Abstract
Ageing is intrinsically complex, being driven by multiple causal mechanisms. Each mechanism tends to be partially supported by data indicating that it has a role in the overall cellular and molecular pathways underlying the ageing process. However, the magnitude of this role is usually modest. The systems biology approach combines (i) data-driven modelling, often using the large volumes of data generated by functional genomics technologies, and (ii) hypothesis-driven experimental studies to investigate causal pathways and identify their parameter values in an unusually quantitative manner, which enables the contributions of individual mechanisms and their interactions to be better understood, and allows for the design of experiments explicitly to test the complex predictions arising from such models. A clear example of the success of the systems biology approach in unravelling the complexity of ageing can be seen in recent studies on cell replicative senescence, revealing interactions between mitochondrial dysfunction, telomere erosion and DNA damage. An important challenge also exists in connecting the network of (random) damage-driven proximate mechanisms of ageing with the higher level (genetically specified) signalling pathways that influence longevity. This connection is informed by actions of natural selection on the determinants of ageing and longevity.
Keywords: ageing, longevity, systems biology, evolution, complexity, models
1. Introduction
Ageing is unquestioningly complex. Despite the seeming simplicity of single gene mutations with major effect on lifespan and of single-mode interventions such as dietary restrictions [1], it is evident that even in these cases the effects on the ageing process are mediated via coordinate influences on a very large number of proximate mechanisms, most of which control components of the organism's network of maintenance and reproductive functions. In nematode worms, for example, the gene daf-16 modulates the expression of literally hundreds of downstream genes of individually small effect [2], whereas rodent dietary restriction upregulates a very broad array of protective processes [3]. Indeed, the salient feature of both these categories of lifespan modifying factors is that they act on the average length of life without substantially altering either the intrinsic process of senescence that causes frailty, disability and disease, or the intrinsic inter-individual variability of lifespan that is seen even when genetic and environmental conditions are made as uniform as possible.
This inherent complexity of ageing is in fact a direct prediction of the evolutionary understanding of ageing, particularly that which derives from the disposable soma theory, which combines, within a single framework, answers to the questions of both why and how ageing occurs [4]. As organisms live their lives, faults arise at all levels of structure and function. This tendency for faults to accumulate is countered by the action of an extensive array of error-preventing and error-correcting systems. However, maintenance and repair are costly. In a multicellular animal, where some cells belong to the reproductive lineage, or ‘germ-line’, but most make up the non-reproductive tissues, or ‘soma’, it is very much more important to prevent damage from accumulating in the germ-line than it is in the soma, which in due course will die by accident, if nothing else. It follows that, under pressure of natural selection, organisms should make only limited investments in maintenance and repair of somatic tissues, with the consequence that damage to the soma serves as the primary cause of the ageing process. There are strong grounds therefore to expect that the mechanisms of ageing will be multiple and inherently stochastic, with the potential for a very diverse set of interactions.
According to the logic of the disposable soma theory, the genetic control of longevity is expected to be mediated through the tuning by natural selection of multiple mechanisms for somatic maintenance and repair to provide the optimal balance between metabolic cost and survival benefit. This predicts a highly polygenic control of longevity (figure 1). It is striking that the major effects of life extending mutations affecting insulin-signalling pathways, such as the daf-2 mutation in Caenorhabditis elegans and others in mice and fruitflies, appear to act via central effects on metabolism. Given the centrality of resource allocation in understanding how trade-offs are mediated between maintenance and other functions, it is unsurprising that such effects are observed via coordinate regulation of multiple downstream genes, such plasticity in resource allocation having evolved presumably in response to unpredictability of the quality of the environment. Equally, it is consistent that externally imposed variations in food supply (dietary restrictions) result in modulation of the same metabolic regulators to produce upregulation of maintenance and postponement not only of ageing itself but also of the general spectrum of age-associated pathology.
