Abstract
Morpholino oligonucleotides, or morpholinos, have emerged as powerful antisense reagents for evaluating gene function in both in vitro and in vivo contexts. However, the constitutive activity of these reagents limits their utility for applications that require spatiotemporal control, such as tissue specific gene disruptions in embryos. In addition, current indirect methods for spatiotemporal regulation of morpholino activity in vivo may have off-target effects. Here we report a novel and efficient synthetic route for directly incorporating photocaged monomeric building blocks into morpholino oligomers, and demonstrate the utility of these caged morpholinos in the light-activated control of gene function in both cell culture and living embryos. We demonstrate that a caged morpholino targeting enhanced green fluorescent protein (EGFP) disrupts EGFP production only after exposure to UV light in both transfected cells and living zebrafish (Danio rerio) and Xenopus frog embryos. Finally, we show that a caged morpholino targeting chordin, a zebrafish gene that yields a distinct phenotype when functionally disrupted by conventional morpholinos, elicits a chordin phenotype in a UV-dependent manner. Our results suggest that directly photocaged morpholinos are readily synthesized and highly efficacious tools for light-activated spatio-temporal control of gene expression in multiple contexts.
Keywords: antisense agents, gene expression, light-activation, morpholino, photocaging
Introduction
Morpholino oligonucleotides were first described in 1997 as novel RNase H resistant antisense reagents applicable to cell culture studies1,2 and since have been used in vivo to block translation3, modify mRNA splicing4 and block microRNA function.5 Morpholinos also have become important for investigating gene function during embryonic development in species as diverse as zebrafish, medaka, carp, chicken, Xenopus, sea lamprey, sea urchin and Drosophila.3,4,6–9 However, these reagents must be injected directly into the cells of an early embryo and are constitutively active. Thus, morpholinos that produce early embryonic lethality eliminate the possibility of assessing gene function at later stages of development. Moreover, because the morpholino is distributed to the progeny of injected cells, spatial control of gene silencing is limited with standard antisense agents.
One strategy to eliminate the early lethality phenotypes and to achieve spatio-temporal control over gene silencing is with the application of photo-responsive morpholinos which are inactive until exposed to a short dose of non-damaging ultraviolet (UV) light.10,11 This strategy is especially relevant to studies employing transparent zebrafish embryos: the transparent nature of the zebrafish embryo permits small regions of the embryo and even single cells to be UV irradiated in vivo. A first generation of light-activated antisense agents for zebrafish embryos has been reported.12,13 Indeed, it was recently shown that a morpholino can be linked to a short complementary blocking morpholino via a photosensitive linker: once irradiated within a zebrafish embryo, the link between the morpholinos is broken and the small number of base pairs is insufficient to maintain a morpholino duplex, releasing the morpholino, which can then base pair with its target mRNA.10 Although effective, this strategy requires the synthesis of two morpholinos, the synthesis of the photo-sensitive linker, and the chemical connection of all three through covalent bond formation followed by purification.14 The length of the linker and the sequence and length of the inhibiting morpholino need to be carefully designed. Moreover, two morpholinos are released after irradiation which leads to a greater risk of off-target effects.4
An alternative strategy has been developed which utilizes a RNA-based inhibitor strand that is fully complementary to the morpholino and contains a photocleavable linkage: once irradiated within a zebrafish embryo, the link joining two short RNA oligomers is broken and, as with the preceding strategy, the small number of RNA base pairs is insufficient for maintaining a duplex with the morpholino.11 However, this approach requires a 5–10 fold molar excess of the inhibitor strand to saturate the morpholino, and since the injection of high amounts of nucleic acids into an embryo can be lethal, this strategy is not feasible for morpholinos that are only effective at high concentration. Furthermore, after UV irradiation this strategy releases two RNA oligomers into an embryo which may induce off-target effects.
Here, we present a different, substantially more direct approach to the generation of light-activatable morpholinos through the incorporation of a photocaged monomer during oligomer polymerization. The direct installation of light-removable (caging) groups on specific morpholino bases efficiently blocks their ability to hybridize to their target mRNA until the caging group is removed by irradiation with UV light of 365 nm. The effectiveness of this caging strategy is fully demonstrated by the UV-dependent activity of these reagents in cell culture and live embryos.
Experimental Section
General information
Chromatography was carried out on silica gel, Merck, 60Å. NMR spectra were obtained on a Varian Oxford 300 MHz spectrometer. Absorbance spectra were obtained on an HP UV/VIS Spectrometer 8453. HPLC and HRMS were obtained using Agilent Technologies 6210 LC-TOF ESI instrument.
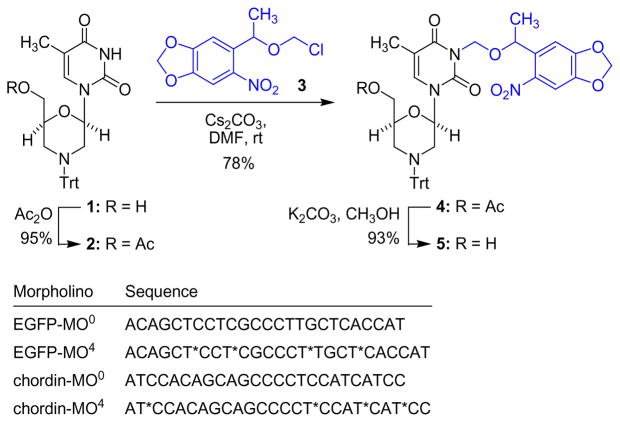
[6-(5-Methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-4-tritylmorpholin-2-yl]methyl acetate (2)
Ac2O (232 mg, 215 μL, 2.28 mmol) was added dropwise over 5 min to a solution of compound 1 (1 g, 2.07 mmol) and DMAP (spatula tip) in pyridine (10 mL) under a N2 atmosphere, at 0 °C. The reaction was allowed to stir for 12 h at r.t. The pyridine was evaporated under vacuum and the residue was purified by silica gel chromatography using hexanes:EtOAc (1:1), containing 1% TEA, affording 2 as a white foam in 95% yield (1.03g, 1.97 mmol). 1H NMR (300 MHz, CDCl3): δ = 1.38–1.48 (m, 2 H), 1.84 (s, 3 H), 2.04 (s, 3 H), 3.09–3.14 (m, 1 H), 3.35–3.39 (m, 1 H), 4.03–4.06 (m, 2 H), 4.38–4.41 (m, 1 H), 6.14–6.18 (m, 1 H), 6.99 (s, 1 H), 7.17–7.49 (m, 15 H). 13C NMR (75 MHz, CDCl3): δ = 12.7, 21.0, 38.4, 49.3, 52.1, 64.7, 74.9, 76.3, 80.6, 110.9, 126.8, 128.2, 129.4, 135.6, 150.2, 164.0, 171.0. HRMS: m/z calcd for C31H31N3O5 [M+H]+: 526.23367; found: 526.27330.
