relative amount of physiological ageing for each of the patients used in the study was measured by examining the morphological and histological appearance of the kidney. A remarkable similarity was found between the transcriptional ageing profiles determined using chronological age and the physiological states of the kidney in different individuals. Some elderly persons displayed an ageing signature similar to that of people who were 30–40 years younger, and these individuals also had relatively healthy kidneys. Others displayed an ageing signature similar to people who were 10–20 years older, and these patients showed poor kidney physiology for their chronological age.
Genes that are age-regulated in all tissues would reveal genes involved in core mechanisms that underlie cellular ageing. Zahn et al. [63] discovered genetic pathways that show common age regulation in human kidney, brain and muscle. They used microarrays to analyse expression in 81 skeletal muscle samples from patients aged 16–86 years and found 250 age-regulated muscle genes [63]. Similar to the ageing expression profile for the kidney, the overall expression behaviour of this set of age-regulated muscle genes correlated with the physiological as well as chronological age of the muscle sample. Next, they compared their muscle-ageing results to previously published data on kidney and brain ageing of similarly large sample size [56,60]. Although most of the age-related changes were tissue specific, they found evidence for common age regulation of six genetic pathways in all three tissues. Specifically, there is an overall increase in expression of the extracellular matrix genes, the ribosomal genes, the cell growth genes and the complement activation genes in all three tissues. Increased overall expression of the extracellular matrix and complement activation gene sets with advancing age may contribute to widespread fibrosis and inflammation in the elderly. There is an overall decrease in expression of the chloride transport genes and the electron transport genes in all three tissues. Decreased overall expression of electron transport chain genes with age might support the mitochondrial free-radical theory of ageing [67], as free-radical generation by mitochondria would preferentially damage the electron transport chain protein complexes. Decreased expression of the electron transport genes (encoded in the nucleus) might be caused by feedback regulation from damage to the electron transport chain protein complexes [63]. However, it is also possible that increased oxidative damage occurs as a consequence of the decreased expression of the electron transport chain genes. In addition, an increasing number of studies in model organisms have critically challenged the mitochondrial free-radical theory of ageing [68].
Importantly, these comparisons of transcriptional networks among different tissues were not done at the single gene level, but rather at the genetic pathway level [63]. With this approach, known as gene set enrichment analysis (GSEA), age regulation for every gene in a pathway is combined to determine whether there is an overall regulation of the entire pathway [69]. Screening for coordinated age regulation of genetic pathways increases the sensitivity of the analysis, as the combined effects of small regulation of many genes in a pathway can be significant. This systems biology method is especially powerful in studies of ageing because of the polygenic nature of the phenotype.
5. Genomic convergence identifies a gene involved in kidney ageing
In genome-wide association studies, hundreds of thousands of SNPs are tested. Thus, the penalty for multiple hypothesis testing is a large obstacle to overcome. A powerful alternative to genome-wide association studies is genomic convergence, which selects candidate genes for a specific phenotype based on genome-wide expression studies [70–72]. Gene expression microarrays can be used to identify genes whose expression levels increase or decrease in affected individuals compared to controls. Differential expression between cases and controls may indicate that the gene is functionally involved in disease pathogenesis. For example, a set of genes that is overexpressed in a type of neural disorder may include the genes that cause that disorder. Therefore, the genes that change expression between cases and controls can be used as candidates in genetic association studies. The power of the genetic association study is increased because a subset of genes rather than the whole genome is tested. Genomic convergence was first used to identify genes associated with Parkinson's disease, schizophrenia and Alzheimer's disease [70–72].