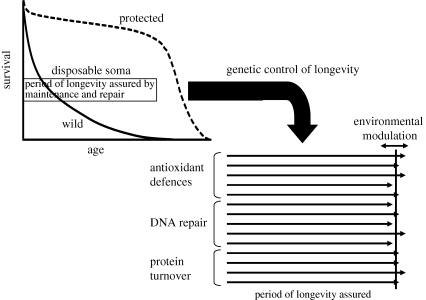
Figure 1.
The genetic regulation of ageing and longevity as explained by the disposable soma theory. Survival in the wild is curtailed by extrinsic mortality and all that is required of the body's maintenance and repair functions is that they should preserve vitality until an age when the chance of remaining alive is small. Genetic control of longevity is effected by setting the many different maintenance and repair functions to provide a sufficient but not excessive period of longevity assured. In some species, there appears to have evolved a capacity to respond to varying resource levels by high-level, nutrient-sensing pathways that co-ordinately alter the investments in maintenance and repair. This can serve to fine-tune optimal resource allocations in order to accommodate environmental fluctuations, such as periods of famine.
To probe the complexity of the ageing process calls for a combination of approaches that can effectively combine the respective strengths of the reductionist and integrative strategies via (i) detailed investigation of the intracellular mechanisms that underlie age-relate damage, (ii) study of how the accumulated damage in cells, which may vary considerably between one individual cell and its neighbours, gives rise to age-related declines in tissue function, and (iii) how these lower level changes both affect the viability of the organism as a whole as well as how systemic factors, such as hormones, provide scope for integration across multiple levels. The necessary integration to deliver this joined-up understanding of ageing is offered by the multi-disciplinary framework of systems biology.
The essence of systems biology is intensely close interaction and collaboration between biologists, mathematicians, computer scientists, engineers and statisticians (e.g. see http://www.bbsrc.ac.uk/web/FILES/Publications/systems_biology.pdf). Systems biology differs from integrative physiology and functional genomics in the focus which is placed on a particularly close coupling between experiment, theory and quantitative modelling. It is thereby capable of generating predictions with high quantitative precision. Although there has long been at least nominal recognition of the importance of interdisciplinary collaboration, the reality is that successful sharing of a common research agenda remains the exception rather than the rule. Fostering the sustained interactions necessary for systems biology requires significant amounts of time spent together, multi-level networking (involving junior as well as senior staff) and active management of integration.
2. The role of models in probing the complexity of ageing
As already noted, a core feature in the systems biology approach is the close coupling of models with experiments. Mathematical and computer modelling is becoming increasingly important in the biological sciences. There are three chief reasons for this. Firstly, biology itself has become much more of an informational science, as a result of the development of genomic (based on advances in gene sequence and expression data) and post-genomic (based on advances in proteomic and functional data) approaches. Secondly, there is growing realization that complex biological processes cannot be understood through the application of reductionist experimental programmes alone. There needs to be some integration of the mass of data and insight from study of the detailed mechanisms at the level of the physiological system. Too often, the contrast between reductionism and systems integration is presented as antagonistic. This is far from true. (Reductionist approaches, aided by rapidly advancing technical sophistication, are absolutely required to generate the detailed data. However, the very power and focus of these techniques take them to a level from which it is necessary to back up and integrate what has been found with what is also being discovered by parallel reductionist efforts on other components of the biological system. Modelling is a potent tool to deliver and validate the relevant integration.) Thirdly, the sophistication and power of desktop computer hardware has increased to a point where the kind of model that, two decades ago, might have required an overnight run on a large mainframe computer can now be done quite fast on a low-cost machine.
Alongside these changes is the development of a different perception of the role and value of computer modelling in biomedical research. To many scientists who work in environments where modelling has not been a part of the scientific toolkit, the nature and scope of computer modelling are still unclear. Many see models as essentially descriptive, begging the question ‘Why bother?’ when the real answer will be revealed in time by experiment. Others adhere to the mistaken belief that as soon as a model has more than two or three parameters, it can ‘explain’ anything, and hence has little predictive value. Fortunately, the increasing dialogue between modellers and experimentalists is beginning to break down these barriers of misunderstanding.