[6-(5-Methyl-3-{[1-(6-nitrobenzo[d][1,3]dioxol-5-yl)ethoxy]methyl}-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-4-tritylmorpholin-2-yl]methyl acetate (4)
NPOM-Cl 3 (609 mg, 2.35 mmol) in DMF (1 mL) was added dropwise over 5 min to a solution of compound 2 (1 g, 1.96 mmol) and Cs2CO3 (1.92 g, 5.88 mmol) in DMF (10 mL), under a N2 atmosphere, at 0 °C. The flask was wrapped in aluminum foil and the reaction was allowed to stir for 12 h at r.t. The reaction was taken up in EtOAc (100 mL) and washed with NaHCO3, water, and brine (100 mL each). The organic layer was dried over Na2SO4, filtered, and the volatiles were evaporated. The product was purified by silica gel chromatography using hexanes:EtOAc (2:1), containing 1% TEA, affording 4 as a yellow foam in 78% yield (1.14 g, 1.52 mmol). 1H NMR (300 MHz, CDCl3): δ = 1.38–1.48 (m, 2 H), 1.52 (d, J = 7.2 Hz, 3 H), 1.78 (s, 3 H), 2.06 (s, 3 H), 3.12 (d, J = 11.7 Hz,1 H), 3.35–3.39 (m, 1 H), 4.03–4.05 (m, 2 H), 4.38–4.41 (m, 1 H), 5.12–5.38 (m, 3 H), 5.95–6.13 (m, 3 H), 6.82–6.88 (m, 1 H), 7.15–7.49 (m, 17 H). 13C NMR (75 MHz, CDCl3): δ = 13.4, 21.2, 24.0, 49.5, 52.6, 64.8, 70.0, 73.2, 75.0, 77.1, 81.2, 103.1, 105.0, 105.3, 106.7, 110.1, 126.7, 128.1, 129.3, 134.2, 137.9, 142.3, 146.9, 150.3, 152.2, 163.0, 170.8. HRMS: m/z calcd for C41H40N4O10 [M+H]+: 749.28174; found: 749.28162. UV/VIS (CH2Cl2): λmax (ε) = 239 (20000), 271 (10000), 344 nm (4400).
1-[6-(Hydroxymethyl)-4-tritylmorpholin-2-yl]-5-methyl-3-{[1-(6-nitrobenzo[d][1,3]dioxol-5-yl)ethoxy]methyl}pyrimidine-2,4(1H,3H)-dione (5)
Compound 4 (1 g, 1.33 mmol) and K2CO3 (552 mg, 4.00 mmol) were dissolved in MeOH (10 mL) at room temperature. The reaction was stirred for 3 h, after which it was filtered and the solvent was evaporated under vacuum. The residue was purified by silica gel chromatography using hexanes:EtOAc (1:1) containing 1% TEA, affording 5 as a yellow foam in 93% yield (874 mg, 1.24 mmol). 1H NMR (300 MHz, CDCl3): δ = 1.38–1.45 (m, 2 H), 1.48–1.52 (m, 3 H), 1.76–1.78 (d, J = 4.5 Hz, 3 H), 1.91–1.95 (m, 0.5 H), 1.99–2.07 (m, 0.5 H), 3.08 (d, J = 11.7 Hz, 1 H), 3.28 (d, J = 12.0 Hz, 1 H), 3.51–3.62 (m, 2 H), 4.22–4.38 (m, 1 H), 5.15–5.39 (m, 3 H), 5.97–6.14 (m, 3 H), 6.85 (d, J = 9.6 Hz, 1 H), 7.15–7.49 (m, 17 H). 13C NMR (75 MHz, CDCl3): δ = 13.2, 23.9, 49.0, 51.9, 64.0, 70.1, 73.1, 73.6, 77.9, 81.1, 103.0, 105.0, 105.3, 106.0, 110.2, 126.7, 128.2, 129.4, 134.2, 138.0, 142.3, 146.9, 150.5, 152.1, 163.1. HRMS: m/z calcd for C39H38N4O9 [M+H]+: 707.27118; found: 707.27130. UV/VIS (CH2Cl2): λmax (ε) = 240 (20000), 267 (10000), 344 nm (3600).
Morpholinos
The caged compound 5 was activated and incorporated into EGFP and chordin morpholino oligomers (sequences indicated in Scheme 2) by Gene-Tools, LLC15–18 (Philomath, OR, USA). Morpholino stocks were resuspended with water, aliquoted and stored at −20 °C.
Scheme 2.
Synthesis of the Caged Morpholino Monomer 5, and Synthesized Morpholino Oligomer Sequences.a
a T* denotes the caged monomer 5. The NPOM caging group is shown in blue.
Morpholino Melting Temperature (Tm) Determination
The melting temperature (Tm) of each morpholino hybridized to its RNA complement target was determine as described previously19 with the following modifications. The morpholino and RNA complement (0.5 μM) were incubated in 0.15 M NaCl, 0.05 M NaH2PO4, pH 7.2. The samples were protected from light or irradiated at 365 nm with an UV transilluminator (3 mW/cm2) for 10 min, heated to 100 °C for 5 min, and then cooled to 25 °C at a rate of 2 °C/min using a Cary 100 Bio UV-vis spectrometer with temperature controller (Varian). The absorbance was recorded at 260 nm every 1 °C. The Tm was determined by the maximum of the first derivative of the absorbance vs. temperature plot. Standard deviations were calculated from three individual experiments.