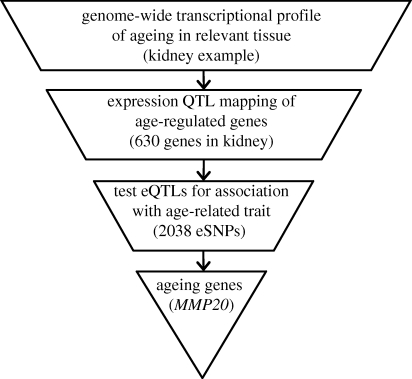
In a recent study, the genomic convergence approach was used to find genes associated with kidney ageing [73]. This genomic convergence approach consisted of three steps (figure 1). First, a genome-wide transcriptional profile of ageing in the human kidney gave an unbiased view of gene expression changes that occur with age [60]. These age-regulated genes also associated with kidney physiological age [60]. Additional genes from age-regulated pathways were included in the subsequent analysis [63]. The 630 age-regulated genes were probably enriched for genes that contribute to kidney ageing. The second step of the genomic convergence approach was mapping of expression quantitative trait loci (eQTLs). If a gene is functionally involved in kidney ageing and if DNA differences in the gene cause variation in expression among individuals, then there may be an association between the specific allele carried by an individual and that individual's physiological ageing trajectory. Wheeler et al. found 101 genes that have SNPs that associate with different expression levels [73]. In the third and final step, the 101 genes with eQTLs were tested for association with glomerular filtration rate (GFR), a phenotype of kidney ageing. Data from the BLSA and InCHIANTI cohorts were used to test SNPs in the 101 eQTLs for association with GFR. SNPs in one of these genes, matrix metalloproteinase 20 (MMP20), showed a statistically significant association with normal kidney ageing. Matrix metalloproteinases degrade extracellular matrix proteins and changes in the extracellular matrix play a key role in ageing of the kidney [74]. Interstitial fibrosis occurs during ageing in the kidney because of an increase in matrix [75] and different activities of MMP20 may play a role in this process. However, this genetic association has not yet been replicated in additional populations. Similar genomic convergence approaches that combine multiple types of genomic information (i.e. gene expression, eQTLs, protein levels) with genetic association studies can be used for all complex phenotypes, including other ageing-related phenotypes to find genes and pathways that affect ageing.
Figure 1.
The genomic convergence approach. Genes are filtered in each step for those the most likely to be functional. In the first step, a genome-wide transcriptional profile defines genes that show age-related changes in expression. In the second step, SNPs are tested for association with the expression of age-regulated genes. In the final step, the expression quantitative trait loci (eQTLs) are tested for association with a phenotype of ageing (e.g. age-dependent decline in kidney glomerular filtration rate in Wheeler et al. [73]).
6. Future directions in the genetics of human ageing
The application of powerful new human genetics technologies to the study of ageing has just begun. Unlike most other biological processes, the genetic factors that influence ageing may not be evolutionarily conserved because wild animals usually die from predation and infection, not ageing [76]. Thus, animal models for ageing may have limited relevance to human ageing. Therefore, genetic studies of ageing in humans are necessary. Luckily, the field of human genetics has experienced rapid advancement. It is now possible to interrogate thousands of samples across the genome in order to find genes associated with a particular phenotype.
Finding new human ageing genes contributes to our understanding of molecular mechanisms underlying the human ageing process. Like other complex human traits, finding common variants that account for the entire genetic component of human lifespan variability has proved difficult. If rare variants rather than common variants explain most of the genetic variation in ageing among humans, new genotyping techniques and new analysis methods must be developed to find genes and pathways involved in ageing. Next-generation sequencing technologies are faster and cheaper than traditional capillary sequencers. Several complete genomes have been sequenced using these next-generation technologies and many rare variants have been identified [77,78].
Methods are needed that can combine rare variants from multiple individuals that are in the same gene into a score that can be tested for association with different phenotypes of ageing and longevity. Information about how that rare variant might affect function could be included in the score. This information could include whether the rare variant causes a stop codon, a nonconservative amino-acid change or affects expression. Several association test methods for rare variants using this logic have been proposed that could be used for any complex trait including ageing [79–81]. In additional to scoring individual genes, combining information from molecular pathways may elucidate additional associations. If differences in a particular gene affect a phenotype, it is probable that other genes in the same pathway also affect the phenotype. Therefore, rare variants in different genes of the same pathway could be combined to test for association of the pathway with an ageing trait of interest. New human genetics technologies may lead to breakthroughs in human ageing in the near future.
