Abstract
Embryonic stem (ES) cells are pluripotent cells that can self renew indefinitely or be induced to differentiate into multiple cell lineages, and thus have the potential to be used in regenerative medicine. Pluripotency transcription factors (TFs), such as Oct4, Sox2, and Nanog, function in a regulatory circuit that silences the expression of key TFs required for differentiation and activates the expression of genes important for maintenance of pluripotency. In addition, proteins that remodel chromatin structure also play important roles in determining the ES cell-specific gene expression pattern. Here we review recent studies demonstrating the roles of enzymes that carry out one facet of chromatin regulation, nucleosome remodeling, in control of ES cell self-renewal and differentiation.
Introduction
Pluripotent ES cells are characterized by higher order chromatin structure that is generally dynamic and permissive to the transcriptional machinery [1]. This specialized chromatin state is unique to ES cells and is established in part by the activity of ATP-dependent nucleosome remodeling complexes. These complexes alter nucleosome-DNA contacts to affect the orientation of DNA around histone octamers, the position of nucleosomes on the DNA, or to exchange histones [2]. Because packaging of DNA into chromatin inhibits transcription by restricting DNA accessibility, the activity of nucleosome remodelers controls the extent to which specific DNA sites are utilized by TFs and other regulatory proteins.
The Swi2/Snf2 superfamily of ATPases is highly conserved throughout eukaryotes, with roles in gene regulation, DNA repair, and recombination, among others [3]. Swi2/Snf2 family members are modified helicase proteins that utilize the energy of ATP hydrolysis to alter the structure or positions of nucleosomes on DNA [4]. Usually, Swi2/Snf2 ATPases function within multisubunit complexes, in which accessory subunits target the complex to specific regions of chromatin, contribute to DNA or histone binding, or provide other regulatory roles [5,6]. In mammals, there are nearly 30 Swi2/Snf2 family members comprising an even larger number of ATP-dependent nucleosome remodeling complexes, the majority of which have not been thoroughly characterized. Recently, a number of ATP-dependent nucleosome remodeling complexes have been shown to play important roles in both ES cell self-renewal and differentiation.
ES cell specific BAF complexes promote self-renewal
The BAF (Brg/Brahma associated factor) complexes contain either the Brg1 or Brm ATPases as their catalytic subunit, with different combinations of accessory subunits. Previously, homozygous deletions of genes encoding BAF subunits Smarca4 (Brg1), Smarcc1 (Baf155), and Smarcb1 (Baf47, Snf5) were found to be early embryonic lethal in mice, consistent with a possible role for BAF complexes in ES cell self-renewal [7–9]. Indeed, several groups have recently reported that ES cells depleted of several known subunits of BAF complexes have defects in gene regulation, self-renewal, and pluripotency. Homozygous deletion of genes encoding either of two related BAF subunits, Arid1a and Arid1b, resulted in partial defects in ES cell self-renewal [10,11]. While Arid1a−/− ES cells exhibited upregulation of markers of primitive endoderm and failure to differentiate into mesoderm when induced to do so, Arid1b−/− ES cells expressed markers of both mesoderm and trophectoderm. These data suggest that differential incorporation of BAF250 isoforms within BAF complexes may alter the spectrum of genes targeted. Similarly, knockdown (KD) of BAF components Brg1 or Baf155 in ES cells results in defects in self-renewal and differentiation, underscoring the importance of BAF complexes in ES cells [12,13].
Several BAF complexes appear to be expressed in ES cells, including an ES cell-specific complex called esBAF that consists of a unique subunit composition relative to BAF complexes in other cell types [10,13,14]. The esBAF complex appears to play a direct role in mediating the gene regulatory functions of several core ES cell TFs, since BAF subunits interact directly with TFs Oct4, Sox2, and Nanog and occupy overlapping regions on chromatin [15,16]. In addition, BAF complexes in ES cells bind to many genes encoding core ES cell TFs: Oct4, Sox2, Nanog, Dppa2, Dppa4, Sall4, and Myc [16]. For some pluripotency genes, BAF complexes function to tonically silence transcription [16], maintaining their expression at adequate, but not excessive, levels. Conversely, the pluripotency TF Myc functions to silence some BAF subunits expressed in differentiated cells but not ES cells, thereby maintaining the correct composition of ES cell-specific BAF complexes [17].
Chd1 regulates ES cell specific heterochromatin organization
Unlike differentiated cells, ES cells lack significant regions of heterochromatin, the relatively condensed, transcriptionally silent chromatin structure enriched for repressive histone modifications and DNA methylation [1,18]. Recently, the Swi/Snf superfamily ATPase Chd1 was found in an RNAi screen of genes expressed in several types of stem cells to be necessary for ES cell self-renewal and pluripotency [19]. ES cells depleted of Chd1 accumulated large blocks of heterochromatin and exhibited upregulation of genes expressed during neural differentiation. Consistent with these data, Chd1 KD ES cells exhibited a high propensity for neural differentiation, with a concomitant defect in formation of primitive endoderm and cardiac mesoderm.