The distinctive advantages of modelling a biological process can be summarized as follows:
— Model building requires that hypotheses be made specific and conceptually rigorous. The investigator must specify each element of the model and how it interacts with other elements.
— The process of model development might lead to the recognition of a gap that needs to be filled by further experimental investigation which may be fundamental to understanding a complex system.
— Computer models yield quantitative as well as qualitative predictions. In ageing, where multiple mechanisms might be at work, it often happens that data are broadly consistent with a hypothesized mechanism but modelling can show that the magnitude of the effect is too small to explain ageing on its own.
— Modelling can result in improved experimental design, especially where the system embodies the potential for complex interactions. Complexity is very hard to deal with experimentally, but relatively straightforward in a computer model.
— Modelling can provide a low-cost, rapid test bed for candidate interventions, thereby enabling a more predictive approach.
Models developed to date to understand the molecular mechanisms of ageing range from those representing specific mechanisms such as telomere erosion [5–10], the accumulation of somatic mutations [11], the accumulation of defective mitochondria [12,13], and breakdown of protein homeostasis [14], to ‘network’ models that have begun to explore how different mechanisms interact synergistically [15–17]. A significant advance has also been the development of the Biology of Ageing e-Science Integration and Simulation (BASIS) system ([18]; see also www.basis.ncl.ac.uk) to support the building of interactive models that can network a variety of individual processes together in a flexible, user-friendly manner.
3. Systems biology of replicative senescence
The contribution of the systems biology approach to understanding ageing is clearly illustrated with reference to the phenomenon of cell replicative senescence—the permanent arrest of cycling in normally proliferating cells such as fibroblasts—which both contributes to age-related loss of mammalian tissue homeostasis and acts as a tumour suppressor mechanism. The pathways leading to establishment of senescence are proving to be more complex than was previously envisaged, when it was widely believed that senescence was caused simply by the erosion of telomeres. A key feature of senescence that has long defied satisfactory explanation is the pronounced stochastic heterogeneity in the division potential of individual cells. Even when a cell population is derived from the multiplication of a single founder cell, the individual cells within the population rapidly diverge to display very different division potentials [19]. This intrinsic variability challenges any idea of a simple cell division counter, such as that the limit on cell division is set by the progressive shortening of telomeres through failure of complete chromosome-end replication in the absence of the enzyme telomerase.
Motivated by evidence that the rate of telomere shortening is strongly affected by oxidative stress, and taking account of the fact that an important source of damage-inducing reactive oxygen species (also known as ‘free radicals’) is the intracellular population of mitochondria, particularly mitochondria which are themselves damaged, Sozou & Kirkwood [17] developed a mathematical model which showed how the heterogeneity of cell senescence could be explained quite naturally by interactions of multiple mechanisms (oxidative damage, telomere shortening and the stochastic nature of mutation to mitochondrial and nuclear DNA). These predictions prompted a search for experimental evidence of a role for mitochondrial dysfunction in senescence. Confirmation of this previously unrecognized role of mitochondria in replicative senescence revealed also that mitochondrial dysfunction could successfully account for the stochastic heterogeneity in cell division potentials [20].
Although it was shown that cell senescence and its intrinsic variability could be explained by an essentially linear process, whereby random defects in mitochondria produced cell to cell variations in oxidative stress, which in turn resulted in cell to cell variation in the timing of telomere-dependent cell division arrest, further observations revealed a more complex picture. Interference with telomere integrity via modification of the telomere-associated protein TRF2 produced a rapid increase in intracellular superoxide and an alteration in mitochondrial morphology, suggestive of dysfunction. Furthermore, irradiation of cells resulting in chromosomal DNA damage also resulted in increased superoxide and mitochondrial dysfunction. These observations suggested some kind of previously unknown connection between telomere erosion, chromosomal DNA damage and mitochondrial dysfunction.