Cell Culture
COS-7 cells were grown to 50–70% confluency at 37 °C and 5% CO2 in complete culture media (DMEM supplemented with 10% FBS, 50 i.u./ml penicillin and 50 μg/ml streptomycin) and split into aliquots containing 106 cells which were pelleted and resuspended in 100 μl of Cell Line Nucleofector® Solution “R” (Amaxa Inc., Gaithersburg, MD, USA) supplemented with 0.10 μg pEGFP-N1 (Clontech Laboratories, Inc, Mountain View, CA, USA), 0.42 μg pDsRed2-N1 (Clontech Laboratories, Inc.) and 150 μM morpholino (EGFP-MO0 or EGFP-MO4). Cell suspensions were then transfected with a Nucleofector® I using program “W-01”. Solutions were incubated at 25 °C for 10 min prior to the addition of 500 μl of complete culture media. A fraction of each cell suspension (100 μl) was then transferred into a single well of a 6-well plate containing 2.9 ml complete culture media. Plates with transfected cells were incubated at 37 °C, 5% CO2 for 1 hr prior to UV exposure. UV exposure was performed for 2 min using a DAPI fluorescence filter (340–380 nm excitation) on a Leica DM5000B compound microscope.20 Fluorescence from transfected cells was imaged 24 hrs after UV exposure with a Zeiss SteREO Lumar.V12 microscope. Fluorescence images were taken at identical exposure times for control (no morpholino), no UV, and UV-exposed cells in the same experiment.
Zebrafish
All experiments involving live zebrafish (Danio rerio) were performed in accordance with relevant institutional and national guidelines and regulations, and were approved by the North Carolina State University Institutional Animal Care and Use Committee. Zebrafish embryos were collected by natural mating, visualized with a Nikon SMZ800 microscope equipped with a 400 nm long-pass filter (GG400, Chroma Technology Corp., Rockingham, VT) and microinjected at the 1–4 cell stage with morpholinos and/or synthetic mRNAs using a pressure microinjector (PV830 picopump, World Precision Instruments, Sarasota, FL) at 60 psi and a mechanical micromanipulator (World Precision Instruments) with pulled glass capillary needles (World Precision Instruments). Embryos were photographed with a Leica DM5000B microscope equipped with a Retiga™ 1300 digital CCD camera and SimplePCI software.
For EGFP morpholino studies, zebrafish embryos were microinjected with in vitro transcribed capped synthetic EGFP mRNA (1.5 fmol, 0.40 ng), synthetic mCherry mRNA (0.7 fmol, 0.16 ng), and EGFP-MO4 (30 fmol, 0.54 ng). All injections were made into the cytoplasm during the one cell stage and some embryos were exposed to 365 nm UV light (2 min, 25 W hand-held UV lamp, 2.1 mW/cm2) immediately following microinjection.21 Embryos were maintained in egg water22 with 0.5 ppm methylene blue at 28 °C degrees for 24 hours before being photographed. A 1:20 molar ratio of EGFP mRNA to EGFP-MO4 allowed for the most efficient photoregulation of EGFP-MO4 activity. Fluorescence images were taken at identical exposure times for all embryos in the same experiment.
For chordin studies, approximately 1 nl of a 500 μM chordin-MO4, 0.15% phenol red solution was microinjected into the yolk of 1–4 cell stage zebrafish embryos. Embryos were either protected from light or exposed to 365nm UV light (25 W hand-held UV lamp, 2.1 mW/cm2) for 2 min at various time points after injection. Embryos were maintained in egg water22 with 0.5 ppm methylene blue at 28 °C degrees for 24–28 hours before being scored and photographed.
Xenopus
All experiments involving live Xenopus laevis were performed in accordance with relevant institutional and national guidelines and regulations, and were approved by the North Carolina State University Institutional Animal Care and Use Committee. Xenopus embryos were obtained by in vitro fertilization as previously described.23 Fertilized embryos were dejellied in 2% cysteine-HCl (pH 7.8–8.1) and cultured in 0.1X Marc’s Modified Ringers (MMR)23. Before injection, embryos were equilibrated in a solution of 3% Ficoll-400 in 0.75X MMR. In each experiment, all embryos were obtained from a single clutch (i.e., eggs were from the same adult female frog). A mixture of in vitro transcribed capped synthetic EGFP mRNA (1.6 fmol, 0.42 ng), synthetic mCherry mRNA (7.0 fmol, 1.6 ng), and EGFP-MO4 (46.8 fmol, 0.84 ng) was injected in a total volume of 0.6 nl in one animal pole blastomere at the 8–16-cell stage using a pressure microinjector (World Precision Instruments) at 60psi and a mechanical micromanipulator (World Precision Instruments) with pulled glass capillary needles (borosilicate with rod, World Precision Instruments). A 1:30 molar ratio of EGFP mRNA to EGFP-MO4 allowed for the most efficient photoregulation of EGFP-MO4 activity. Injected embryos were recovered in the dark in 1.5% Ficoll in 0.5X MMR for 20–30 min at r.t, before being gradually transferred to 0.1X MMR. Embryos were either protected from light or exposed to UV light for 2 min at 25 °C at the 32–64 cell stage, using a Zeiss Lumar fluorescent Stereomicroscope equipped with a DAPI filter and a 1.2X objective at 120X magnification, focusing on the animal pole. Control and UV irradiated embryos were then cultured at 16 °C until stage 24.24 Photographs were taken using a Zeiss Lumar fluorescent stereomicroscope with an AxioCam MRc camera and Axiovision image capture software. Fluorescence images were taken at identical exposure times for all embryos in the same experiment.
Results and Discussion
Development of NPOM caged morpholinos
We hypothesized that the incorporation of a photo-sensitive group directly on a morpholino base would block the ability of a morpholino to base pair with its target mRNA until the caging group is removed by UV irradiation (Scheme 1). By installing the caging group directly on the base of a phosphoramidite morpholino building block, it can be included in the direct synthesis of a morpholino and site-specifically incorporated into the oligomer, providing an efficient strategy for synthesizing caged morpholinos. This strategy is based upon earlier successes in using a novel 6-nitropiperonyloxymethyl caging group (NPOM) to disrupt Watson-Crick base pairing in DNA:DNA and DNA:RNA duplexes.25–28 This caging group is particularly effective at caging nitrogen heterocycles and has the advantage of disrupting hydrogen bonding between nucleotides while providing the stability needed for use in conventional DNA or morpholino syntheses.26 While stable under physiological conditions, oligonucleotides harboring these caging groups are returned to near full activity following a brief irradiation with 365 nm UV light.25–28 We adopted this caging approach for the synthesis of the caged morpholino monomer 5 (Scheme 2). Compound 1 (gift from GeneTools, LLC) was acetylated in 95% yield to deliver 2. The NPOM caging group was then installed on 2 using the chloromethyl ether 3 (synthesized in 3 steps29) and Cs2CO3 in DMF at room temperature, providing 4 in 78% yield. The free 5′ hydroxyl group in 5 was obtained through saponification of the acetyl ester with K2CO3 in methanol (93% yield). Using standard polymerization chemistry,16,30 the caged morpholino monomer 5 was then incorporated into morpholino sequences shown to block the translation of EGFP31 and Chordin3 (Scheme 2). The morpholino sequence selected to target EGFP expression (EGFP-MO0) has been shown to effectively knock down EGFP expression in both cell culture and in vivo.31 The morpholino sequence selected to target the expression of endogenous zebrafish chordin (chordin-MO0) routinely induces a specific morphological phenotype when injected into zebrafish embryos.3,32,33 Four caged monomers were included in each morpholino since a common control for morpholino experiments is to include a non-active morpholino with four mismatched bases1,3,34 and we have shown that three to four NPOM caging groups can functionally disrupt DNA:DNA and phosphorothioate DNA:RNA duplexes.27,28 A morpholino nomenclature was adopted in which a superscript numeral represents the number of caging groups within a morpholino (e.g. EGFP-MO4 includes four caging groups and EGFP-MO0 includes zero caging groups).
