Abstract
European eels (Anguilla anguilla) undertake spawning migrations of more than 5000 km from continental Europe and North Africa to frontal zones in the Sargasso Sea. Subsequently, the larval offspring are advected by large-scale eastward ocean currents towards continental waters. However, the Sargasso Sea is oligotrophic, with generally low plankton biomass, and the feeding biology of eel larvae has so far remained a mystery, hampering understanding of this peculiar life history. DNA barcoding of gut contents of 61 genetically identified A. anguilla larvae caught in the Sargasso Sea showed that even the smallest larvae feed on a striking variety of plankton organisms, and that gelatinous zooplankton is of fundamental dietary importance. Hence, the specific plankton composition seems essential for eel larval feeding and growth, suggesting a linkage between eel survival and regional plankton productivity. These novel insights into the prey of Atlantic eels may furthermore facilitate eel larval rearing in aquaculture, which ultimately may replace the unsustainable use of wild-caught glass eels.
Keywords: Sargasso Sea, European eel, Anguilla anguilla, diet, barcoding
1. Introduction
The European eel (Anguilla anguilla) exhibits one of the most remarkable and yet enigmatic life histories in the Animalia Kingdom. The spawning areas were unreported until 1922 (Schmidt 1922) and the biology of the peculiar leaf-like larval stage (leptocephalus) still remains largely unknown (Tesch 2003). The Sargasso Sea is generally oligotrophic, but the spawning by both European and American eels (Anguilla rostrata) is associated with relatively productive frontal zones (Kleckner & McCleave 1988), which could enhance feeding opportunities for newly hatched larvae. However, the lack of knowledge about the feeding biology of eel larvae prevents an understanding of the eel life cycle (Miller 2009). Moreover, recent drastic declines in the European eel have led to its listing in Convention on International Trade in Endangered Species (CITES'), appendix SII. These declines have been ascribed to overfishing, habitat degradation and changes in ocean currents (Van Ginneken & Maes 2005), the latter of which could decrease productivity and larval feeding opportunities in the spawning region (Friedland et al. 2007). Evaluation of this hypothesis first requires a better understanding of larval feeding biology.
Early studies reported empty guts in eel larvae (Miller 2009), while more recent studies based on visual identification of gut contents proposed dissolved and particulate organic matter as sources of eel larval nutrition, possibly in the form of marine snow, larvacean houses, or zooplankton faecal pellets (Otake et al. 1993; Mochioka & Iwamizu 1996). Unfortunately, these reports mostly concerned other eel species and were based on large (35–129 mm) larvae beyond the first feeding stage. Artificially reproduced larvae of the Japanese eel (Anguilla japonica) can survive on a diet based on shark egg yolk (Tanaka et al. 2003; Miller 2009), but this is unlikely to be a food source in nature.
Determining diets by traditional gut content analysis is often problematic because most prey become unrecognizable once partly digested. However, molecular barcoding enables species assignment of prey through sequencing of short DNA strands surviving in the digestive systems (King et al. 2008). We applied this approach to gain insights into the feeding ecology of small (4.5–14.5 mm) European eel larvae associated with thermal fronts in the southern Sargasso Sea in March–April 2007. Guts excised from 61 genetically identified European eel larvae were analysed using sequencing of 18S ribosomal RNA genes isolated from polymerase chain reaction (PCR) amplicons, separated by denaturing gradient gel electrophoresis (DGGE).
2. Material and methods
Details on capture, molecular analyses and technical literature references are provided in the electronic supplementary material. Eel larvae were caught on three transects, along the longitudes 64° W, 67° W and 70° W (electronic supplementary material, figure S1). Larvae were sorted and stored in RNALATER (QIAGEN) or 96 per cent ethanol. In the laboratory, the guts were excised and stored individually in 96 per cent ethanol until DNA extraction. DNA was extracted from the remaining larval tissue for species identification of European eel larvae, based on analysis of the mitochondrial cytochrome b gene, the nuclear 5S rRNA gene and microsatellite genotyping. Subsequently, DNA was extracted from guts from 61 randomly selected A. anguilla larvae.