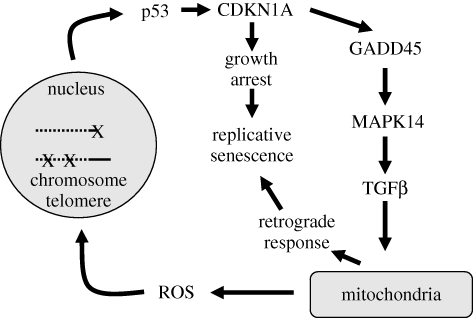
To address this complexity, bioinformatic analysis was performed on the interactome of genes and gene products previously shown to be associated with the state of cell replicative senesecence. Interactome analysis allows the integration of large amounts of data from a wide range of sources, even if they are available only in a dispersed set of databases. Possible relationships between genes were analysed by standard graph-theoretic algorithms for shortest-path analysis. Edges (connections between nodes) were assigned log-likelihood scores [21] providing a measure of the strength of individual interactions and permitting a ranking of the possible paths in order of biological plausibility. A limitation, however, of a raw interactome analysis is the restricted directionality of the inferred edges. To begin to address this, we first combined Biogrid data with Phospho.ELM, a genome-wide protein phosphorylation database [22] and overlaid the results with a query-specific set of gene expression data on genes showing expression level changes with senescence. These procedures identified a candidate pathway involving a feedback loop that is responsible for initiating and maintaining growth arrest in senescent cells (figure 2). The loop connects the DNA damage response, which is consequent to either DNA damage foci or uncapping of critically shortened telomeres, and mitochondrial dysfunction/ROS production via signalling through TP53-CDKN1A-GADD45A-MAPK14-GRB2-TGFBRII-TGFβ. The existence and operation of this feedback loop was then confirmed experimentally by functional target gene inhibition [23].
Figure 2.
Feedback loop responsible for initiating and maintaining growth arrest in senescent cells (adapted from Passos et al. [23]). Damage may arise in mitochondria, by telomere erosion, or by induction of lesions in non-telomeric chromosomal DNA. Once initiated, feedback proceeds (see text), eventually locking the cell in a state of permanent arrest.
To understand further the kinetics of the feedback loop, we formalized the feedback loop in a stochastic computational model in order to see whether it could sufficiently explain the kinetics of senescence induction and maintenance. The model was derived by extension of a previous model of the p53/Mdm2 circuit [24] to include reactions for synthesis/activation and degradation/deactivation/repair of CDKN1A, GADD45, MAPK14, ROS and DNA damage. Realistic values were identified for reaction rate constants, and the initial amounts of the variables and stochastic simulations were run to represent periods of up to a week following irradiation of cells. The model displayed close concordance with the experimental data and it was then used to predict the effects of intervening in the feedback loop. Again, close concordance was found between the model predictions and subsequent experimental tests, resulting in the clear prediction and inference that the feedback loop is both necessary and sufficient for the stability of growth arrest after induction of senescence by DNA damage [23]. The model was also able to predict with considerable accuracy the kinetics of appearance and disappearance of DNA damage foci in young proliferating, irradiated and senescent human fibroblasts, as revealed by live cell imaging. Interestingly, it appeared that once the dynamic feedback loop is triggered by molecular damage, there follows a delay of several days before the cell becomes locked into an actively maintained state of ‘deep’ cellular senescence. The essential feature of the loop is that long-term activation of the checkpoint gene CDKN1A (p21) induces mitochondrial dysfunction and production of reactive oxygen species (ROS). These ROS in turn replenish short-lived DNA damage foci and maintain an ongoing DNA damage response.