Once key ageing genes and pathways are found, an individual's genotype could predict the rate of overall health decline. Genotypes of tissue-specific ageing genes could predict which tissues are likely to decline rapidly and which are likely to decline more gradually. Among young individuals, an unfavourable SNP genotype may indicate risk for rapid decline in a particular organ or tissue and this information could be extremely useful to identify patients who may require early intervention. Among older individuals, a favourable SNP genotype may indicate that they may still be eligible as organ donors even though they are currently over the upper age limit. For some ageing genes, one allele may adversely affect tissue function in old age because it increases the activity of the gene above a healthy level. In these cases, one could develop a drug to target the gene or pathway in individuals carrying the overactive allele, and thereby preserve function in old age. The goal of human ageing research is not necessarily to extend lifespan, but instead to extend the healthy years of life.
Acknowledgements
This work was supported by the National Institutes of Health (R01 AG025 941). H.E.W. was funded by a NIH grant to the Stanford Genetics and Developmental Biology Training Programme.
Footnotes
One contribution of 15 to a Discussion Meeting Issue ‘The new science of ageing’.
References
- 1.Oeppen J., Vaupel J. W.2002Demography. Broken limits to life expectancy. Science 296, 1029–1031 10.1126/science.1069675 (doi:10.1126/science.1069675) [DOI] [PubMed] [Google Scholar]
- 2.Wilmoth J. R.1998The future of human longevity: a demographer's perspective. Science 280, 395–397 10.1126/science.280.5362.395 (doi:10.1126/science.280.5362.395) [DOI] [PubMed] [Google Scholar]
- 3.Sonnega A.2006The future of human life expectancy: have we reached the ceiling or is the sky the limit? Research highlights in the demography and economics of aging 8, 1–4 [Google Scholar]
- 4.Herskind A. M., McGue M., Holm N. V., Sorensen T. I., Harvald B., Vaupel J. W.1996The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum. Genet. 97, 319–323 10.1007/BF02185763 (doi:10.1007/BF02185763) [DOI] [PubMed] [Google Scholar]
- 5.McGue M., Vaupel J. W., Holm N., Harvald B.1993Longevity is moderately heritable in a sample of Danish twins born 1870–1880. J. Gerontol. 48, B237–B244 [DOI] [PubMed] [Google Scholar]
- 6.Atzmon G., Rincon M., Schechter C. B., Shuldiner A. R., Lipton R. B., Bergman A., Barzilai N.2006Lipoprotein genotype and conserved pathway for exceptional longevity in humans. PLoS Biol. 4, e113. 10.1371/journal.pbio.0040113 (doi:10.1371/journal.pbio.0040113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perls T. T., et al. 2002Life-long sustained mortality advantage of siblings of centenarians. Proc. Natl Acad. Sci. USA 99, 8442–8447 10.1073/pnas.122587599 (doi:10.1073/pnas.122587599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puca A. A., Daly M. J., Brewster S. J., Matise T. C., Barrett J., Shea-Drinkwater M.2001A genome-wide scan for linkage to human exceptional longevity identifies a locus on chromosome 4. Proc. Natl Acad. Sci. USA 98, 10 505–10 508 10.1073/pnas.181337598 (doi:10.1073/pnas.181337598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geesaman B. J., Benson E., Brewster S. J., Kunkel L. M., Blanche H., Thomas G., Perls T. T., Daly M. J., Puca A. A.2003Haplotype-based identification of a microsomal transfer protein marker associated with the human lifespan. Proc. Natl