Universal primers were used to PCR amplify 18S rRNA genes from gut samples for DGGE (electronic supplementary material, figure S2). A total of 79 bands representative of prey were successfully sequenced. By including two to three samples with sequenced prey bands on each DGGE gel, 14 additional bands could be identified from their vertical alignments with sequenced bands (Quantity One 4.6.3; BioRad; tolerance level set to 0.5%, electronic supplementary material, table S1).
Sequences were aligned using Clustal W (MegAlign, Lasergene 7) and compared against DNA sequences in GenBank using BLASTN. Selected nearest relatives were retrieved and included with the sequences in a phylogenetic tree constructed in MEGA4 (Tamura et al. 2007). Sequences of prey items are deposited in GenBank under accession numbers GU188286–GU188364.
3. Results and discussion
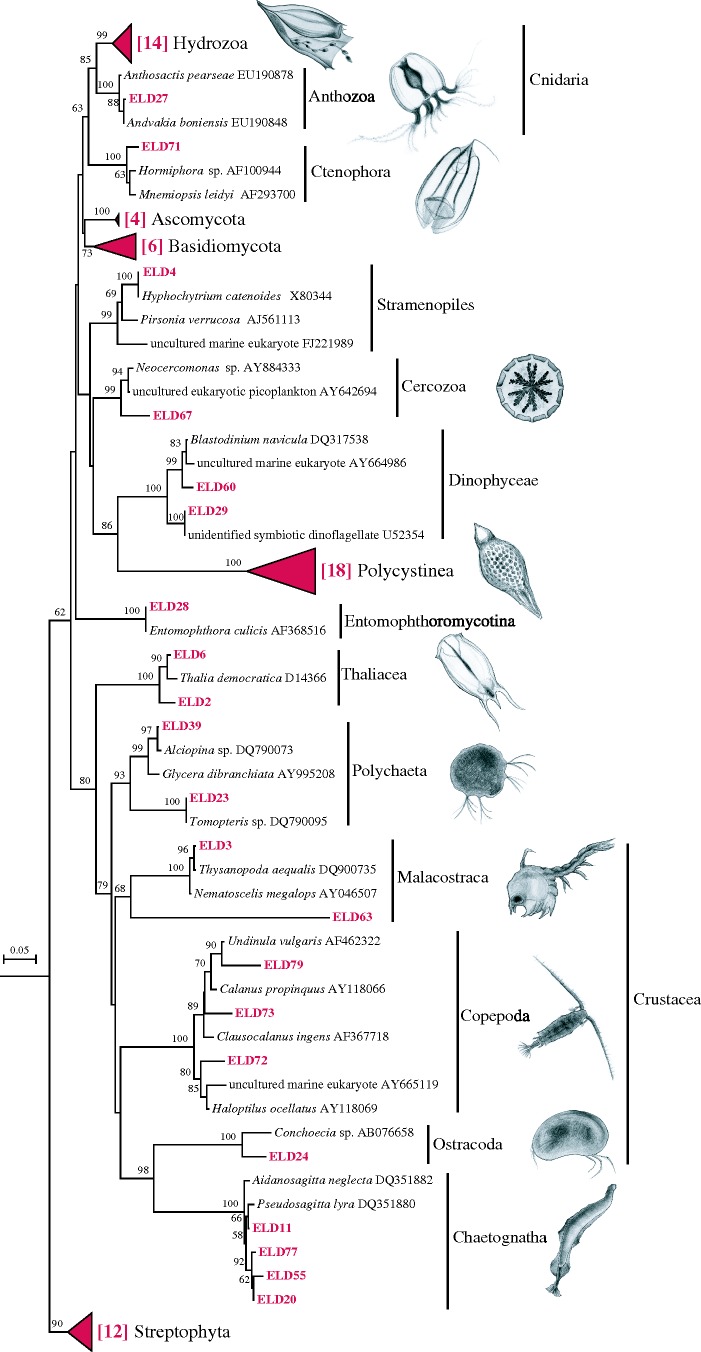
Prey bands, at an average of three per larvae (range 1–17), were discernible in 42 of the 61 European eel larvae analysed (electronic supplementary material, table S1). Based on their topological position within a phylogenetic tree (figure 1), the 75 sequences were assigned to 17 taxonomic lineages supported by high bootstrap values. The gut analyses showed consumption of a wide range of plankton taxa, and a number of the nearest relatives identified in GenBank represented species or DNA sequences obtained from the Sargasso Sea. Sequences representing Hydrozoa or Polycystinea species were found in 55 and 40 per cent of larvae with detectable prey (figure 2). Less frequently occurring prey included Chaetognatha, Copepoda, Malacostraca, Thaliacea, Polychaeta, Dinophyceae, Ostracoda, Anthozoa, Stramenopiles, Cercozoa, Ctenophora and Entomophthoromycotina. Notably, since almost a third of the discernible DGGE bands were unidentified (electronic supplementary material, table S1), the true diversity of prey DNA fragments in the larval guts is probably underestimated. Sequences within