The significance of apparently active management of a cell's entry into senescence is that it places this outcome on a similar footing to apoptosis. Both apoptosis and senescence are responses invoked in response to damage (figure 3). Apoptosis serves to delete the cell entirely; senescence places it into a state of irreversible cell cycle arrest. Which option is invoked appears to depend on cell type and, presumably, on the hazard it acts to minimize. The fact that these alternative cell fates are subject to active management should occasion no surprise. Cells have been exposed to intrinsic damage for literally billions of years. Their contributions to ageing of the organism are, however, unlikely to reflect the outcome of natural selection, since it is generally believed that selection acts only very weakly, if at all, on the later stages of the life history; rather, the damage management ‘strategies’ represented by apoptosis/senescence evolved to deal with cells becoming damaged earlier in life. The fact that more cells undergo apoptosis or senescence in older organisms merely reflects the fate that the burden of damage increases with age.
Figure 3.
Alternative cellular responses to damage. Depending on cell type, the normal response to detection of damage will be either to delete the cell by apoptosis or to arrest cell division permanently by senescence. Both of these outcomes will result in impaired tissue homeostasis, contributing to ageing of the organism. Failure to eliminate or arrest damaged cells will leave them vulnerable to accumulating further damage, which may result in cancer.
4. Systems, ageing and disease
For a very wide range of clinically important conditions, age is much the biggest risk factor. Yet exactly how ageing renders the older individual more vulnerable to pathology is as yet little understood, but, far less explicably, is not yet the focus of much investigation. Given that ageing is driven by damage, and that this is also true for many age-associated diseases, there is likely to be considerable overlap between the underlying causative pathways. Systems biology approaches are likely to be crucial in advancing understanding of the links between ageing and disease, resulting in novel routes to intervention.
Connections with ageing are particularly evident in human progeroid conditions, such as Werner's syndrome, where affected individuals experience many features of ageing at nearly twice the normal rate. Werner's syndrome is accompanied by greatly increased risk of osteoporosis and cancer, and results in early death, usually by age 50. The WRN gene that is mutated in Werner's syndrome codes for a protein that plays multiple roles in DNA maintenance. Defective WRN function results in failure of a part of the normal DNA maintenance network, and this drives the rapid genome instability that then causes multiple pathology, which develops through effects on cell survival and senescence, as well as resulting in perturbations of tissue homeostasis [25]. A similar scenario is found in many other inherited human DNA repair deficiencies, and can be replicated in mouse models where a gene modification results in accelerated accumulation of lesions and shortened lifespan [26]. Mouse models with increased rates of intrinsic damage show that cells display a complex capacity to react to changes in their internal state. This is well illustrated in a study of a mouse model with mutation in the Ercc1 gene associated with DNA repair, which showed altered expression in more than a thousand genes, including a general down-regulation of the hormonal pathways associated with regulation of metabolism, including growth hormone/IGF1 signalling, and an upregulation of antioxidant and DNA repair pathways [27]. Intriguingly, the pattern of altered gene expression in this short-lived mutant was the same as has been reported in the context of gene mutations that increase lifespan in the nematode C. elegans and in long-lived, calorie-restricted mice. It seems plausible that the detection of widespread damage in this model triggers pathways that invoke heightened protection against such damage and can thus be understood within the systems biology framework of understanding why and how ageing occurs.
The response to damage is also centrally important in probing the links between cancer and ageing. Ageing and cancer are connected at many levels: (i) age is the biggest risk factor for the great majority of cancers; (ii) cancer incidence rates scale with longevity; and (iii) age-related changes in the tissue micro-environment, including replicative senescence, may favour cancer development and growth. There is evidence that cell senescence serves to reduce risk of malignancy by taking the damaged cell permanently out of cycle [28–30]. Also, manipulation of the pathways that determine whether or not damaged cells undergo apoptosis suggests that the balance between allowing damaged cells to remain (perhaps entering senescence) or deleting them has been optimized through natural selection. Increasing the likelihood of apoptosis protects against cancer but at the cost of accelerating age-related declines of tissue cellularity [31].