Scheme 1.
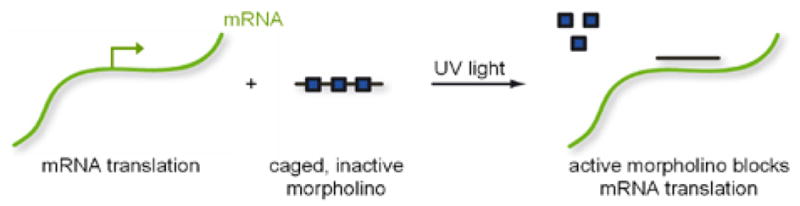
Photochemical Gene Regulation with Caged Morpholinos.a
a Squares represent the 6-nitropiperonyloxymethyl (NPOM) caging group.
NPOM caging groups disrupt morpholino:RNA duplexes
In order to evaluate the effectiveness of NPOM caging groups to disrupt morpholinos base pairing, we determined the Tm for morpholino:RNA duplexes before and after 365 nm UV exposure. We evaluated EGFP-MO0, chordin-MO0, EGFP-MO4 and chordin-MO4. As indicated in Table 1 (and Supporting Figure S1), the presence of four NPOM caging groups on EGFP-MO4 resulted in a ~30 °C decrease in its Tm as compared to the non-caged EGFP-MO0. This observation is consistent with a 27 °C decrease in Tm when four mismatches are included in a 25mer morpholino:RNA duplex.1 Exposure of EGFP-MO4 to UV resulted in increasing its Tm to be virtually the same as EGFP-MO0. Similar experiments demonstrate that the presence of four caging groups on chordin-MO4 decreases the Tm of its morpholino:RNA duplex by ~10 °C and that exposure of chordin-MO4 to UV results in increasing its Tm to be nearly identical to that of chordin-MO0. This smaller change in Tm is likely because the caging groups are clustered near the ends of the chordin-MO4 leaving more contiguous non-caged internal bases that might weakly base pair with its target RNA. However, the number of contiguous hybridizing bases in chordin-MO4 is fewer than the reported number required for a morpholino to block gene function.35 In addition, gel shift experiments clearly demonstrate that the NPOM caging groups on both EGFP-MO4 and chordin-MO4 effectively disrupt the stable formation of morpholino:RNA duplexes (Supporting Figures S2 and S3); morpholino:RNA duplexes are restored with as little as a two minute exposure to UV (Supporting Figure S4). It must be noted that changes in UV source, morpholino concentration, and biological context may alter the efficiency of decaging.
Table 1.
Morpholino melting temperatures (Tm).a
| Morpholino | Irradiation | Tm (°C) |
|---|---|---|
| EGFP-MO0 | −UV | 82.4 ± 1.0 |
| EGFP-MO4 | −UV | 53.8 ± 1.1 |
| EGFP-MO4 | +UV | 84.5 ± 1.0 |
| chordin-MO0 | −UV | 80.1 ± 0.6 |
| chordin-MO4 | −UV | 70.8 ± 0.5 |
| chordin-MO4 | +UV | 79.9 ± 1.1 |
Experimentally determined melting temperatures (Tm) of duplexes of non-caged and caged morpholinos with their targeted RNA sequences. The distribution of the four caging groups has a substantial influence on the melting temperature of the morpholino:RNA duplexes, as discovered earlier for DNA:DNA duplexes.27 When the caging groups are evenly distributed along the length of the morpholino (e.g. EGFP-MO4), a greater change in Tm is observed. Standard deviations were determined from three independent experiments.
Spatial control of EGFP-MO4 in transfected cells
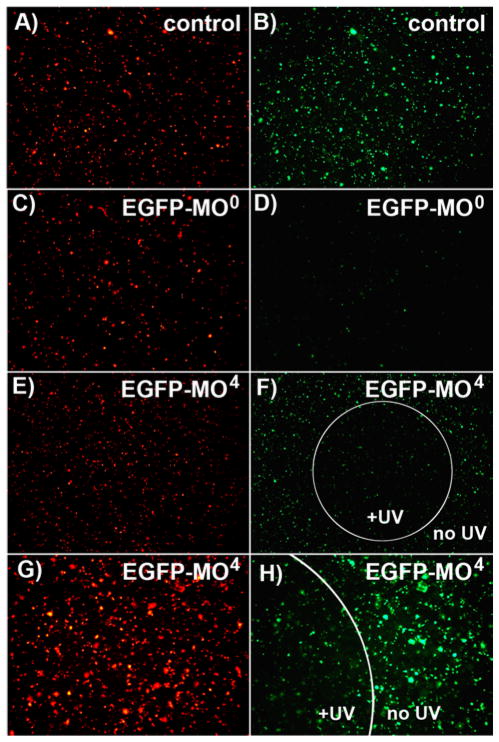
EGFP is an easily visualized protein, and thus a highly useful target for evaluating antisense agents.3,11,31 To assess the photosensitivity of EGFP-MO4 in cells, COS-7 cells (a monkey kidney cell line) were co-transfected with a plasmid encoding EGFP, a plasmid encoding a control red fluorescent reporter (DsRed2) and either EGFP-MO0 or EGFP-MO4. Then, a small region of the cell culture plate was briefly irradiated with UV light. The EGFP-MO0 effectively blocks EGFP translation whereas, EGFP-MO4 only blocks EGFP translation in the region which had been exposed to UV light (Figure 1). This loss of EGFP is not the result of UV quenching since the cells were exposed to UV light immediately after transfection, prior to EGFP production. Neither EGFP-MO0 nor uncaged EGFP-MO4 resulted in a complete knock-down of EGFP fluorescence in transfected cells; this is likely the result of the high levels of EGFP mRNA generated from the expression plasmid and/or relatively inefficient transfection of morpholinos. Nevertheless, this experiment demonstrates that, in agreement with previous observations regarding disruption of oligonucleotide duplex formation25–28 and our Tm and gel-shift experiments, the installation of four NPOM caging groups on a morpholino oligomer effectively inhibits antisense activity in cultured mammalian cells, and that this activity can be readily restored using UV irradiation, even in a spatially controlled fashion.