Stramenopiles and Dinophyceae were related to parasites of larger organisms while other sequences (e.g. within Cercozoa) showed lower similarity (less than 90%) to published sequences; however, in general the identified prey categories all showed size and reproductive timing in the Sargasso Sea (Moore 1949; Deevey 1971) that make them or their offspring suitable prey for eel larvae. For instance, the abundance of larval stages of Polychaeta, Chaetognatha, and the relevant groups within Crustacea, peaks in spring (Deevey 1971), when Anguilla eels spawn and also the time of our sampling.
Figure 1.
Neighbour-joining tree showing prey sequences (approx. 500 bp) obtained from A. anguilla gut samples (marked in red, eel larvae diet (ELD)) and 18S rRNA gene sequences from GenBank (in italics followed by the accession number). For collapsed groups, the number of sequences per group is given within brackets. Bootstrap values greater than 50% (2000 replications) are shown above branches. Drawings illustrate potential free-living prey within each group. Scale bar indicates nucleotide substitutions per site.
Figure 2.
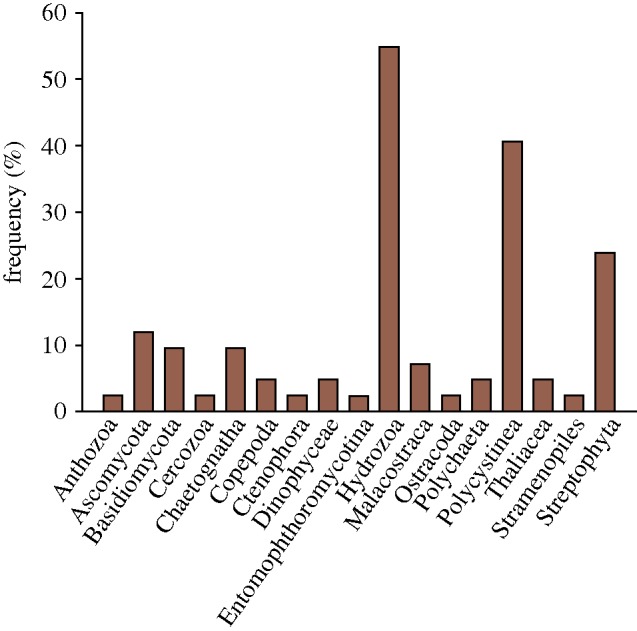
Frequency of prey taxa in guts of 42 larvae of A. anguilla.
The data suggest that gelatinous zooplankton (Hydrozoa, Thaliacea and Ctenophora) are of particular dietary importance for eel larvae (figure 2). Further, colonial forms of Polycystinea also have a gelatinous matrix (Michaels et al. 1995). Both Hydrozoa and Polycystinea, a subgroup of the radiolaria, are abundant in the Sargasso Sea and of suitable sizes as prey (Moore 1949). Broad diets as well as specialized gelatinous zooplankton-based diets are known from first-feeding larvae of other fish species with diets affected by prey availability and the width of the larval mouth (Last 1978). We found no evidence of larvacean DNA in the examined eel guts, but this does not exclude the possibility that eels feed on their polysaccharide-based houses (Körner 1952).