5. Discussion
The biology of ageing is sometimes described as being burdened by an excess of competing theories [32]. This view serves only to perpetuate a way of investigating ageing that has long been shown to be inadequate. The molecular and cellular mechanisms underlying ageing are multiple and complex. This does not, however, mean that they are intractable. Although there has been substantial progress from exploring single gene mutations that have major effects on lifespan and single-mode interventions such as dietary restriction, and from probing the associated functional genetic pathways [33,34], significant further integration is likely to be necessary in order to understand the ageing process itself. Single gene mutations with major effects on lifespan, as well as dietary restriction, appear to act via wholesale adjustment of metabolic investments in the hundreds of specific maintenance and repair pathways that collectively result in the ageing of the organism, as manifest in the form of age-related frailty, disability and disease. Indeed, although there are questions of considerable interest in probing from a systems biology perspective the regulation of insulin-signalling and related pathways (e.g. [35]), there has as yet been little attempt to connect the high-level metabolic factors from the ‘many downstream targets’ [36] on which these factors act. Since even the most optimistic view of the extent to which it may prove possible through metabolic intervention to extend human healthspan does not foresee the abolition of ageing, it is imperative that a more detailed understanding of the molecular and cellular mechanisms responsible both for ageing and disease should be attained. This will not happen without the use of systems biology.
Acknowledgements
The author's research on systems biology of ageing has been supported through multiple grants from the UK Biotechnology and Biological Sciences Research Council (BBSRC) including funding for the Centre for Integrated Systems Biology of Ageing and Nutrition (CISBAN). Former colleagues and members of CISBAN are warmly acknowledged for their many contributions.
Footnotes
One contribution of 15 to a Discussion Meeting Issue ‘The new science of ageing’.
References
- 1.Partridge L.2010The new biology of ageing. Phil. Trans. R. Soc. B 365, 147–154 10.1098/rstb.2009.0222 (doi:10.1098/rstb.2009.0222) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., Ahringer J., Li H., Kenyon C.2003Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277–284 10.1038/nature01789 (doi:10.1038/nature01789) [DOI] [PubMed] [Google Scholar]
- 3.Shanley D. P., Kirkwood T. B. L.2000Calorie restriction and aging: a life-history analysis. Evolution 54, 740–750 10.1554/0014-3820(2000)054[0740:CRAAAL]2.3.CO;2 (doi:10.1554/0014-3820(2000)054[0740:CRAAAL]2.3.CO;2) [DOI] [PubMed] [Google Scholar]
- 4.Kirkwood T. B. L.2005Understanding the odd science of ageing. Cell 120, 437–447 10.1016/j.cell.2005.01.027 (doi:10.1016/j.cell.2005.01.027) [DOI] [PubMed] [Google Scholar]
- 5.Rubelj I., Vondracek Z.1999Stochastic mechanism of cellular aging—abrupt telomere shortening as a model for stochastic nature of cellular aging. J. Theoret. Biol. 197, 425–438 10.1006/jtbi.1998.0886 (doi:10.1006/jtbi.1998.0886) [DOI] [PubMed] [Google Scholar]
- 6.Proctor C. J., Kirkwood T. B. L.2002Modelling telomere shortening and the role of oxidative stress. Mech. Ageing Dev. 123, 351–363 10.1016/S0047-6374(01)00380-3 (doi:10.1016/S0047-6374(01)00380-3) [DOI] [PubMed] [Google Scholar]