Chromatin localization of Chd1 mirrors that of RNA Polymerase II and histone H3K4me3, suggesting a role for Chd1 in transcriptional activation [19]. However, transcriptional profiling experiments showed that Chd1 KD ES cells exhibited upregulation of far more genes than downregulation. Interestingly, Chd1 binds the promoter of the Oct4 gene (Pou5f1), and is required for Oct4 expression in ES cells. Conversely, Oct4 binds the regulatory region of Chd1 gene, suggesting the existence of a regulatory loop.
Chromatin remodelers are required for differentiation
While not required for ES cell self-renewal, two other ATP-dependent nucleosome remodeling factors, Lsh and NURF, appear to be required for proper ES cell differentiation. KD of Lsh, a Swi2/Snf2 superfamily ATPase that functions in retrotransposon silencing, resulted in failure to methylate and silence the promoters of pluripotency TFs during differentiation [20]. NURF is a multisubunit nucleosome remodeling complex involved in gene regulation in metazoans. ES cells lacking NURF subunit Bptf exhibit defects in differentiation of all three germ layers [21].
Two bi-functional chromatin regulatory complexes, the Tip60-p400 and NURD complexes, have also been shown to play important roles in ES cell self-renewal and pluripotency [15,22–26]. The NURD complex exhibits two chromatin regulatory activities: ATP-dependent nucleosome remodeling, catalyzed by the Mi-2 Swi2/Snf2 superfamily ATPase, and histone deacetylase activity, catalyzed by Hdac1 and Hdac2. Homozygous deletion of the non-catalytic NURD subunit Mbd3 allows mutant ES cells to self-renew in the absence of leukemia-inhibitory factor (LIF) [23]. Furthermore, loss of Mbd3 impairs ES cell differentiation in general and alters the spectrum of cell types produced during differentiation [23,25]. Knockout or KD of Mbd3 in ES cells causes misregulation of a number of genes [23,25], including upregulation of genes expressed in trophectoderm (TE) [25]. Consistent with these data, in vitro differentiation of Mbd3−/− ES cells in embryoid bodies (EBs) results in induction of markers of TE [23], a cell type not normally produced during differentiation of ES cells in vitro or in vivo.
In addition, a second form of the NURD complex lacking Mbd3 and Rbbp7, has recently been identified in ES cells [15]. This complex, called NODE (Nanog and Oct4 associated deacetylase), appears to form a high molecular weight complex with both Nanog and Oct4 that functions to repress expression of developmentally regulated genes in ES cells [15]. Similarly, one or both NURD-like complexes has been shown to interact with ES cell TF Sall4 [32]. KD of Mta1, a subunit shared by the NURD and NODE complexes, caused different changes in gene expression than Mbd3 KD or KO, confirming the different functions of these complexes [15]. Unlike Mbd3 loss, which inhibits ES cell differentiation, Mta1 KD resulted in upregulation of differentiation genes of multiple lineages, as well as ES cell differentiation. It remains to be determined how the levels, assembly and activities of the two NURD-like complexes are regulated in ES cells.
Tip60-p400 functions in the same pathway as Nanog to promote self-renewal
Like NURD, Tip60-p400 also exhibits two chromatin regulatory activities. The p400 protein is a Swi2/Snf2 superfamily member that functions in exchange of dimers of histones H2AZ-H2B (or other H2A variants) within nucleosomes [27–30]. Tip60-p400 complex has a second catalytic subunit, Tip60, which functions as a protein acetyltransferase [31]. KD of p400, Tip60, or any other member of the Tip60-p400 complex results in partial differentiation of ES cells, coincident with induction of markers of all three germ layers [26]. In ES cells, p400 was found to bind to the promoters of both highly expressed genes and silent genes, many of which are induced during differentiation. However, upon KD of either Tip60 or p400, many targets of Tip60-p400 that are normally silent in ES cells became upregulated, while expression very few of the highly-expressed targets of the complex were significantly affected [26]. These data were surprising, given the established functions of Tip60 as a histone acetyltransferase and transcriptional co-activator [31].
In addition, Tip60-p400 complex was found to functionally overlap with the core pluripotency TF Nanog. Although Nanog and Tip60-p400 were not found to interact physically, they share a significantly overlapping set of target genes, and have similar affects on gene expression upon KD in ES cells. Epistasis analysis suggests that for some common targets, Nanog and Tip60-p400 complex function in a common pathway to repress differentiation-induced genes in ES cells, although the nature of this pathway remains unclear [26].