Figure 1.
Spatial control of EGFP-MO4 activity in mammalian cell culture. In a 96-well plate, COS-7 cells were co-transfected with plasmids encoding EGFP (pEGFP-N1) and DsRed2 (pDsRed2-N1) (A–B) with no morpholino, or (C–D) with EGFP-MO0, or (E–F) with EGFP-MO4. Cells transfected with EGFP-MO4 were briefly irradiated in a small circle at the center of the well. Expression levels of EGFP and DsRed were evaluated 24 hr after transfection and irradiation and representative images are shown on the right and left respectively. (G–H) A magnified view of (E–F) is shown.
EGFP-MO4 regulation of EGFP expression in zebrafish embryos is UV dependent
We then assessed whether the caged EGFP-MO4 is effective in regulating gene expression in vivo. We utilized the zebrafish embryo, a developmental model organism with transparent embryos in which morpholinos are routinely employed.3,34 Embryos were co-injected at the 1–4 cell stage with in vitro transcribed, synthetic mRNA encoding EGFP, EGFP-MO4 and synthetic mRNA encoding the red fluorescent protein, mCherry (as a control). Figure 2 demonstrates that in the absence of UV irradiation, injected embryos express both EGFP and mCherry, and that UV exposure specifically disrupts EGFP expression. Importantly, zebrafish embryos exposed to UV light display a normal phenotype (also see Figure 4F).36 As expected, embryos co-injected with EGFP mRNA and EGFP-MO0 display little or no EGFP expression regardless of UV exposure (data not shown). In addition, this loss of EGFP expression is not the result of UV quenching as the embryos are exposed to UV light immediately after injection, well prior to EGFP protein production. These results demonstrate that NPOM-caged morpholinos can be used to achieve effective light-controlled regulation of gene expression in live embryos.
Figure 2.
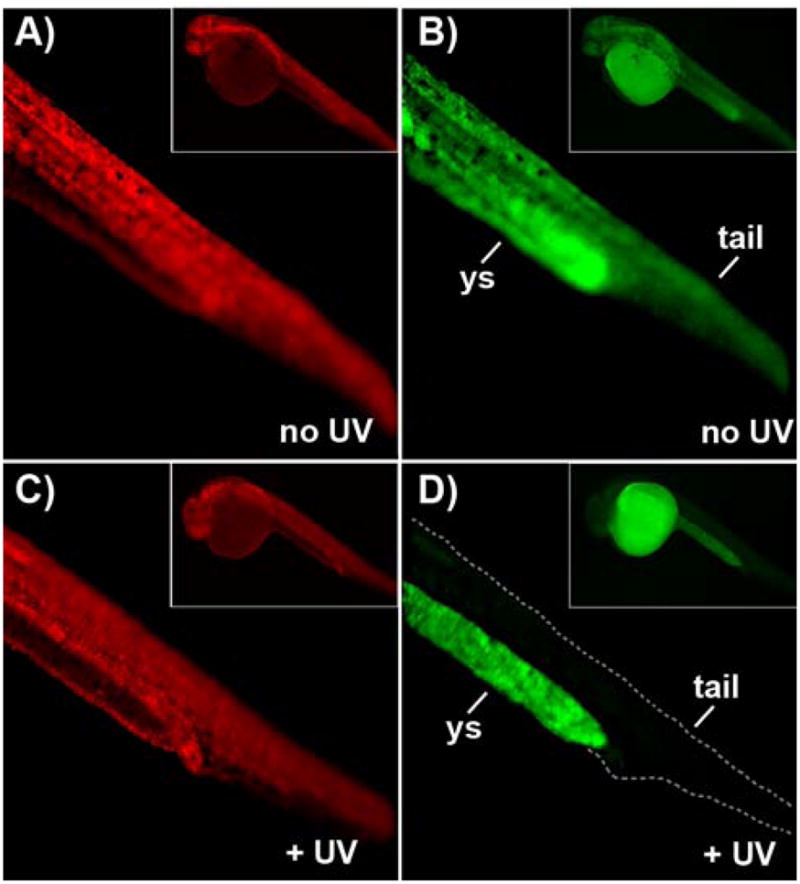
Light-activation of EGFP-MO4 in zebrafish embryos. Zebrafish embryos at the 1–4 cell stage were microinjected with synthetic mRNA encoding EGFP, EGFP-MO4 (1:20 molar ratio) and synthetic mRNA encoding mCherry and were (A–B) protected from light or (C–D) immediately exposed to UV light. Expression levels of EGFP and mCherry were evaluated 24 hr after injection and irradiation and representative images of zebrafish tails are shown on the right and left, respectively. The fluorescence observed in (D) is not derived from EGFP, but is autofluoresence of the yolk sac (ys) as previously observed.37 Photos of entire embryos are shown in insets.
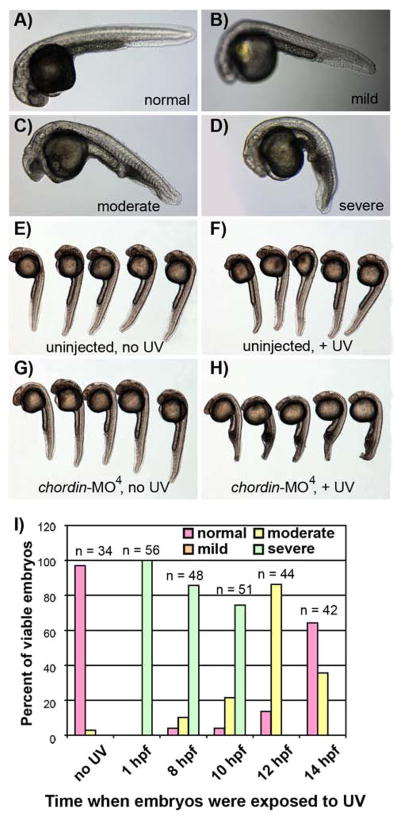
Figure 4.