Acad. Sci. USA 100, 14 115–14 120 10.1073/pnas.1936249100 (doi:10.1073/pnas.1936249100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nebel A., Croucher P. J., Stiegeler R., Nikolaus S., Krawczak M., Schreiber S.2005No association between microsomal triglyceride transfer protein (MTP) haplotype and longevity in humans. Proc. Natl Acad. Sci. USA 102, 7906–7909 10.1073/pnas.0408670102 (doi:10.1073/pnas.0408670102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corder E. H., Saunders A. M., Strittmatter W. J., Schmechel D. E., Gaskell P. C., Small G. W., Roses A. D., Haines J. L., Pericak-Vance M. A.1993Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261, 921–923 10.1126/science.8346443 (doi:10.1126/science.8346443) [DOI] [PubMed] [Google Scholar]
- 12.Kervinen K., Savolainen M. J., Salokannel J., Hynninen A., Heikkinen J., Ehnholm C., Koistinen M. J., Kesäniemi Y. A.1994Apolipoprotein E and B polymorphisms–longevity factors assessed in nonagenarians. Atherosclerosis 105, 89–95 10.1016/0021-9150(94)90011-6 (doi:10.1016/0021-9150(94)90011-6) [DOI] [PubMed] [Google Scholar]
- 13.Zuo L., van Dyck C. H., Luo X., Kranzler H. R., Yang B. Z., Gelernter J.2006Variation at APOE and STH loci and Alzheimer's disease. Behav. Brain Funct. 2, 13. 10.1186/1744-9081-2-13 (doi:10.1186/1744-9081-2-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang Q., et al. 2008ApoE promotes the proteolytic degradation of Aβ. Neuron 58, 681–693 10.1016/j.neuron.2008.04.010 (doi:10.1016/j.neuron.2008.04.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schachter F., Faure-Delanef L., Guenot F., Rouger H., Froguel P., Lesueur-Ginot L., Daniel C.1994Genetic associations with human longevity at the APOE and ACE loci. Nat. Genet. 6, 29–32 10.1038/ng0194-29 (doi:10.1038/ng0194-29) [DOI] [PubMed] [Google Scholar]
- 16.Aviv A.2006Telomeres and human somatic fitness. J. Gerontol. A Biol. Sci. Med. Sci. 61, 871–873 [DOI] [PubMed] [Google Scholar]
- 17.Njajou O. T., et al. 2007Telomere length is paternally inherited and is associated with parental lifespan. Proc. Natl Acad. Sci. USA 104, 12 135–12 139 10.1073/pnas.0702703104 (doi:10.1073/pnas.0702703104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong J. M., Collins K.2003Telomere maintenance and disease. Lancet 362, 983–988 10.1016/S0140-6736(03)14369-3 (doi:10.1016/S0140-6736(03)14369-3) [DOI] [PubMed] [Google Scholar]
- 19.Atzmon G., et al. 2010Evolution in health and medicine Sackler colloquium: genetic variation in human telomerase is associated with telomere length in Ashkenazi centenarians. Proc. Natl Acad. Sci. USA 107(Suppl. 1), 1710–1717 10.1073/pnas.0906191106 (doi:10.1073/pnas.0906191106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R.1993A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464 10.1038/366461a0 (doi:10.1038/366461a0) [DOI] [PubMed] [Google Scholar]
- 21.Clancy D. J., Gems D., Harshman L. G., Oldham S., Stocker H., Hafen E., Leevers S. J., Partridge L.2001Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292, 104–106 10.1126/science.1057991 (doi:10.1126/science.1057991) [DOI] [PubMed] [Google Scholar]
- 22.Tatar M., Kopelman A., Epstein D., Tu M. P., Yin C. M., Garofalo R. S.2001A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292, 107–110 10.1126/science.1057987 (doi:10.1126/science.1057987) [DOI] [PubMed] [Google Scholar]
- 23.Bluher M., Kahn B. B., Kahn C. R.2003Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299, 572–574 10.1126/science.1078223 (doi:10.1126/science.1078223) [DOI] [PubMed] [Google Scholar]