One noteworthy feature of ES cell chromatin is that specific regulatory sites, particularly those at lineage specific transcription factor loci, are silenced but remain poised for activation. This specialized chromatin state is promoted by the incorporation of the histone variant, H2AZ, which is incorporated into chromatin by Tip60-p400 [33]. H2AZ is enriched at regions flanking transcriptional start sites, and can promote both transcriptional activation and repression [34]. H2AZ incorporation can influence nucleosome positioning, H1 linker binding, and chromatin remodeling enzyme activity [35]. In ES cells, H2AZ is enriched at silent developmentally regulated promoters, as are repressive TFs and repressive covalent histone modifications [33]. In addition, depletion of H2AZ causes increased expression of H2AZ occupied promoters, indicating that H2AZ contributes to silencing of developmental regulators. In differentiated cells, H2AZ is enriched at active promoters. These data suggest that H2AZ incorporation may be one of several mechanisms that contribute to keeping genes that encode developmental regulators in a silent but poised state in ES cells.
What mechanisms ensure the correct regulation of chromatin remodelers during differentiation?
The studies summarized here show that ATP-dependent nucleosome remodeling enzymes are required for ES cell self-renewal, for pluripotency, and for differentiation into specific lineages. A general theme that emerges from these studies is that the activities of chromatin remodelers are dynamically regulated to maintain pluripotency and that their function can change upon differentiation. In many instances a direct interaction between remodeling complexes and pluripotency TFs in ES cells suggests that the TFs may direct remodelers to their sites of action, which in turn promotes self-renewal. For example, in ES cells, members of the BAF class of Swi/Snf complexes interact with pluripotency TFs to inhibit expression of developmental regulators [15,16]. In contrast, Tip60-p400 lies in the same pathway as Nanog, but does not directly interact with this TF [26]. Upon ES cell differentiation, BAF activity turns off pluripotency specific genes, such as Nanog, and activates expression of developmental regulators. How is BAF function altered from a role in repression of differentiation to that of an activator? BAF subunit composition plays an important role in modulating its function (Figure 1), but the mechanisms that trigger alterations in the make up of BAF complexes have not been defined, nor have the mechanisms by which they alter function. Also, how is the specialized chromatin structure that characterizes distinct ES cell developmental states established, and how does this chromatin structure affect chromatin remodeling at specific loci and on a genome wide basis? Lastly, the mechanisms by which the activities of the different chromatin remodeling enzymes are integrated to regulate chromatin structure in ES cells are not well understood. While some nucleosome remodeling enzymes are likely recruited to specific pluripotency and differentiation genes through interactions with pluripotency TFs, other factors appear to localize more broadly, perhaps via interactions with specific histone modifications. The relative contributions of TF-specific recruitment of nucleosome remodelers to localized regions and widespread binding of nucleosome remodelers to large chromatin domains in ES cells remain unclear. A better understanding of how ATP dependent chromatin remodeling enzymes are targeted and regulated in ES cells will help clarify the mechanisms by which chromatin is prepared to achieve a developmentally appropriate gene expression program.
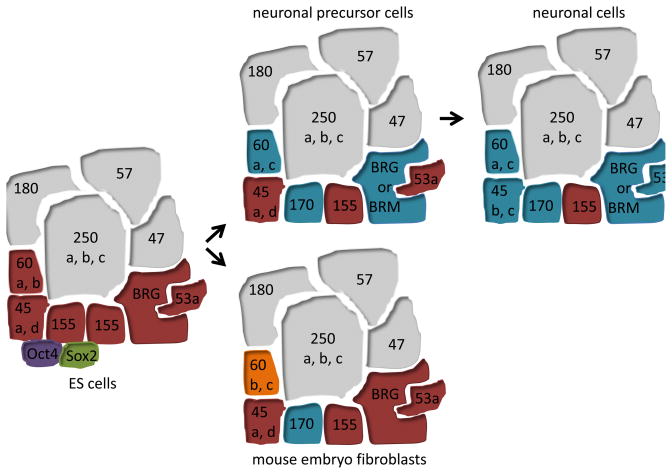
Figure 1.
The composition of the BAF complexes is regulated in a cell type specific fashion. In ES cells, the BAF complex consists of an ES specific combination of subunits and interacts with the pluripotency TFs Oct4 (purple) and Sox2 (green). It remains to be determined whether the BAF complex associates with master regulatory TFs that direct cell type specification in other cell types. BAF subunits that are present in all cell types indicated are coloured gray. Subunits that change between cell types are burgundy, teal and orange.
