Abstract
DNA damage can induce a tumor suppressive response termed cellular senescence. Damaged senescent cells permanently arrest growth, secrete inflammatory cytokines and other proteins and harbor persistent nuclear foci that contain DNA damage response (DDR) proteins. To understand how persistent damage foci differ from transient foci that mark repairable DNA lesions, we identify sequential events that differentiate transient foci from persistent foci, which we term ‘DNA segments with chromatin alterations reinforcing senescence’ (DNA-SCARS). Unlike transient foci, DNA-SCARS associate with PML nuclear bodies, lack the DNA repair proteins RPA and RAD51, lack single-stranded DNA and DNA synthesis and accumulate activated forms of the DDR mediators CHK2 and p53. DNA-SCARS form independently of p53, pRB and several other checkpoint and repair proteins but require p53 and pRb to trigger the senescence growth arrest. Importantly, depletion of the DNA-SCARS-stabilizing component histone H2AX did not deplete 53BP1 from DNA-SCARS but diminished the presence of MDC1 and activated CHK2. Furthermore, depletion of H2AX reduced both the p53-dependent senescence growth arrest and p53-independent cytokine secretion. DNA-SCARS were also observed following severe damage to multiple human cell types and mouse tissues, suggesting that they can be used in combination with other markers to identify senescent cells. Thus, DNA-SCARS are dynamically formed distinct structures that functionally regulate multiple aspects of the senescent phenotype.
Keywords: Aging, Cancer, Cellular senescence, DNA repair, Homologous recombination, Interleukin 6 (IL6), Promyelocytic leukemia protein (PML)
Introduction
Cellular senescence limits the proliferation (growth) of damaged cells that are at risk for neoplastic transformation by imposing an essentially irreversible growth arrest. Cells senesce in response to many potentially oncogenic stressors, including dysfunctional telomeres, DNA damage, chromatin alterations and strong mitogenic signals such as those delivered by some oncogenes (Ben-Porath and Weinberg, 2004; Campisi and d'Adda di Fagagna, 2007). The senescence response depends crucially on the cellular tumor antigen p53 (also known as tumor suppressor TP53) and the retinoblastoma-associated protein (pRb) tumor suppressor pathways and is now accepted as a potent cell-autonomous mechanism for suppressing the development of cancer (Braig and Schmitt, 2006; Campisi, 2005; Dimri, 2005; Prieur and Peeper, 2008). Accordingly, loss of the senescence response increases the incidence of cancer in humans and mice.
Unlike apoptotic cells, which rapidly disintegrate, senescent cells remain viable in culture for long intervals and are found with increasing frequency in aged tissues and at sites of age-related pathology, including preneoplastic lesions (Collado et al., 2005; Dimri et al., 1995; Erusalimsky and Kurz, 2005; Jeyapalan et al., 2007; Price et al., 2002). In addition, they develop a senescence-associated secretory phenotype (SASP) with potent autocrine and paracrine activities. The SASP includes numerous cytokines, growth factors and proteases, and develops several days after cells receive a senescence stimulus and cease growth (Coppe et al., 2010; Coppe et al., 2008; Rodier et al., 2009). Some SASP components reinforce the growth arrest (Acosta et al., 2008; Kuilman et al., 2008; Wajapeyee et al., 2008). Others disrupt epithelial differentiation (Parrinello et al., 2005) or promote cancer cell growth and invasion in culture and in vivo (Bavik et al., 2006; Coppe et al., 2008; Krtolica et al., 2001; Liu and Hornsby, 2007). Because senescent cells can strongly influence nearby cells, it is important to understand how the SASP develops.
Several signaling cascades are associated with the establishment and maintenance of senescence-associated phenotypes, including growth arrest and SASP (Campisi and d'Adda di Fagagna, 2007; Kuilman and Peeper, 2009). Many senescence-inducing stimuli generate a persistent DNA damage response (DDR), normally associated with DNA double-strand breaks (DSBs) (d'Adda di Fagagna, 2008). Recent findings show that DDR signaling is essential for establishing and maintaining senescent phenotypes. Thus, loss of DDR checkpoint kinases such as ATM or the serine/threonine-protein kinase CHK2, which phosphorylate and activate p53, not only prevents the p53-dependent senescence growth arrest (Bartkova et al., 2006; Beausejour et al., 2003; Di Micco et al., 2006; Gire et al., 2004; Herbig et al., 2004) but also prevents the p53-independent inflammatory cytokine secretion that comprises the SASP (Rodier et al., 2009).
DDR signaling is initiated at DSBs by sensor proteins such as the phosphoinositide 3-kinase-like kinases (PIKKs) ATM and ATR, and amplified by the MRN (MRE11–RAD50–NBS1) complex. These proteins help recruit and further activate PIKKs, and participate in DNA repair. PIKKs promote local chromatin remodeling, which spreads for megabases surrounding the DSB and facilitates repair. PIKKs also transduce the DDR signal to downstream mediators, such as CHK2 and p53, which integrate the signal with cellular physiology and coordinate DNA repair with cell cycle checkpoints (Bartek and Lukas, 2007; Berkovich et al., 2007; Rodier et al., 2007). Many of these DDR signaling and repair proteins assemble rapidly (within minutes) around DSBs and can be detected in the nuclei of fixed or living cells as focal aggregates termed DNA damage foci. Two components are typically used to detect these foci by fluorescence microscopy: the PIKK-phosphorylated form of the histone variant H2A.x (γH2AX), and the adaptor protein tumor suppressor p53-binding protein 1 (53BP1) (Celeste et al., 2003; Huyen et al., 2004; Lobrich et al., 2010; Meier et al., 2007; Rogakou et al., 1999).