Light activation of chordin-MO4 in zebrafish embryos. (A–H) Zebrafish embryos were microinjected with chordin-MO4 during the 1–4 cell stage and either protected from light or immediately irradiated with UV light and their phenotype assessed 24–28 hr post fertilization (hpf). Phenotypes include (A) normal, (B) mild, (C) moderate, and (D) severe. (E–F) Uninjected control embryos display a normal phenotype regardless of UV exposure. (G) Embryos injected with chordin-MO4 and not exposed to UV light of 365 nm display normal or mild phenotypes. (H) Embryos injected with chordin-MO4 and immediately exposed to UV light display moderate and severe phenotypes. (I) Embryos were injected with chordin-MO4 as described above and divided into groups that were irradiated at various time points after fertilization and their phenotype assessed at 24–28 hpf. Note that light-activation of chordin-MO4 during or after the developmental stage of gastrulation (~10 hpf) fails to generate the severe chordin phenotype.
EGFP-MO4 regulation of EGFP expression in Xenopus embryos is UV dependent
To test the broader applicability of this approach for species beyond zebrafish, the caged EGFP morpholino was also investigated in Xenopus frog embryos. Although not as optically transparent, caged compounds have successfully been used in Xenopus.37,38 One advantage of Xenopus is the ability to target specific tissues by injecting individual cells of the early embryo to limit the distribution of injected reagents.39 Xenopus embryos were co-injected in a single cell at the 8–16 cell stage with synthetic EGFP mRNA, EGFP-MO4, and synthetic mCherry mRNA. All injected embryos were phenotypically normal and Figure 3 clearly demonstrates that, although mCherry expression was unaffected, EGFP-MO4 blocks EGFP production only after UV exposure. As in zebrafish, this loss of EGFP expression cannot be the result of UV quenching as the embryos are exposed to UV light immediately after injection which is prior to EGFP production. In conclusion, the direct caging approach can be used for the light-regulation of morpholino activity in cell culture, zebrafish embryos, and Xenopus embryos.
Figure 3.
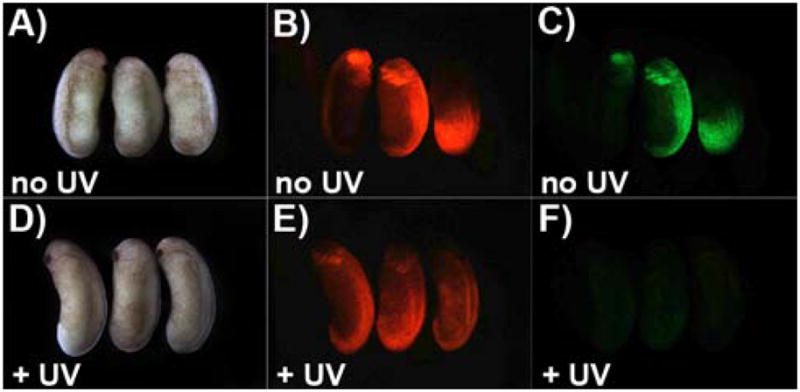
Light activation of EGFP-MO4 in Xenopus embryos. Xenopus embryos were microinjected with synthetic mRNA encoding EGFP, EGFP-MO4 (1:30 molar ratio) and synthetic mRNA encoding mCherry and (A–C) protected from light or (D–F) immediately exposed to UV light. Expression levels of EGFP (right), and mCherry (middle) were evaluated at embryonic stage 24 (~ 1 day after injection) and are shown in comparison to bright field images (left).
chordin-MO4 regulation of the chordin gene in zebrafish embryos is UV dependent
The above results show that our NPOM-caged EGFP morpholino enables light-regulated control of EGFP expression in cell culture and in vivo. In order to determine if NPOM-caged morpholinos can effectively regulate endogenous genes, we chose to target the zebrafish chordin gene. We selected a well characterized morpholino sequence (chordin-MO0) which, when injected into 1–4 cell zebrafish embyros, induces a shrunken head and a ventralized tail by 24–28 hours post fertilization (hpf), characteristic of the chordin phenotype.3,32,33 We routinely observe >90% of chordin-MO0 injected embryos displaying these phenotypes (data not shown). To evaluate the light-dependent efficacy of a NPOM-caged chordin morpholino (chordin-MO4), it was injected into 1–4 cell stage zebrafish embryos which were then either protected from light or immediately subjected to a brief exposure to UV. Embryos were allowed to develop until 24–28 hpf at which time their phenotype was assessed and the severity of the chordin phenotype was scored as normal, mild, moderate, or severe (Figure 4A–D). The moderate and severe phenotypes are deformities known to be induced by disruption of the chordin gene,3,32,33 and the mild phenotype is characterized by a slight blunting in the animal’s tail. We suggest that this latter phenotype is caused by a small amount of partial decaging of chordin-MO4 through handling under ambient light or by the stretch of contiguous non-caged bases within chordin-MO4 weakly associating with its target mRNA. Note that the mild phenotype is not caused merely by the presence of a NPOM-caged morpholino as this phenotype is not observed when embryos are injected with EGFP-MO4 (data not shown). One hundred percent of control embryos that are not injected with a morpholino displayed a normal phenotype at 24–28 hpf independent of UV exposure, confirming that irradiation does not induce any observable developmental defect (Figure 4E–F; no UV, n = 26; +UV, n = 32). The observation that 99% of chordin-MO4 injected embryos not exposed to UV displayed normal or mild phenotypes at 24–28 hpf, confirms that the NPOM-caged morpholinos are stable in the zebrafish embryo (Figure 4G; n = 74 embryos). In contrast, 90% of chordin-MO4 injected embryos that are immediately exposed to UV light displayed moderate and severe phenotypes (Figure 4H; n = 88 embryos), suggesting an efficient removal of the NPOM caging groups in vivo after UV exposure, activation of the morpholino antisense agent, and subsequent knock-down of chordin function.
In order to determine if NPOM-caged morpholinos can be employed for temporal control of gene expression, zebrafish embryos were injected with chordin-MO4 and irradiated at successively later stages of development (Figure 4I). Interestingly, exposure of chordin-MO4 injected embryos to UV at any time point prior to the developmental stage of gastrulation (~10 hpf) reliably induced a severe chordin mutant phenotype, while disruption of chordin during mid to late stage gastrulation elicited milder anomalies. These results are highly consistent with the known expression patterns and the timing of the dorsalizing function of chordin in the zebrafish embryo.3 These results confirm that the NPOM-caged morpholino technology is applicable to the light-regulation of different genes and allows for the temporal investigation of gene function in live zebrafish embryos.