- 7.Proctor C. J., Kirkwood T. B. L.2003Modelling cellular senescence as a result of telomere state. Aging Cell 2, 151–157 10.1046/j.1474-9728.2003.00050.x (doi:10.1046/j.1474-9728.2003.00050.x) [DOI] [PubMed] [Google Scholar]
- 8.Aviv A., Levy D., Mangel M.2003Growth, telomere dynamics and successful and unsuccessful human aging. Mech. Ageing Dev. 124, 829–837 10.1016/S0047-6374(03)00143-X (doi:10.1016/S0047-6374(03)00143-X) [DOI] [PubMed] [Google Scholar]
- 9.den Buijs J. O., van den Bosch P. P. J., Musters M., van Riel N. A. W.2004Mathematical modeling confirms the length-dependency of telomere shortening. Mech. Ageing Dev. 125, 437–444 10.1016/j.mad.2004.03.007 (doi:10.1016/j.mad.2004.03.007) [DOI] [PubMed] [Google Scholar]
- 10.Sidorov I. A., Gee D., Dimitrov D. S.2004A kinetic model of telomere shortening in infants and adults. J. Theoret. Biol. 226, 169–175 10.1016/j.jtbi.2003.08.009 (doi:10.1016/j.jtbi.2003.08.009) [DOI] [PubMed] [Google Scholar]
- 11.Kirkwood T. B. L., Proctor C. J.2003Somatic mutations and ageing in silico. Mech. Ageing Dev. 124, 85–92 10.1016/S0047-6374(02)00177-X (doi:10.1016/S0047-6374(02)00177-X) [DOI] [PubMed] [Google Scholar]
- 12.Kowald A., Kirkwood T. B. L.2000Accumulation of defective mitochondria through delayed degradation of damaged organelles and its possible role in the ageing of post-mitotic and dividing cells. J. Theoret. Biol. 202, 145–160 10.1006/jtbi.1999.1046 (doi:10.1006/jtbi.1999.1046) [DOI] [PubMed] [Google Scholar]
- 13.Elson J. L., Samuels D. C., Turnbull D. M., Chinnery P. F.2001Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. Am. J. Hum. Genet. 68, 802–806 10.1086/318801 (doi:10.1086/318801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Proctor C. J., Soti C., Boys R. J., Gillespie C. S., Shanley D. P., Wilkinson D. J., Kirkwood T. B. L.2005Modelling the actions of chaperones and their role in ageing. Mech. Ageing Dev. 126, 119–131 10.1016/j.mad.2004.09.031 (doi:10.1016/j.mad.2004.09.031) [DOI] [PubMed] [Google Scholar]
- 15.Kowald A., Kirkwood T. B. L.1994Towards a network theory of ageing: a model combining the free radical theory and the protein error theory. J. Theoret. Biol. 168, 75–94 10.1006/jtbi.1994.1089 (doi:10.1006/jtbi.1994.1089) [DOI] [PubMed] [Google Scholar]
- 16.Kowald A., Kirkwood T. B. L.1996A network theory of ageing: the interactions of defective mitochondria, aberrant proteins, free radicals and scavengers in the ageing process. Mutat. Res. 316, 209–236 [DOI] [PubMed] [Google Scholar]
- 17.Sozou P. D., Kirkwood T. B. L.2001A stochastic model of cell replicative senescence based on telomere shortening, oxidative stress, and somatic mutations in nuclear and mitochondrial DNA. J. Theoret. Biol. 213, 573–586 10.1006/jtbi.2001.2432 (doi:10.1006/jtbi.2001.2432) [DOI] [PubMed] [Google Scholar]
- 18.Kirkwood T. B. L., Boys R. J., Gillespie C. S., Proctor C. J., Shanley D. P., Wilkinson D. J.2003Towards an e-biology of ageing: integrating theory and data. Nat. Rev. Mol. Cell Biol. 4, 243–249 10.1038/nrm1051 (doi:10.1038/nrm1051) [DOI] [PubMed] [Google Scholar]
- 19.Smith J. R., Whitney R. G.1980Intraclonal variation in proliferative potential of human diploid fibroblasts: stochastic mechanism for cellular aging. Science 207, 82–84 10.1126/science.7350644 (doi:10.1126/science.7350644) [DOI] [PubMed] [Google Scholar]