When DNA lesions are repairable, DNA damage foci are transient. They typically resolve within 24 hours, during which time cells transiently arrest growth, presumably to allow time for repair. However, severe or irreparable DNA damage, such as complex breaks or uncapped telomeres, causes many cells to senesce with persistent DNA damage foci, constitutive DDR signaling and chronic p53 activation. These persistent changes precede establishment of senescence-associated phenotypes, including growth arrest (Beausejour et al., 2003; d'Adda di Fagagna et al., 2003; Herbig et al., 2004) and SASP (Coppe et al., 2008; Rodier et al., 2009).
Much is known about the hierarchy and temporal recruitment of DDR signaling and repair proteins that aggregate immediately after DNA damage and disaggregate as transient damage foci resolve (Bekker-Jensen et al., 2006; Lisby et al., 2004; Paull et al., 2000). Many proteins that associate with transient foci also occur in the persistent foci that mark senescent cells (d'Adda di Fagagna et al., 2003; Herbig et al., 2004; Kim et al., 2004; Takai et al., 2003). However, DNA damage foci per se might not be accurate senescence markers. For example, telomere dysfunction-induced foci (TIF) – γH2AX or 53BP1 foci localized to telomeres – can identify senescent cells in culture and tissues (Herbig et al., 2006; Herbig et al., 2004; Takai et al., 2003), but TIF can also occur in pre-senescent cells (Beliveau et al., 2007; Verdun et al., 2005) and the damage foci in senescent cells are not always telomere associated (Nakamura et al., 2008; Passos et al., 2010; Wang et al., 2009). Furthermore, it is not known whether the persistent foci associated with senescence contain components that are distinct from those in transient foci. In addition, while the persistent foci clearly contain active DDR components, it is not known whether these structures maintain the DDR activity that is required for the senescence-associated growth arrest or cytokine secretion.
Here, we investigate the spatiotemporal dynamics of persistent DNA damage foci and the impact these structures have on establishing and maintaining senescence-associated phenotypes. We use X-irradiation to initiate damage-induced senescence synchronously in normal human cells, follow DNA damage foci over time and identify characteristics that distinguish transient from persistent foci. Because markers of persistent foci are recruited to locally modified chromatin, we termed these persistent foci ‘DNA segments with chromatin alterations reinforcing senescence’ or DNA-SCARS. Finally, we show that DNA-SCARS are important for maintaining the senescence-associated growth arrest and secretion of IL-6, an important SASP component.
Results
Kinetics of transient versus persistent DNA damage foci
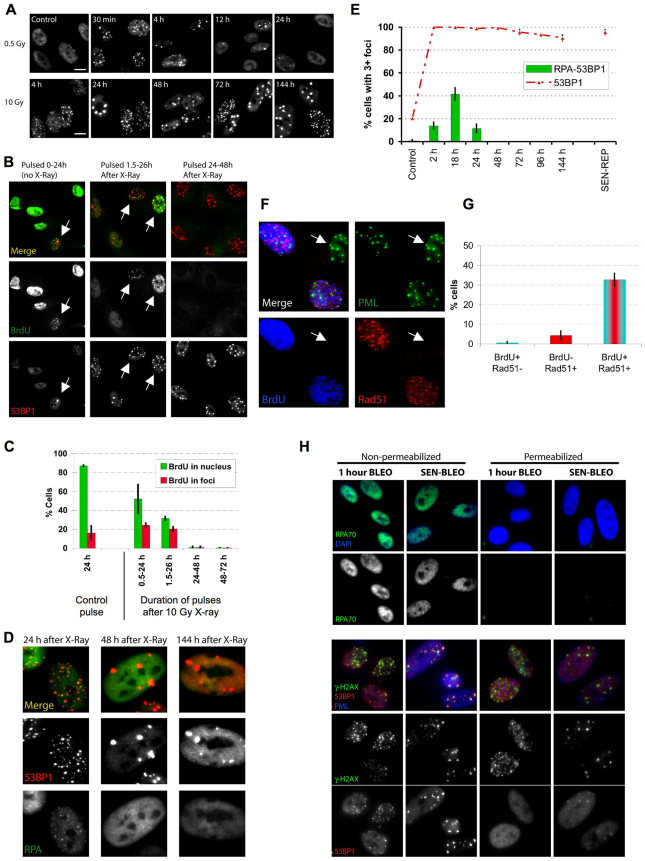
To investigate the kinetics of damage foci formation and resolution, we exposed proliferating normal human fibroblasts (strain HCA2) to 0.5 Gy ionizing radiation (IR; X-ray), which causes a transient growth arrest that reverses within 24 hours; alternatively, we used 10 Gy, which causes a (permanent) senescence arrest (Rodier et al., 2009). We monitored damage foci by 53BP1 immunofluorescence. As expected, 0.5 Gy rapidly produced many small foci, which resolved almost completely within 12 hours; only a few residual foci remained 24 hours later (Fig. 1A; supplementary material Fig. S1A). 10 Gy also rapidly produced many small foci, but a subfraction failed to resolve, became enlarged and persisted for days (Fig. 1A,E) and months (data not shown). These IR-induced persistent DNA damage foci contained many proteins that are also present in transient foci or foci that mark dysfunctional telomeres (d'Adda di Fagagna et al., 2003; Herbig et al., 2004; Lisby et al., 2004; Meier et al., 2007; Rogakou et al., 1999; Takai et al., 2003). Such proteins included the modified chromatin component γH2AX (see Fig. 1H), the repair or adaptor proteins MDC1 (see below), NBS1 and MRE11 (data not shown), the activated DDR signaling protein ATM-pSer-1981 (data not shown) and proteins containing the ATM/ATR-pSer/Thr substrate motif (supplementary material Fig. S1B). Virtually all the 10-Gy-irradiated proliferating cells developed persistent foci, demonstrating that the cell cycle phase at the time of irradiation did not influence their formation. A senescence-inducing dose of bleomycin, a radiomimetic used in cancer therapy (Regulus et al., 2007), also produced both small transient and large persistent foci (Fig. 1H, nonpermeabilized panels). Not surprising, the higher IR and bleomycin doses produced more small transient foci, and possibly some larger early foci (although difficult to distinguish from coincident foci), than lower doses, but, in all cases, the foci largely resolved after low doses, whereas large foci persisted after the higher doses.