Conclusion
We have developed a caged morpholino monomer and incorporated it in the synthesis of two different morpholino antisense reagents. These caged antisense agents were inactive in cell culture, zebrafish embryos, and Xenopus embryos until irradiated with UV light of 365 nm. Using these reagents, we demonstrated photochemical gene silencing in mammalian cells and in live aquatic animals and observed the expected embryonic phenotypes. No detectable toxic effects of the irradiation or the caging groups were observed. The developed direct caging approach can be easily applied to any morpholino oligomer, since the straightforward design only requires insertion of caged building blocks in the oligomer synthesis. We are currently developing the chemistry to synthesize caged morpholino monomers for the other three bases which will provide greater flexibility in the design of caged morpholino oligonucleotides. It is likely that this methodology will find broad application in the cell and developmental biology research community, since it is generally applicable to the spatial and temporal regulation of gene function in multiple biological contexts.
Supplementary Material
Supporting Figure S1. Melt curves of duplexes of non-caged and caged morpholinos and their complementary RNA. (A) chordin-MO0 (B) chordin-MO4, no UV (C) chordin-MO4, 10 min UV (D) EGFP-MO0 (E) EGFP-MO4, no UV (F) EGFP-MO4, 10 min UV. Error bars represent standard deviations from three independent measurements.
Supporting Figure S2. Representative gel shift assay of (A) EGFP-MO0, (B) EGFP-MO4, (C) chordin-MO0 and (D) chordin-MO4 with corresponding target RNA. RNA (10 μM) complementary to each morpholino was 5′ end labeled with γ-32P ATP. 100 nM of radiolabeled RNA was incubated with increasing concentrations of morpholino (0, 5, 12.5, 25, 50, and 100 nM) at 80 °C for 5 min and cooled to 37 °C for 1 hour. RNA monomers were separated from morpholino:RNA duplexes by a 20% native polyacrylamide gel electrophoresis and imaged with a Storm Phosphorimager. The non-caged morpholino fully binds the RNA substrate at equimolar concentration, while the caged morpholino leaves almost all RNA free. Gel shifts were performed in triplicate.
Supporting Figure S3. Quantification of morpholino gel shift assays. Gel images (Supporting Figure S2) were quantified with ImageQuant, and the bound morpholino:RNA fraction was plotted against the concentration of morpholino. Gel shifts were performed in triplicate and error bars represent the standard deviation of three individual experiments.
Supporting Figure S4. Irradiation time course gel shift assay of EGFP morpholino:RNA duplex. Gel shifts were performed using 100 nM radiolabeled RNA and 100 nM morpholino as in Supporting Figure S2. Complete RNA binding can be observed after a 2 min UV irradiation (25 W hand-held UV lamp, 2.1 mW/cm2). Lane (1) EGFP RNA; (2) EGFP-MO0:EGFP RNA; (3) EGFP-MO4:EGFP RNA, no UV; (4) EGFP-MO4:EGFP RNA, 2 min UV; (5) EGFP-MO4:EGFP RNA, 5 min UV; (6) EGFP-MO4:EGFP RNA, 10 min UV.
Acknowledgments
We thank Gene-Tools, LLC for synthesizing caged morpholino oligomers from our caged building block and R. Read Tull for assistance with Xenopus microinjections. Financial support from the National Institutes of Health (R01GM079114 and R01DK085300) is acknowledged. AD is the recipient of a Beckman Young Investigator Award and a Cottrell Scholar Award.
Footnotes
Supporting Information Available: Melting curves for caged morpholinos and morpholino gel shift assay results: this material is available free of charge via the Internet at http://pubs.acs.org.
Reference List
- 1.Summerton J, Stein D, Huang SB, Matthews P, Weller D, Partridge M. Antisense Nucleic Acid Drug Dev. 1997;7:63–70. doi: 10.1089/oli.1.1997.7.63. [DOI] [PubMed] [Google Scholar]
- 2.Summerton J. Biochim Biophys Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- 3.Nasevicius A, Ekker SC. Nature Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 4.Eisen JS, Smith JC. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- 5.Pase L, Layton JE, Kloosterman WP, Carradice D, Waterhouse PM, Lieschke GJ. Blood. 2008 doi: 10.1182/blood-2008-05-155812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osorio J, Retaux S. Dev Genes Evol. 2008;218:221–235. doi: 10.1007/s00427-008-0208-1. [DOI] [PubMed] [Google Scholar]
- 7.Xia JH, Liu JX, Zhou L, Li Z, Gui JF. Int J Dev Biol. 2008;52:1089–1098. doi: 10.1387/ijdb.072519jx. [DOI] [PubMed] [Google Scholar]
- 8.Sauka-Spengler T, Barembaum M. Methods Cell Biol. 2008;87:237–256. doi: 10.1016/S0091-679X(08)00212-4. [DOI] [PubMed] [Google Scholar]
- 9.Ekker SC, Larson JD. Genesis. 2001;30:89–93. doi: 10.1002/gene.1038. [DOI] [PubMed] [Google Scholar]
- 10.Shestopalov IA, Sinha S, Chen JK. Nat Chem Biol. 2007 doi: 10.1038/nchembio.2007.30. [DOI] [PubMed] [Google Scholar]
- 11.Tomasini AJ, Schuler AD, Zebala JA, Mayer AN. Genesis. 2009;47:736–743. doi: 10.1002/dvg.20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blidner RA, Svoboda KR, Hammer RP, Monroe WT. Mol Biosyst. 2008;4:431–440. doi: 10.1039/b801532e. [DOI] [PubMed] [Google Scholar]
- 13.Tang X, Maegawa S, Weinberg ES, Dmochowski IJ. J Am Chem Soc. 2007;129:11000–11001. doi: 10.1021/ja073723s. [DOI] [PubMed] [Google Scholar]
- 14.Ouyang X, Shestopalov IA, Sinha S, Zheng G, Pitt CL, Li WH, Olson AJ, Chen JK. J Am Chem Soc. 2009;131:13255–13269. doi: 10.1021/ja809933h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Summerton JE, Weller DD. Uncharged morpholino-based polymers having achiral intersubunit linkages. 5,034,506. US Patent. 1991 Jul 23;
- 16.Summerton JE, Weller DD. Uncharged morpholino-based polymers having phosphorous containing chiral intersubunit linkages. 5,185,444. US Patent. 1993 Feb 9;
- 17.Summerton JE, Weller DD, Stirchak EP. Alpha-morpholino ribonucleoside derivatives and polymers thereof. 5,378,841. US Patent. 1995 Jan 3;
- 18.Summerton JE, Weller DD. Morpholino-subunit combinatorial library and method. 5,698,685. US Patent. 1997 Dec 16;
- 19.Stein D, Foster E, Huang SB, Weller D, Summerton J. Antisense Nucleic Acid Drug Dev. 1997;7:151–157. doi: 10.1089/oli.1.1997.7.151. [DOI] [PubMed] [Google Scholar]
- 20.UV exposure of one minute was not as effective at reducing EGFP expression in transfected cells.