- 24.Holzenberger M., Dupont J., Ducos B., Leneuve P., Geloen A., Even P. C., Cervera P., Le Bouc Y.2003IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421, 182–187 10.1038/nature01298 (doi:10.1038/nature01298) [DOI] [PubMed] [Google Scholar]
- 25.Guarente L., Kenyon C.2000Genetic pathways that regulate ageing in model organisms. Nature 408, 255–262 10.1038/35041700 (doi:10.1038/35041700) [DOI] [PubMed] [Google Scholar]
- 26.Willcox B. J., et al. 2008FOXO3A genotype is strongly associated with human longevity. Proc. Natl Acad. Sci. USA 105, 13 987–13 992 10.1073/pnas.0801030105 (doi:10.1073/pnas.0801030105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suh Y., Atzmon G., Cho M. O., Hwang D., Liu B., Leahy D. J., Barzilai N., Cohen P.2008Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc. Natl Acad. Sci. USA 105, 3438–3442 10.1073/pnas.0705467105 (doi:10.1073/pnas.0705467105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anselmi C. V., Malovini A., Roncarati R., Novelli V., Villa F., Condorelli G., Bellazzi R., Puca A. A.2009Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenat. Res. 12, 95–104 10.1089/rej.2008.0827 (doi:10.1089/rej.2008.0827) [DOI] [PubMed] [Google Scholar]
- 29.Flachsbart F., Caliebe A., Kleindorp R., Blanche H., von Eller-Eberstein H., Nikolaus S., Schreiber S., Nebel A.2009Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc. Natl Acad. Sci. USA 106, 2700–2705 10.1073/pnas.0809594106 (doi:10.1073/pnas.0809594106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., et al. 2009Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum. Mol. Genet. 18, 4897–4904 10.1093/hmg/ddp459 (doi:10.1093/hmg/ddp459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pawlikowska L., et al. 2009Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell 8, 460–472 10.1111/j.1474-9726.2009.00493.x (doi:10.1111/j.1474-9726.2009.00493.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borkan G. A., Norris A. H.1980Assessment of biological age using a profile of physical parameters. J. Gerontol. 35, 177–184 [DOI] [PubMed] [Google Scholar]
- 33.Karasik D., Demissie S., Cupples L. A., Kiel D. P.2005Disentangling the genetic determinants of human aging: biological age as an alternative to the use of survival measures. J. Gerontol. A Biol. Sci. Med. Sci. 60, 574–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrucci L.2008The Baltimore Longitudinal Study of Aging (BLSA): a 50-year-long journey and plans for the future. J. Gerontol. A Biol. Sci. Med. Sci. 63, 1416–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindeman R. D., Tobin J. D., Shock N. W.1984Association between blood pressure and the rate of decline in renal function with age. Kidney Int. 26, 861–868 10.1038/ki.1984.229 (doi:10.1038/ki.1984.229) [DOI] [PubMed] [Google Scholar]
- 36.Gerstenblith G., Frederiksen J., Yin F. C., Fortuin N. J., Lakatta E. G., Weisfeldt M. L.1977Echocardiographic assessment of a normal adult aging population. Circulation 56, 273–278 [DOI] [PubMed] [Google Scholar]
- 37.Lindeman R. D., Tobin J., Shock N. W.1985Longitudinal studies on the rate of decline in renal function with age. J. Am. Geriatr. Soc. 33, 278–285 [DOI] [PubMed] [Google Scholar]
- 38.Sorkin J. D., Andres R., Muller D. C., Baldwin H. L., Fleg J. L.1992Cholesterol as a risk factor for coronary heart disease in elderly men. The Baltimore longitudinal study of aging. Ann. Epidemiol. 2, 59–67 10.1016/1047-2797(92)90038-R (doi:10.1016/1047-2797(92)90038-R) [DOI] [PubMed] [Google Scholar]