Fig. 1.
Persistent DNA damage foci lack evidence of ssDNA and DNA synthesis. (A) HCA2 [population doubling 25 (PD25)] cells were irradiated with 0.5 or 10 Gy X-rays and followed for the indicated intervals before being fixed and stained for 53BP1. Scale bars: 10 μm. (B) Cells, either untreated or irradiated with 10 Gy, were pulsed with BrdU for the indicated intervals after irradiation before being fixed and stained for 53BP1 (red) and BrdU (green). Arrows indicate cells with BrdU-positive 53BP1 foci. (C) Cell populations in B were analyzed for the percentage of cells synthesizing DNA (BrdU positive throughout the nucleus) and the percentage that harbor BrdU-positive 53BP1 foci (BrdU in foci). Shown are the means ± s.d. from three or more independent measurements. (D) Cells were irradiated with 10 Gy and followed for the indicated intervals before being fixed and stained for 53BP1(red) and RPA70 (RPA, green). Note the RPA70 foci 24 hours after irradiation. (E) Cells treated and stained in D were analyzed for the percentage harboring three or more 53BP1 foci and the percentage with three or more foci positive for both RPA and 53BP1. Shown are the means ± s.d. from three or more independent measurements. (F) Cells were irradiated with 10 Gy. Three hours later, they were pulsed for 4 hours with BrdU, then fixed and stained for Rad51 (red), PML (green) and BrdU (blue). The arrow indicates a BrdU-Rad51-negative cell. (G) Single cells in the cell populations in F were analyzed simultaneously for the presence of nuclear RAD51 foci and nucleoplasmic BrdU. Shown are the means ± s.d. from three or more independent measurements. (H) Cells were given 10 μg/ml bleomycin (BLEO) for 30 minutes and processed 30 minutes later (1 hour BLEO), or induced to senesce by an exposure for 2 hours to 20 μg/ml BLEO and processed 12 days later (SEN-BLEO). Cells were either fixed (‘Non-permeabilized’) or permeabilized before fixation (‘Permeabilized’), then stained for RPA70 (green) and DNA (DAPI, blue) in the top panels or 53BP1 (red), γ-H2AX (green) and PML (blue) in the lower panels.
Persistent damage foci lack evidence of the active DNA repair that occurs in transient foci
To determine whether transient and/or persistent foci are sites of active DNA synthesis, indicative of DNA replication or repair, we exposed proliferating cells to 10 Gy IR, pulsed them with bromodeoxyuridine (BrdU) for 24 hours at varying times after IR and analyzed single cells.
In unirradiated cells, >80% showed uniform nuclear staining, consistent with passage through S phase. ~15% of unirradiated cells contained BrdU-positive foci, indicating DNA synthesis and presumably repair of spontaneous damage at these sites (Fig. 1B,C).
As expected (Rodier et al., 2009), 10 Gy IR arrested cell cycle progression within 24 hours. Thus, between 0.5 and 24 hours after IR, only ~50% of cells showed uniform BrdU labeling; this fraction declined to <30% 1.5–26 hours after IR, and <1% 24–48 hours after IR (Fig. 1B,C). DNA synthesis was also evident in foci, but only those that were pulsed early after IR. Thus, 20–25% of cells had BrdU-positive foci when pulsed 0.5–24 hours or 1.5–26 hours after IR. However, 48 hours after IR, few if any cells had BrdU-positive foci, despite numerous persistent foci (Fig. 1B,C). Even long BrdU pulses, from 3–8 days, failed to label persistent foci in irradiated senescent cells; the same was true for replicatively senescent cells, which contain persistent damage foci localized to dysfunctional telomeres (TIF) (data not shown). We conclude that persistent damage foci are not sites of replicative or repair DNA synthesis.