- 21.In trial experiments with chordin-MO4 injected zebrafish embryos, a two minute UV exposure was found to be the minimum required to elicit a severe chordin phenotype in 80–100% of the embryos.
- 22.Westerfield M. Guide for the Laboratory Use of Zebrafish (Danio rerio) University of Oregon Press; Eugene: 2000. The Zebrafish Book. [Google Scholar]
- 23.Sive HL, Grainger RM, Harland RM. Early Development of Xenopus Laevis. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1998. [Google Scholar]
- 24.Nieuwkoop PD, Faber J. External and internal stage criteria in the development of Xenopus laevis. In: Nieuwkoop PD, Faber J, editors. Normal Table of Xenopus laevis (Daudin) North-Holland Publishing Co; Amsterdam: 1956. pp. 162–168. [Google Scholar]
- 25.Lusic H, Lively MO, Deiters A. Mol Biosyst. 2008;4:508–511. doi: 10.1039/b800166a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lusic H, Young DD, Lively MO, Deiters A. Org Lett. 2007;9:1903–1906. doi: 10.1021/ol070455u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young DD, Edwards WF, Lusic H, Lively MO, Deiters A. Chem Commun(Camb) 2008:462–464. doi: 10.1039/b715152g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young DY, Lusic H, Lively MO, Yoder JA, Deiters A. ChemBioChem. 2008;9:2937–2940. doi: 10.1002/cbic.200800627. [DOI] [PubMed] [Google Scholar]
- 29.Lusic H, Deiters A. Synthesis. 2006;8:2147–2150. [Google Scholar]
- 30.Fox CM, Reeves MD, Weller DD. Method of synthesis of morpholino oligomers. WO/2009/064471. International patent application. 2009 May 22;
- 31.Schnapp E, Tanaka EM. Dev Dyn. 2005;232:162–170. doi: 10.1002/dvdy.20203. [DOI] [PubMed] [Google Scholar]
- 32.Fisher S, Amacher SL, Halpern ME. Development. 1997;124:1301–1311. doi: 10.1242/dev.124.7.1301. [DOI] [PubMed] [Google Scholar]
- 33.Hammerschmidt M, Pelegri F, Mullins MC, Kane DA, van Eeden FJ, Granato M, Brand M, Furutani-Seiki M, Haffter P, Heisenberg CP, Jiang YJ, Kelsh RN, Odenthal J, Warga RM, Nusslein-Volhard C. Development. 1996;123:95–102. doi: 10.1242/dev.123.1.95. [DOI] [PubMed] [Google Scholar]
- 34.Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC. Zebrafish. 2009;6:69–77. doi: 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Summerton JE. Curr Top Med Chem. 2007;7:651–660. doi: 10.2174/156802607780487740. [DOI] [PubMed] [Google Scholar]
- 36.Dong Q, Svoboda K, Tiersch TR, Monroe WT. J Photochem Photobiol B. 2007;88:137–146. doi: 10.1016/j.jphotobiol.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cambridge SB, Geissler D, Calegari F, Anastassiadis K, Hasan MT, Stewart AF, Huttner WB, Hagen V, Bonhoeffer T. Nat Methods. 2009;6:527–531. doi: 10.1038/nmeth.1340. [DOI] [PubMed] [Google Scholar]
- 38.Minden J, Namba R, Mergliano J, Cambridge S. Sci STKE. 2000;2000:11. doi: 10.1126/stke.2000.62.pl1. [DOI] [PubMed] [Google Scholar]
- 39.Moody SA, Kline MJ. Anat Embryol(Berl) 1990;182:347–362. doi: 10.1007/BF02433495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figure S1. Melt curves of duplexes of non-caged and caged morpholinos and their complementary RNA. (A) chordin-MO0 (B) chordin-MO4, no UV (C) chordin-MO4, 10 min UV (D) EGFP-MO0 (E) EGFP-MO4, no UV (F) EGFP-MO4, 10 min UV. Error bars represent standard deviations from three independent measurements.
Supporting Figure S2. Representative gel shift assay of (A) EGFP-MO0, (B) EGFP-MO4, (C) chordin-MO0 and (D) chordin-MO4 with corresponding target RNA. RNA (10 μM) complementary to each morpholino was 5′ end labeled with γ-32P ATP. 100 nM of radiolabeled RNA was incubated with increasing concentrations of morpholino (0, 5, 12.5, 25, 50, and 100 nM) at 80 °C for 5 min and cooled to 37 °C for 1 hour. RNA monomers were separated from morpholino:RNA duplexes by a 20% native polyacrylamide gel electrophoresis and imaged with a Storm Phosphorimager. The non-caged morpholino fully binds the RNA substrate at equimolar concentration, while the caged morpholino leaves almost all RNA free. Gel shifts were performed in triplicate.
Supporting Figure S3. Quantification of morpholino gel shift assays. Gel images (Supporting Figure S2) were quantified with ImageQuant, and the bound morpholino:RNA fraction was plotted against the concentration of morpholino. Gel shifts were performed in triplicate and error bars represent the standard deviation of three individual experiments.
Supporting Figure S4. Irradiation time course gel shift assay of EGFP morpholino:RNA duplex. Gel shifts were performed using 100 nM radiolabeled RNA and 100 nM morpholino as in Supporting Figure S2. Complete RNA binding can be observed after a 2 min UV irradiation (25 W hand-held UV lamp, 2.1 mW/cm2). Lane (1) EGFP RNA; (2) EGFP-MO0:EGFP RNA; (3) EGFP-MO4:EGFP RNA, no UV; (4) EGFP-MO4:EGFP RNA, 2 min UV; (5) EGFP-MO4:EGFP RNA, 5 min UV; (6) EGFP-MO4:EGFP RNA, 10 min UV.