- 39.Zonderman A. B., Giambra L. M., Arenberg D., Resnick S. M., Costa P. T., Jr, Kawas C. H.1995Changes in immediate visual memory predict cognitive impairment. Arch. Clin. Neuropsychol. 10, 111–123 [PubMed] [Google Scholar]
- 40.Ferrucci L., Bandinelli S., Benvenuti E., Di Iorio A., Macchi C., Harris T. B., Guralnik J. M.2000Subsystems contributing to the decline in ability to walk: bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J. Am. Geriatr. Soc. 48, 1618–1625 [DOI] [PubMed] [Google Scholar]
- 41.Melzer D., et al. 2008A genome-wide association study identifies protein quantitative trait loci (pQTLs). PLoS Genet. 4, e1000072. 10.1371/journal.pgen.1000072 (doi:10.1371/journal.pgen.1000072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka T., et al. 2009Genome-wide association study of vitamin B6, vitamin B12, folate, and homocysteine blood concentrations. Am. J. Hum. Genet. 84, 477–482 10.1016/j.ajhg.2009.02.011 (doi:10.1016/j.ajhg.2009.02.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka T., et al. 2010A genome-wide association analysis of serum iron concentrations. Blood 115, 94–96 10.1182/blood-2009-07-232496 (doi:10.1182/blood-2009-07-232496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin I., Grotewiel M. S.2006Oxidative damage and age-related functional declines. Mech. Ageing Dev. 127, 411–423 [DOI] [PubMed] [Google Scholar]
- 45.Pilia G., et al. 2006Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2, e132. 10.1371/journal.pgen.0020132 (doi:10.1371/journal.pgen.0020132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Bree A., Verschuren W. M., Blom H. J., Nadeau M., Trijbels F. J., Kromhout D.2003Coronary heart disease mortality, plasma homocysteine, and B-vitamins: a prospective study. Atherosclerosis 166, 369–377 [DOI] [PubMed] [Google Scholar]
- 47.Giovannucci E., Stampfer M. J., Colditz G. A., Hunter D. J., Fuchs C., Rosner B. A., Speizer F. E., Willett W. C.1998Multivitamin use, folate, and colon cancer in women in the Nurses' Health Study. Ann. Intern. Med. 129, 517–524 [DOI] [PubMed] [Google Scholar]
- 48.Tanaka T., et al. 2009Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 5, e1000338. 10.1371/journal.pgen.1000338 (doi:10.1371/journal.pgen.1000338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corella D., et al. 2007The -256T > C polymorphism in the apolipoprotein A-II gene promoter is associated with body mass index and food intake in the genetics of lipid lowering drugs and diet network study. Clin. Chem. 53, 1144–1152 10.1373/clinchem.2006.084863 (doi:10.1373/clinchem.2006.084863) [DOI] [PubMed] [Google Scholar]
- 50.Sun Q., et al. 2007A prospective study of trans fatty acids in erythrocytes and risk of coronary heart disease. Circulation 115, 1858–1865 10.1161/CIRCULATIONAHA.106.679985 (doi:10.1161/CIRCULATIONAHA.106.679985) [DOI] [PubMed] [Google Scholar]
- 51.Kerner B., North K. E., Fallin M. D.2009Use of longitudinal data in genetic studies in the genome-wide association studies era: summary of Group 14. Genet. Epidemiol. 33(Suppl. 1), S93–S98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luan J., Kerner B., Zhao J. H., Loos R. J., Sharp S. J., Muthen B. O., Wareham N. J.2009A multilevel linear mixed model of the association between candidate genes and weight and body mass index using the Framingham longitudinal family data. BMC Proc. 3(Suppl. 7), S115. 