Because we irradiated proliferating cells, we hypothesized that cells irradiated during S phase incorporated BrdU into foci owing to homologous recombination repair (HRR), the preferred mode of DSB repair during S phase. Indeed, immunostaining revealed the presence of RPA70, a single-strand DNA (ssDNA) binding and HRR protein, in some 53BP1 foci. A fraction of RPA70 also remained nucleoplasmic. However, focal coincidence between RPA70 and 53BP1 was limited to 24 hours after IR, with a peak ~18 hours after IR (Fig. 1D,E). RPA70 and 53BP1 coincided at frequencies similar to the number of cells in S phase (compare Fig. 1C and 1E), but foci that persisted for >48 hours were devoid of RPA70. RPA70 was also absent from 53BP1 foci in replicatively senescent cells (Fig. 1E), suggesting that persistent foci, regardless of origin, lack ssDNA. We also exposed proliferating cells to 10 Gy IR, allowed recovery for 3 hours, pulsed with BrdU for 4 hours, then stained for BrdU and RAD51, another ssDNA binding or HRR protein. Cells with RAD51 foci were almost exclusively positive for BrdU (Fig. 1F,G). Thus, in contrast to early transient foci, the persistent foci that remain 24–48 hours after senescence-inducing damage lack evidence of DNA synthesis, ssDNA and HRR.
All proliferating cells exposed to 10 Gy developed persistent foci, suggesting that S phase (or repair associated with S phase) is not required for their formation or maintenance. To test this idea, we arrested cell proliferation using a lentivirus to overexpress the cyclin-dependent kinase inhibitor 2A p16INK4A. As expected, cells arrested with a G1 DNA content ~24 hours after infection. Forty eight hours after infection, we irradiated the cells with 10 Gy. Dual immunostaining showed that the p16INK4A-arrested cells resolved early transient foci in a manner similar to that of pre-senescent cells. Thus, at least for cells induced to senesce by p16INK4A, early repair is not strongly affected, although other senescent cells might repair DSBs less efficiently (Gorbunova et al., 2007). Importantly, 5 days later, most irradiated p16INK4A-positive cells harbored persistent 53BP1 foci, whereas few unirradiated p16INK4A-positive cells had foci (supplementary material Fig. S1C,D). Because persistent foci do not incorporate BrdU or harbor certain repair proteins (RPA70, RAD51) found in early foci, we suggest that they might not be sites of damaged DNA per se but instead are stable chromatin alterations resulting from damage. Indeed, live-cell imaging of damage foci labeled with MDC1–eGFP showed that a significant fraction of the persistent foci were stable during at least 6 hours (supplementary material Fig. S1E), a period that was largely sufficient to resolve most of the transient foci induced by 0.5 Gy IR (Fig. 1A; supplementary material Fig. S1A). Similar observations were recently made by others using a 53BP1–eGFP fusion protein (Passos et al., 2010). For this and the reasons explained below, we refer to persistent foci as DNA-SCARS.
53BP1 is less soluble in DNA-SCARS
To evaluate how proteins associate with transient foci and DNA-SCARS, we treated cells with bleomycin and either fixed before permeabilization to detect both soluble and non-soluble proteins or permeabilized before fixation to extract soluble proteins and detect only non-soluble proteins. Pre-permeabilization completely removed RPA70 from the nucleus (Fig. 1H, upper panels), indicating that RPA70 is readily soluble, regardless of whether it is nucleoplasmic or in early foci. By contrast, pre-permeabilization failed to remove γH2AX (lower panels), regardless of whether it was in early foci or DNA-SCARS (Fig. 1H), as expected for a core histone tightly integrated into chromatin. 53BP1, however, extracted differentially, depending on the type of foci. Pre-permeabilization removed 53BP1 from foci that formed early after damage but failed to remove 53BP1 from DNA-SCARS. This difference in extractability could be due to the greater amount of 53BP1 in DNA-SCARS (Fig. 1H) but nonetheless identifies a notable difference between early transient foci and DNA-SCARS. This finding supports the idea that early foci differ from DNA-SCARS, one characteristic being decreased 53BP1 solubility.