Though most of the identified prey items represent known marine plankton, DNA barcoding also identified Streptophyta and fungi (Basidiomycota and Ascomycota). The nearest relatives of the Streptophyta sequences included not only marine sequences but also genes from higher plants, suggesting potential contamination. However, reports of similar sequences from whale faeces (Jarman et al. 2004) and guts of lobster larvae (Suzuki et al. 2006) indicate that they represent hitherto unknown organisms in marine plankton. Similarly, some of the fungal sequences were related to sequences obtained from marine plankton.
A total of 19 of the 61 analysed larvae did not contain amplifiable prey items. Eel larvae greater than or equal to 5 mm perform diel vertical migrations (Castonguay & McCleave 1987), possibly related to predator avoidance and/or feeding. However, the frequency of empty guts among our examined larvae did not differ significantly between larvae caught at night (n = 33) or day (n = 28; Fisher's test, p = 1.0). This may suggest feeding independent of light conditions, although the unknown retention time of prey in larval guts precludes firm conclusions. No significant correspondence between the number of different prey per larva and phytoplankton biomass, latitude, longitude, day/night capture, or larva size were detected, possibly reflecting the limited data available (electronic supplementary material, table S2). Nevertheless, for the arbitrary size groups of small (4.0–7.0 mm) and large larvae (10.0–14.5 mm), the frequency of empty guts was higher among small larvae (40%) than among large larvae (24%), and the diet complexity was tentatively higher for the large relative to the small larvae (4.3 ± 4.3 versus 2.0 ± 1.4 different prey items per larvae, average ± s.d.). This may suggest more continuous feeding in larger larvae, consistent with increased diel vertical migration in this size group (Castonguay & McCleave 1987).
The present results represent, to our knowledge, the first qualitative assessment of the diet of European eel larvae, even if several factors may have affected the relative representation of prey sequences in our PCR amplicons (King et al. 2008). For instance, prey organisms probably differ in their respective retention times in the gut and the condition of the prey when entering the gut is unknown. Further, findings of Dinophyceae sequences similar to symbiotic dinoflagellates of Copepoda and Polycystinea in the eel guts, where Copepoda and Polycystinea were also found, illustrate that some prey items may not be the primary prey of the eel larvae.
Our study documents for the first time, to our knowledge, that even the smallest European eel larvae feed on plankton organisms found in the Sargasso Sea, and highlights gelatinous zooplankton as particularly common prey. The finding of a variety of prey items in the larval guts shows that the frontal zones provide ample feeding opportunities. The results may therefore ultimately pave the way for future identification of linkages between regional plankton biology and recruitment variation of the European eel (Friedland et al. 2007). Finally, aquaculture production of eels currently relies exclusively on wild-caught glass eels. This new information on potentially important prey of eel larvae, notably gelatinous zooplankton, could facilitate efforts to identify suitable prey for rearing eel larvae, e.g.Tanaka et al. (2003), eventually leading to sustainable aquaculture production of anguillid eels.
Acknowledgements
The experiments using killed eel larvae were approved by the Linnaeus University and met ethics rules at our university and those of the involved co-authors of this research project.
We thank the captain and crew of HMS Vædderen for assistance with the sampling, C. Havel for drawings, and T. Bataillon for help with statistics. The study was funded by the Knud Højgaards Foundation, the Danish Natural Science Research Council, the Villum Kann Rasmussen Foundation, the Elisabeth and Knud Petersens Foundation, the Nordea Foundation and the Danish Expedition Foundation. This is paper no. P62 of the Danish Galathea 3 Expedition.