10.1186/1753-6561-3-s7-s115 (doi:10.1186/1753-6561-3-s7-s115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frayling T. M., et al. 2007A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894 10.1126/science.1141634 (doi:10.1126/science.1141634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scuteri A., et al. 2007Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 3, e115. 10.1371/journal.pgen.0030115 (doi:10.1371/journal.pgen.0030115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fraser H. B., Khaitovich P., Plotkin J. B., Paabo S., Eisen M. B.2005Aging and gene expression in the primate brain. PLoS Biol. 3, e274. 10.1371/journal.pbio.0030274 (doi:10.1371/journal.pbio.0030274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu T., Pan Y., Kao S. Y., Li C., Kohane I., Chan J., Yanker B. A.2004Gene regulation and DNA damage in the ageing human brain. Nature 429, 883–891 10.1038/nature02661 (doi:10.1038/nature02661) [DOI] [PubMed] [Google Scholar]
- 57.Tan Q., Christensen K., Christiansen L., Frederiksen H., Bathum L., Dahlgaard J., Kruse T. A.2005Genetic dissection of gene expression observed in whole blood samples of elderly Danish twins. Hum. Genet. 117, 267–274 10.1007/s00439-005-1308-x (doi:10.1007/s00439-005-1308-x) [DOI] [PubMed] [Google Scholar]
- 58.Segev F., Mor O., Segev A., Belkin M., Assia E. I.2005Downregulation of gene expression in the ageing lens: a possible contributory factor in senile cataract. Eye (Lond) 19, 80–85 [DOI] [PubMed] [Google Scholar]
- 59.Melk A., Mansfield E. S., Hsieh S. C., Hernandez-Boussard T., Grimm P., Rayner D. C., Halloran P. F., Sarwal M. M.2005Transcriptional analysis of the molecular basis of human kidney aging using cDNA microarray profiling. Kidney Int. 68, 2667–2679 10.1111/j.1523-1755.2005.00738.x (doi:10.1111/j.1523-1755.2005.00738.x) [DOI] [PubMed] [Google Scholar]
- 60.Rodwell G. E., et al. 2004A transcriptional profile of aging in the human kidney. PLoS Biol. 2, e427. 10.1371/journal.pbio.0020427 (doi:10.1371/journal.pbio.0020427) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giresi P. G., Stevenson E. J., Theilhaber J., Koncarevic A., Parkington J., Fielding R. A., Kandarian S. C.2005Identification of a molecular signature of sarcopenia. Physiol. Genom. 21, 253–263 [DOI] [PubMed] [Google Scholar]
- 62.Welle S., Brooks A. I., Delehanty J. M., Needler N., Bhatt K., Shah B., Thornton C. A.2004Skeletal muscle gene expression profiles in 20–29 year old and 65–71 year old women. Exp. Gerontol. 39, 369–377 10.1016/j.exger.2003.11.011 (doi:10.1016/j.exger.2003.11.011) [DOI] [PubMed] [Google Scholar]
- 63.Zahn J. M., et al. 2006Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2, e115. 10.1371/journal.pgen.0020115 (doi:10.1371/journal.pgen.0020115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lener T., Moll P. R., Rinnerthaler M., Bauer J., Aberger F., Richter K.2006Expression profiling of aging in the human skin. Exp. Gerontol. 41, 387–397 10.1016/j.exger.2006.01.012 (doi:10.1016/j.exger.2006.01.012) [DOI] [PubMed] [Google Scholar]
- 65.Kim S. K.2008Genome-wide views of aging gene networks. Molecular Biology of Aging Monograph 9. Cold Spring Harbor, CT: Cold Spring Harbor Laboratory Press [Google Scholar]
- 66.Zahn J. M., Kim S. K.2007Systems biology of aging in four species. Curr. Opin. Biotechnol. 18, 355–359 10.1016/j.copbio.2007.07.004 (doi:10.1016/j.copbio.2007.07.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harman D.1956Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 11, 298–300 [DOI] [PubMed] [Google Scholar]