Activated p53 resides in DNA-SCARS
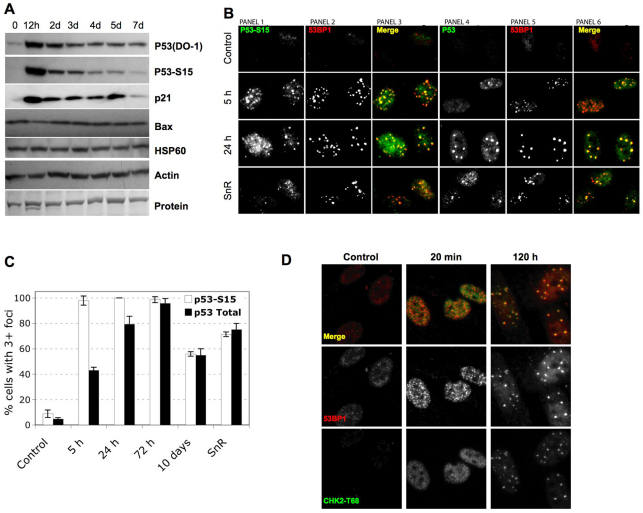
In many human cells, active p53 is required for both establishment and maintenance of the senescence growth arrest (Beausejour et al., 2003; Gire and Wynford-Thomas, 1998; Rodier et al., 2009), suggesting that there is a reservoir of active p53 in senescent cells. p53 activation occurs through stabilization and posttranslational modifications, particularly Ser15 phosphorylation (p53-pS15) by DDR kinases such as ATM (Appella and Anderson, 2001). After 10 Gy IR, HCA2 cells showed the expected rapid rise in p53 and p53-pS15 levels, followed by a decline, as detected by western analysis (Fig. 2A). However, p53 and p53-pS15 typically declined to levels slightly above those of pre-senescent controls (Fig. 2A). This finding raised the possibility that DNA-SCARS, which contain activated ATM (d'Adda di Fagagna et al., 2003; Herbig et al., 2004; Rodier et al., 2009), might also harbor activated p53 and act as reservoirs to maintain its activity.
Fig. 2.
Persistent DNA damage foci accumulate activated DDR mediators. (A) HCA2 cells were either untreated or irradiated (10 Gy). At the indicated intervals thereafter, protein lysates were prepared. 25 μg were analyzed by SDS-PAGE and probed for the indicated proteins by western analysis. Membranes were exposed for different intervals to obtain optimal signals. β-Actin, HSP60 and ponceaux staining (protein) were used as loading controls. (B) Cells were either untreated (‘Control’), cultured to replicative senescence (SnR) or irradiated with 10 Gy and followed for 5 or 24 hours. Cells were fixed and stained for p53-pS15 (panels 1 and 3, green), total p53 (panels 4 and 6, green) or 53BP1 (panels 2 and 5, and panels 3 and 6, red). Yellow indicates merged red and green signals. (C) Early-passage (PD25) HCA2 cells were irradiated with 10 Gy. At the indicated intervals thereafter, the irradiated cells and replicatively senescent (SnR) cells were stained as in B. Cells were scored for three or more p53 or p53-pS15 foci. Shown are the means ± s.d. from three or more independent measurements. (D) Cells were either untreated (‘Control’) or irradiated with 10 Gy and followed for the indicated times before being fixed and stained for 53BP1 (red) and CHK2-pT68 (green) using a rabbit antibody (Cell Signaling #2661, lot #7). Yellow indicates merged red and green signals.
Depending on the type of cell and damage, p53 has been reported present in (Al Rashid et al., 2005) or absent (Bakkenist and Kastan, 2003; Bekker-Jensen et al., 2006) from early DNA damage foci. We therefore asked whether p53 was present in early foci or DNA-SCARS in normal human fibroblasts. Both p53 and p53-pS15 appeared in situ at 53BP1 foci within 5 hours of 10 Gy IR; moreover, both remained associated with DNA-SCARS for at least 10 days after IR and in replicatively senescent (SnR) cells (Fig. 2B,C). The specificities of the p53 antibodies were confirmed by immunostaining bleomycin-damaged human cancer cells that were either wild-type or null for p53 (supplementary material Fig. S1F).
These results demonstrate that at least a fraction of the low level of p53-pS15 detected in senescent cells by western analysis (Fig. 2A) is contained in DNA-SCARS, suggesting that they serve as reservoirs for the p53 activity that maintains the growth arrest.
Activated CHK2 resides in DNA-SCARS
CHK2 is a downstream DDR effector that phosphorylates p53 and is required to maintain a senescence growth arrest (Gire et al., 2004). Activated CHK2 (phosphorylated on threonine 68, pT68) was reported absent from early damage foci in human cancer cells (Lukas et al., 2003) but present at TIF in normal replicatively senescent cells (Herbig et al., 2004). We immunostained early damage foci and DNA-SCARS in normal human fibroblasts for CHK2-pT68. Minutes after 10 Gy IR, CHK2-pT68 staining intensity increased, but it did so throughout the nucleus and without discernible enrichment at foci (Fig. 2D). This nucleoplasmic staining generally declined within 24 hours, after which focal staining began to appear. Many hours later, CHK2-pT68 clearly localized to DNA-SCARS (Fig. 2D). The specificity of the phospho-CHK2 antibody was confirmed by immunostaining irradiated wild-type and CHK2-null cancer cells (supplementary material Fig. S1G) and using a different antibody against phospho-CHK2 (supplementary material Fig. S1H). We also detected CHK2-pT68 in the DNA-SCARS (TIF) of replicatively senescent cells (data not shown). These results reconcile earlier results (Herbig et al., 2004; Lukas et al., 2003) and suggest that, although activated CHK2 diffuses through the nucleus immediately after damage, a fraction accumulates at DNA-SCARS in senescent cells. They also suggest that TIF are selectively localized DNA-SCARS.