References
- Castonguay M., McCleave J. D.1987Vertical distributions, diel and ontogenetic vertical migrations and net avoidance of leptocephali of Anguilla and other common species in the Sargasso Sea. J. Plankton Res. 9, 195–214 (doi:10.1093/plankt/9.1.195) [Google Scholar]
- Deevey G. B.1971The annual cycle in quantity and composition of the zooplankton of the Sargasso Sea off Bermuda. I. The upper 500 m. Limnol. Oceanogr. 16, 219–240 (doi:10.4319/lo.1971.16.2.0219) [Google Scholar]
- Friedland K. D., Miller M. J., Knights B.2007Oceanic changes in the Sargasso Sea and declines in recruitment of the European eel. ICES J. Mar. Sci. 64, 519–530 (doi:10.1093/icesjms/fsm022) [Google Scholar]
- Jarman S. N., Deagle B. E., Gales N. J.2004Group-specific polymerase chain reaction for DNA-based analysis of species diversity and identity in dietary samples. Mol. Ecol. 13, 1313–1322 (doi:10.1111/j.1365-294X.2004.02109.x) [DOI] [PubMed] [Google Scholar]
- King R. A., Read D. S., Traugott M., Symondson W. O. C.2008Molecular analysis of predation: a review of best practise for DNA-based approaches. Mol. Ecol. 17, 947–963 (doi:10.1111/j.1365-294X.2007.03613.x) [DOI] [PubMed] [Google Scholar]
- Kleckner R. C., McCleave J. D.1988The northern limit of spawning by Atlantic eels (Anguilla spp.) in the Sargasso Sea in relation to thermal fronts and surface water masses. J. Mar. Res. 46, 647–667 (doi:10.1357/002224088785113469) [Google Scholar]
- Körner W. F.1952Untersuchungen über die Gehäusebilbung bei Appendicularien (Oikopleura dioica Fol). Z. Morphol. Oekol. Tiere 41, 1–53 (doi:10.1007/BF00407623) [Google Scholar]
- Last J. M.1978The food of four species of pleuronectiform larvae in the Eastern English Channel and Southern North Sea. Mar. Biol. 45, 359–368 (doi:10.1007/BF00391822) [Google Scholar]
- Michaels A. F., Caron D. A., Swanberg N. R., Howse F. A., Michaels C. M.1995Planktonic sarcodines (Acantharia, Radiolaria, Foraminifera) in surface waters near Bermuda: abundance, biomass and vertical flux. J. Plankton Res. 17, 131–163 (doi:10.1093/plankt/17.1.131) [Google Scholar]
- Miller M. J.2009Ecology of anguilliform leptocephali: remarkable transparent fish larvae of the ocean surface layer. Aqua BioSci. Monogr. 2, 1–94 [Google Scholar]
- Mochioka N., Iwamizu M.1996Diet of anguilloid larvae: leptocephali feed selectively on larvacean houses and fecal pellets. Mar. Biol. 125, 447–452 [Google Scholar]
- Moore H. B.1949The zooplankton of the upper waters of the Bermuda area of the N. Atlantic. Bull. Bingham Ocean. Coll. 12, 1–97 [Google Scholar]
- Otake T., Nogami K., Maruyama K.1993Dissolved and particulate organic matter as possible food sources for eel leptocephali. Mar. Ecol. Prog. Ser. 92, 27–34 (doi:10.3354/meps092027) [Google Scholar]
- Schmidt J.1922The breeding places of the eel. Phil. Trans. R. Soc. Lond. B 211, 179–208 (doi:10.1098/rstb.1923.0004) [Google Scholar]
- Suzuki N., Murakami K., Takeyama H., Chow S.2006Molecular attempt to identify prey organisms of lobster phyllosoma larvae. Fish. Sci. 72, 342–349 (doi:10.1111/j.1444-2906.2006.01155.x) [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S.2007MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4. Mol. Biol. Evol. 24, 1596–1599 (doi:10.1093/molbev/msm092) [DOI] [PubMed] [Google Scholar]
- Tanaka H., Kagawa H., Ohta H., Unuma T., Nomura K.2003The first production of glass eels in captivity: fish reproductive physiology facilitates great progress in aquaculture. Fish. Physiol. Biochem. 28, 493–497 (doi:10.1023/B:FISH.0000030638.56031.ed) [Google Scholar]
- Tesch F.2003The Eel. Oxford, UK: Blackwell [Google Scholar]
- Van Ginneken V. J. T., Maes G. E.2005The European eel (Anguilla anguilla, Linnaeus), its lifecycle, evolution and reproduction: a literature review. Rev. Fish. Biol. Fisheries 15, 367–398 [Google Scholar]