- 68.Lapointe J., Hekimi S.2010When a theory of aging ages badly. Cell Mol. Life Sci. 67, 1–8 10.1007/s00018-009-0138-8 (doi:10.1007/s00018-009-0138-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Subramanian A., et al. 2005Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15 545–15 550 10.1073/pnas.0506580102 (doi:10.1073/pnas.0506580102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hauser M. A., et al. 2003Genomic convergence: identifying candidate genes for Parkinson's disease by combining serial analysis of gene expression and genetic linkage. Hum. Mol. Genet. 12, 671–677 10.1093/hmg/12.6.671 (doi:10.1093/hmg/12.6.671) [DOI] [PubMed] [Google Scholar]
- 71.Le-Niculescu H., et al. 2007Towards understanding the schizophrenia code: an expanded convergent functional genomics approach. Am. J. Med. Genet. B Neuropsychiatr. Genet. 144, 129–158 [DOI] [PubMed] [Google Scholar]
- 72.Liang X., et al. 2009Genomic convergence to identify candidate genes for Alzheimer disease on chromosome 10. Hum. Mutat. 30, 463–471 10.1002/humu.20953 (doi:10.1002/humu.20953) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wheeler H. E., et al. 2009Sequential use of transcriptional profiling, expression quantitative trait mapping, and gene association implicates MMP20 in human kidney aging. PLoS Genet. 5, e1000685. 10.1371/journal.pgen.1000685 (doi:10.1371/journal.pgen.1000685) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jormsjo S., Whatling C., Walter D. H., Zeiher A. M., Hamsten A., Eriksson P.2001Allele-specific regulation of matrix metalloproteinase-7 promoter activity is associated with coronary artery luminal dimensions among hypercholesterolemic patients. Arterioscler. Thromb. Vasc. Biol. 21, 1834–1839 10.1161/hq1101.098229 (doi:10.1161/hq1101.098229) [DOI] [PubMed] [Google Scholar]
- 75.Abrass C. K., Adcox M. J., Raugi G. J.1995Aging-associated changes in renal extracellular matrix. Am. J. Pathol. 146, 742–752 [PMC free article] [PubMed] [Google Scholar]
- 76.Kirkwood T. B., Austad S. N.2000Why do we age? Nature 408, 233–238 10.1038/35041682 (doi:10.1038/35041682) [DOI] [PubMed] [Google Scholar]
- 77.Pushkarev D., Neff N. F., Quake S. R.2009Single-molecule sequencing of an individual human genome. Nat. Biotechnol. 27, 847–852 10.1038/nbt.1561 (doi:10.1038/nbt.1561) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wheeler D. A., et al. 2008The complete genome of an individual by massively parallel DNA sequencing. Nature 452, 872–876 10.1038/nature06884 (doi:10.1038/nature06884) [DOI] [PubMed] [Google Scholar]
- 79.Zhu X., Feng T., Li Y., Lu Q., Elston R. C.2009Detecting rare variants for complex traits using family and unrelated data. Genet. Epidemiol. 34, 171–187 10.1002/gepi.20449 (doi:10.1002/gepi.20449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li B., Leal S. M.2008Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 83, 311–321 10.1016/j.ajhg.2008.06.024 (doi:10.1016/j.ajhg.2008.06.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo W., Lin S.2009Generalized linear modeling with regularization for detecting common disease rare haplotype association. Genet. Epidemiol. 33, 308–316 10.1002/gepi.20382 (doi:10.1002/gepi.20382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Novelli V., Viviani Anselmi C., Roncarati R., Guffanti G., Malovini A., Piluso G., Puca A. A.2008Lack of replication of genetic associations with human longevity. Biogerontology 9, 85–92 10.1007/s10522-007-9116-4 (doi:10.1007/s10522-007-9116-4) [DOI] [PubMed] [Google Scholar]