DNA-SCARS associate with PML nuclear bodies
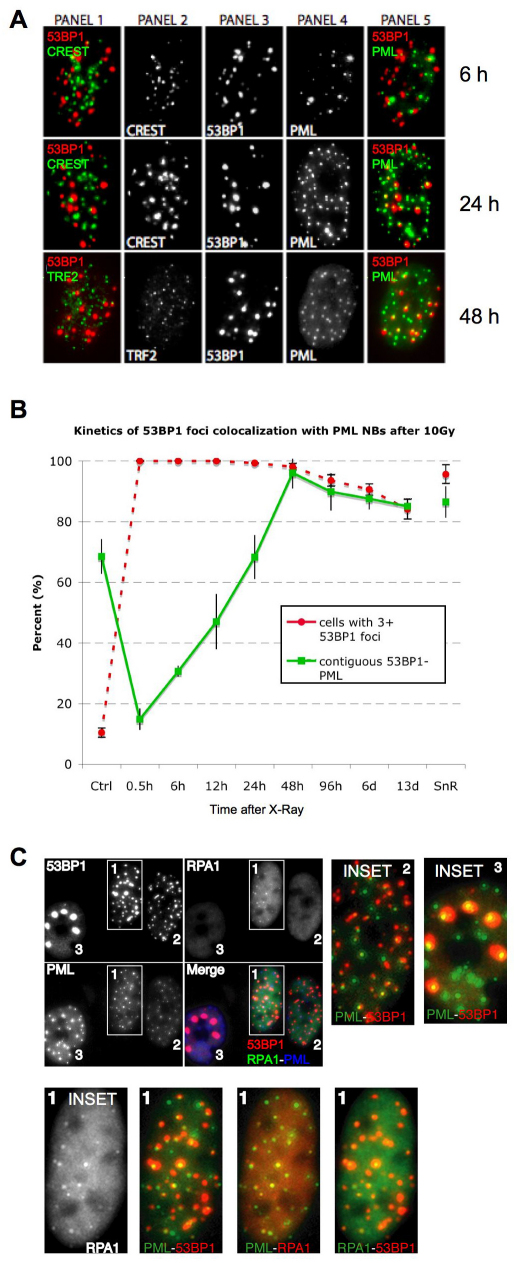
PML (promyelocytic leukemia protein) defines a dynamic, heterogeneous subnuclear domain (nuclear body, NB) that participates in cellular responses to stress, including genotoxic stress (Bernardi and Pandolfi, 2007). PML NBs can be found at DNA damage foci (Carbone et al., 2002; Xu et al., 2003) and act as general sensors of genomic damage (Varadaraj et al., 2007). Because PML also facilitates p53 activation and the senescence growth arrest (Ferbeyre et al., 2000; Pearson et al., 2000), we evaluated its relationship to damage foci during the transition to senescence after IR.
Senescence-inducing IR (10 Gy) generated 53BP1 foci that initially showed no preferential localization to PML NBs (Fig. 3A). However, as DNA-SCARS formed, most of them associated with a PML NB. This association began ~24 hours after IR, was complete 48 hours later (Fig. 3A) and persisted for many days (Fig. 3B; note, ~10% of control cells had damage foci, most of which were PML associated (70%), suggesting that they were DNA-SCARS or TIF in the senescent cells that are present in most normal human cell populations). Unlike 53BP1 and γH2AX, which completely colocalize, PML NBs were often at the periphery of DNA-SCARS. This was true for IR-induced (Fig. 3A,B), bleomycin-induced and replicative senescence (supplementary material Fig. S2A,B).
Fig. 3.
DNA-SCARS associate with a subset of PML NBs. (A) HCA2 cells were irradiated with 10 Gy and followed for 6 or 24 hours before being fixed and stained for CREST (panel 2), 53BP1 (panel 3) and PML (panel 4). At 6 and 24 hours, panel 1 shows CREST (green)–53BP1 (red) colocalization (yellow), and panel 5 shows 53BP1 (red)–PML (green) colocalization (yellow). The same cells were also stained for TRF2 (panel 2), 53BP1 (panel 3) and PML (panel 4) 48 hours after irradiation. Panel 1 shows TRF2 (green)–53BP1 (red) colocalization (yellow), and panel 5 shows 53BP1 (red)–PML (green) colocalization (yellow). (B) Cells were irradiated with 10 Gy, then fixed and stained at the indicated intervals thereafter. Cells were scored for the percentage of cells with three or more 53BP1 foci (red line) and the extent to which individual 53BP1 foci totally or partially overlapped with a PML NB (green line). Shown are the means ± s.d. from three or more independent measurements. (C) Cells were irradiated with 10 Gy. Twenty four hours later, they were fixed and stained for 53BP1 (red in merge), RPA (green in merge) and PML (blue in merge). Cells in the selected field, also shown in Fig. 1A for 53BP1 only, were labeled 1–3, and individual colocalizations are shown in the insets (one selected protein is displayed in green, the other red and colocalization in yellow). In cells 2 and 3, colocalization between 53BP1 and PML is shown. For cell 1, which displays RPA foci, all combinations are shown.