- 20.Passos J. F., et al. 2007Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol. 5, e110. 10.1371/journal.pbio.0050110 (doi:10.1371/journal.pbio.0050110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee I., Date S. V., Adai A. T., Marcotte E. M.2004A probabilistic functional network of yeast genes. Science 306, 1555–1558 10.1126/science.1099511 (doi:10.1126/science.1099511) [DOI] [PubMed] [Google Scholar]
- 22.Diella F., Gould C. M., Chica C., Via A., Gibson T. J.2008Phospho.ELM: a database of phosphorylation sites—update. Nucleic Acids Res. 36, D240–D244 10.1093/nar/gkm772 (doi:10.1093/nar/gkm772) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Passos J. F., et al. 2010Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol. Syst. Biol. 6, 347. 10.1038/msb.2010.5 (doi:10.1038/msb.2010.5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Proctor C. J., Gray D. A.2008Explaining oscillations and variability in the p53-Mdm2 system. BMC Syst. Biol. 2, 75. 10.1186/1752-0509-2-75 (doi:10.1186/1752-0509-2-75) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kipling D., Davis T., Ostler E. L., Faragher R. G.2004What can progeroid syndromes tell us about human aging? Science 305, 1426–1431 10.1126/science.1102587 (doi:10.1126/science.1102587) [DOI] [PubMed] [Google Scholar]
- 26.Hasty P., Campisi J., Hoeijmakers J., van Steeg H., Vijg J.2003Aging and genome maintenance: lessons from the mouse? Science 299, 1355–1359 10.1126/science.1079161 (doi:10.1126/science.1079161) [DOI] [PubMed] [Google Scholar]
- 27.Niedernhofer L. J., et al. 2006A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature 444, 1038–1043 10.1038/nature05456 (doi:10.1038/nature05456) [DOI] [PubMed] [Google Scholar]
- 28.Braig M., et al. 2005Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436, 660–665 10.1038/nature03841 (doi:10.1038/nature03841) [DOI] [PubMed] [Google Scholar]
- 29.Michaloglou C., et al. 2005BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436, 720–724 10.1038/nature03890 (doi:10.1038/nature03890) [DOI] [PubMed] [Google Scholar]
- 30.Chen Z., et al. 2005Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436, 725–730 10.1038/nature03918 (doi:10.1038/nature03918) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyner S. D., et al. 2002p53 mutant mice that display early ageing-associated phenotypes. Nature 415, 45–53 10.1038/415045a (doi:10.1038/415045a) [DOI] [PubMed] [Google Scholar]
- 32.Medvedev Z. A.1990An attempt at a rational classification of theories of ageing. Biol. Rev. Camb. Phil. Soc. 65, 375–398 10.1111/j.1469-185X.1990.tb01428.x (doi:10.1111/j.1469-185X.1990.tb01428.x) [DOI] [PubMed] [Google Scholar]
- 33.Southworth L. K., Owen A. B., Kim S. K.2009Aging mice show a decreasing correlation of gene expression within genetic modules. PLoS Genet. 5, e1000776. 10.1371/journal.pgen.1000776 (doi:10.1371/journal.pgen.1000776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapahi P., Chen D., Rogers A. N., Katewa S. D., Li P. W., Thomas E. L., Kockel L.2010With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 11, 453–465 10.1016/j.cmet.2010.05.001 (doi:10.1016/j.cmet.2010.05.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith G. R., Shanley D. P.2010Modelling the response of FOXO transcription factors to multiple post-translational modifications made by ageing-related signalling pathways. PLoS ONE 5, e11092. 10.1371/journal.pone.0011092 (doi:10.1371/journal.pone.0011092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christensen K., Johnson T. E., Vaupel J. W.2006The quest for genetic determinants of human longevity: challenges and insights. Nat. Rev. Genet. 7, 436–448 10.1038/nrg1871 (doi:10.1038/nrg1871) [DOI] [PMC free article] [PubMed] [Google Scholar]