Acknowledgments
The authors are funded in part by grants (R01GM085186 to B.P. and 4R00CA140854-02 to T.F.) from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2*.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 463:474–484. doi: 10.1038/nature08911. Thorough and thoughtful review on chromatin remodelers during development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varga-Weisz P. ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene. 2001;20:3076–3085. doi: 10.1038/sj.onc.1204332. [DOI] [PubMed] [Google Scholar]
- 4.Racki LR, Narlikar GJ. ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Curr Opin Genet Dev. 2008;18:137–144. doi: 10.1016/j.gde.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryme J, Asp P, Bohm S, Cavellan E, Farrants AK. Variations in the composition of mammalian SWI/SNF chromatin remodelling complexes. J Cell Biochem. 2009;108:565–576. doi: 10.1002/jcb.22288. [DOI] [PubMed] [Google Scholar]
- 7.Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JK, Huh SO, Choi H, Lee KS, Shin D, Lee C, Nam JS, Kim H, Chung H, Lee HW, et al. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol Cell Biol. 2001;21:7787–7795. doi: 10.1128/MCB.21.22.7787-7795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 10.Yan Z, Wang Z, Sharova L, Sharov AA, Ling C, Piao Y, Aiba K, Matoba R, Wang W, Ko MS. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–1165. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci U S A. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12*.Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–328. doi: 10.1634/stemcells.2008-0710. Shows an important role for Brg1 in regulating self-renewal and pluripotency, as Brg1 occupies the promoters of and is necessary for normal expression Oct4, Nanog and other pluripotency related genes. [DOI] [PubMed] [Google Scholar]
- 13*.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. Demonstrates that the ES cell-specific composition of the SWI/SNF-Brg1 complex is necessary for self-renewal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaeser MD, Aslanian A, Dong MQ, Yates JR, 3rd, Emerson BM. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J Biol Chem. 2008;283:32254–32263. doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15**.Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. Shows that the TFs Nanog and Oct4 interact with several repression complexes, including NURD, Sin3A, and Pml, in ES cells. An Hdac1/2- and Mta1/2-containing complex, termed NODE (for Nanog and Oct4 associated deacetylase), is identified and found to be necessary for self-renewal. [DOI] [PubMed] [Google Scholar]
- 16**.Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. Shows that the ES cell specific form of the BAF remodeling complex colocalizes extensively with the TFs Oct4, Sox2 and Nanog. In addition esBAF colocalizes with Stat3 and Smad1 genome-wide, suggesting that esBAF may link LIF and BMP signaling pathways with the pluripotency TFs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin CH, Lin C, Tanaka H, Fero ML, Eisenman RN. Gene regulation and epigenetic remodeling in murine embryonic stem cells by c-Myc. PLoS One. 2009;4:e7839. doi: 10.1371/journal.pone.0007839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 19**.Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. Demonstrates that Chd1 is essential for open chromatin and pluripotency of ES cells, and for somatic cell reprogramming to the pluripotent state. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xi S, Zhu H, Xu H, Schmidtmann A, Geiman TM, Muegge K. Lsh controls Hox gene silencing during development. Proc Natl Acad Sci U S A. 2007;104:14366–14371. doi: 10.1073/pnas.0703669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landry J, Sharov AA, Piao Y, Sharova LV, Xiao H, Southon E, Matta J, Tessarollo L, Zhang YE, Ko MS, et al. Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet. 2008;4:e1000241. doi: 10.1371/journal.pgen.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deshpande AM, Dai YS, Kim Y, Kim J, Kimlin L, Gao K, Wong DT. Cdk2ap1 is required for epigenetic silencing of Oct4 during murine embryonic stem cell differentiation. J Biol Chem. 2009;284:6043–6047. doi: 10.1074/jbc.C800158200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 24.Kaji K, Nichols J, Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007;134:1123–1132. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- 25.Zhu D, Fang J, Li Y, Zhang J. Mbd3, a component of NuRD/Mi-2 complex, helps maintain pluripotency of mouse embryonic stem cells by repressing trophectoderm differentiation. PLoS One. 2009;4:e7684. doi: 10.1371/journal.pone.0007684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. Shows a role for the Tip60-p400 complex in ES cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krogan NJ, Baetz K, Keogh MC, Datta N, Sawa C, Kwok TC, Thompson NJ, Davey MG, Pootoolal J, Hughes TR, et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc Natl Acad Sci U S A. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 30.Gevry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 2007;21:1869–1881. doi: 10.1101/gad.1545707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Lu J, Jeong HW, Kong N, Yang Y, Carroll J, Luo HR, Silberstein LE, Yupoma, Chai L. Stem cell factor SALL4 represses the transcriptions of PTEN and SALL1 through an epigenetic repressor complex. PLoS One. 2009;4:e5577. doi: 10.1371/journal.pone.0005577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. Demonstrates that the histone H2A variant H2AZ is enriched on developmental genes that are silenced but poised in ES cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci U S A. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]