The association between DNA-SCARS and PML NBs was specific and did not occur with other nuclear structures such as centromeres or telomeres. Twenty four hours after 10 Gy IR, when many 53BP1 foci (DNA-SCARS) associated with PML NBs, they were largely excluded from centromeres, as determined by staining for CREST (Fig. 3A), and telomeres, as determined by staining for TRF2 (Fig. 3A). Because PML NB association began ~24 hours after IR, we asked whether RPA70, which showed a peak of association with damage foci ~18 hours after IR, was excluded from PML-associated foci. Triple immunofluorescence showed that, 24 hours after IR, before RPA70 foci completely declined, it was possible to detect foci containing RPA70, 53BP1 and PML (Fig. 3C). Thus, ssDNA-binding proteins do not preclude association of DNA-SCARS with PML NBs.
DNA-SCARS do not depend on several DNA repair or signaling proteins or PML organization
The formation and resolution of damage foci are linked to damage signaling and repair pathways. We therefore asked whether important components of these pathways altered DNA-SCAR formation, as determined by association with PML NBs.
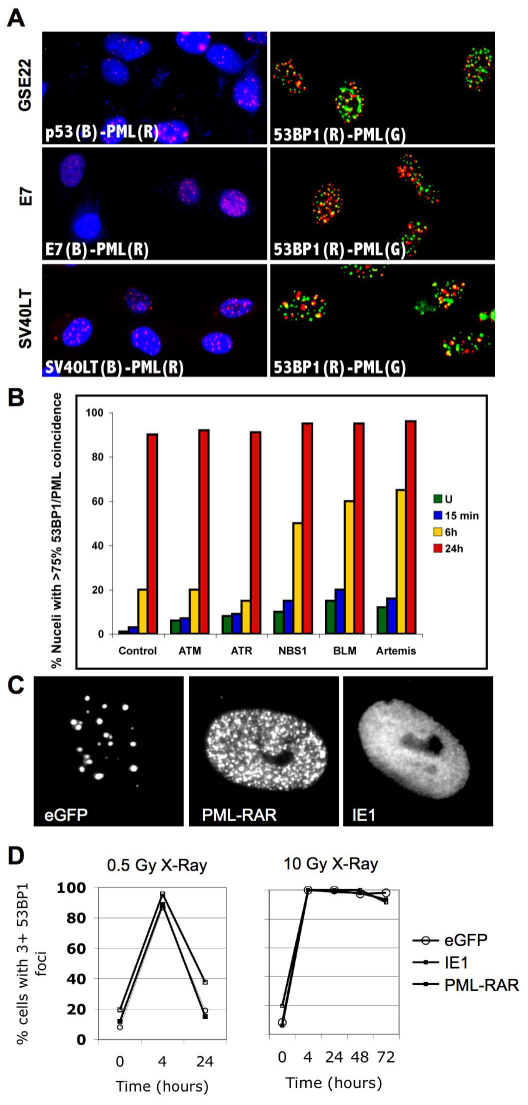
To test the requirement for p53, we infected proliferating HCA2 cells with a retrovirus encoding GSE22, an often-used peptide that stabilizes p53 but prevents formation of active tetramers (Ossovskaya et al., 1996). These cells develop spontaneous damage foci after 5–10 doublings (Rodier et al., 2009). Despite loss of p53 function, 53BP1 foci associated with PML NBs, suggesting that they are DNA-SCARS and do not require p53 for formation (Fig. 4A). Similar results were obtained with cells infected with a retrovirus encoding the viral oncogene E7, which inactivates pRb, or a lentivirus encoding SV40LT, which inactivates p53 and pRb (Fig. 4A). Thus, formation of DNA-SCARS did not depend on functional p53 or pRb pathways. Importantly, cells deficient for either p53 or pRB harbored senescence levels of DNA-SCARS but proliferated, indicating that DNA-SCARS can be uncoupled from the senescence growth arrest when cell cycle checkpoints are defective.
Fig. 4.
DNA-SCARS do not depend on selected DNA repair or signaling proteins. (A) HCA2 cells were infected at PD25 with a lentivirus expressing GSE22, then stained at PD35 for p53 (blue) and PML (red) (upper-left panel) or 53BP1 (red) and PML (green) (upper-right panel). HCA2 cells were infected at PD35 with a retrovirus expressing E7, then stained at PD50 for E7 (blue) and PML (red) (middle-left panel) or 53BP1 (red) and PML (green) (middle-right panel). HCA2 cells were infected at PD40 with a lentivirus expressing SV40LT, then stained at PD55 for SV40LT (blue) and PML (red) (lower-left panel) or for 53BP1 (red) and PML (green) (lower-right panel). (B) Early-passage primary human fibroblasts deficient for ATM, ATR, NBS1, BLM or Artemis were irradiated with 8 Gy. At the indicated intervals thereafter, they were fixed and stained for 53BP1 and PML. Cell populations were scored for the percentage of cells harboring three or more 53BP1 foci, most of which (>75%) colocalized with PML NBs. (C) HCA2 cells at PD25 were infected with retroviruses expressing eGFP (control, left panel), iE1 (middle panel) or PML-RAR (right panel). At PD29, the infected cells were stained for PML. (D) HCA2 cells expressing eGFP, iE1 or PMLRAR (PD30) were irradiated with 0.5 or 10 Gy. At the indicated intervals thereafter, they were stained for 53BP1 foci. Populations were scored for the percentage of cells having three or more 53BP1 foci.
We also examined human fibroblasts deficient for the ATM or ATR signaling kinases, the Artemis repair-associated nuclease, the NBS1 DNA damage signaling transducer or the BLM HRR helicase. All these cells developed DNA-SCARS (53BP1 foci associated with PML NBs) ~24 hours after 8 Gy IR. Thus, DNA-SCARS did not require ATM, ATR, Artemis, NBS1 or BLM for formation. Furthermore, cells deficient in NBS1, BLM or Artemis formed DNA-SCARS faster than wild-type cells (Fig. 4B), suggesting that DNA-SCARS might result from faulty DNA repair.
We also asked whether PML NBs were essential for DNA-SCARS formation or maintenance. We used lentiviruses to express eGFP (control), the PML-RAR oncogene, or the human cytomegalovirus protein iE1 (also known as ICP0) in human fibroblasts. PML-RAR disperses PML into microspeckles (Fig. 4C) and is a dominant inhibitor of wild-type PML function (Dyck et al., 1994). iE1 prevents PML sumoylation, causing diffuse nucleoplasmic organization (Fig. 4C), an absence of PML NBs and loss of PML function (Ahn and Hayward, 1997). We irradiated the infected cells and followed the appearance, resolution and retention of 53BP1 foci over time. The rapid appearance and resolution of early 53BP1 foci after 0.5 or 10 Gy IR were unaffected by PML-RAR or iE1 (Fig. 4D). Likewise, the appearance of DNA-SCARS was unaffected by PML-RAR or iE1 (Fig. 4D). Thus, although DNA-SCARS associate with PML-NBs, PML function is not required for formation of DNA-SCARS.
DNA-SCARS integrity regulates senescence-associated phenotypes
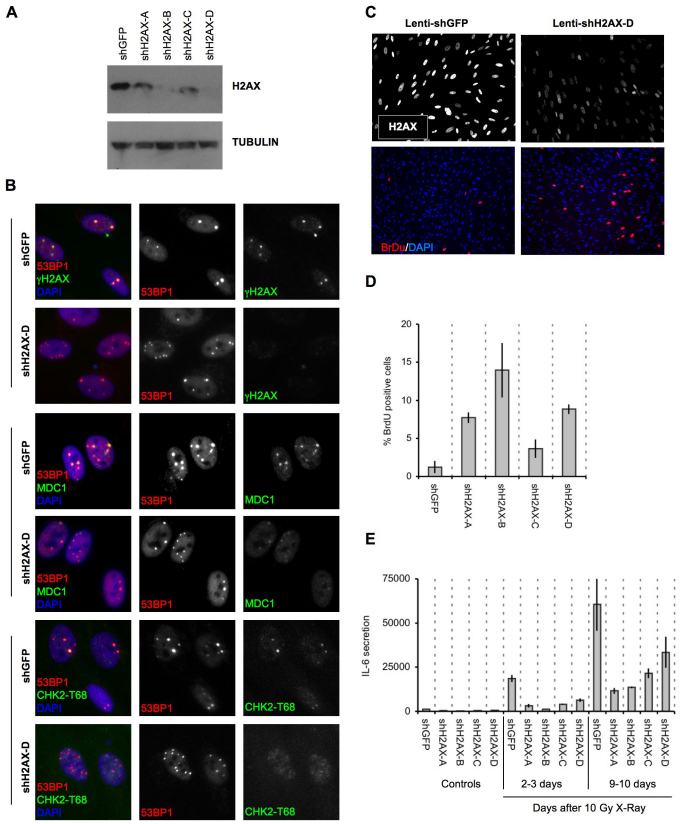
We previously showed that persistent damage foci correlate with the DDR signaling that is essential for maintaining the senescence-associated growth arrest and inflammatory cytokine secretion (Rodier et al., 2009), consistent with our finding here that DNA-SCARS contain active CHK2 and p53 (Fig. 2). Histone H2AX is dispensable for the initial formation of DNA damage foci but is required for their stabilization (Celeste et al., 2003). We therefore tested the idea that depletion of H2AX might disrupt DNA-SCARS and hence senescence-associated phenotypes.
We depleted H2AX by RNA interference (RNAi) using lentiviruses encoding short-hairpin (sh) RNAs against eGFP (control) or H2AX. Western blotting confirmed the decrease in levels of H2AX (Fig. 5A). We then followed the formation of 53BP1 foci after wild-type and H2AX-deficient cells were irradiated with 10 Gy. Forty eight hours after IR, 53BP1 efficiently formed persistent foci in both cell types. However, the DDR adaptor protein MDC1 and CHK2-pT68 were either absent or present at much reduced levels in H2AX-depleted 53BP1 foci (Fig. 5B). Thus, H2AX deficiency interfered with the efficient assembly of some DDR proteins into DNA-SCARS.
Fig. 5.
Histone H2AX is required for senescence-associated phenotypes. (A) HCA2 cells were infected with lentiviruses expressing short hairpin RNAs directed against eGFP (control, shGFP) or H2AX (shH2AX-A to shH2AX-D). Forty eight hours later, cells were selected for 4 days and allowed to recover for 3 days. Whole-cell lysates were analyzed by western blotting for the indicated proteins. (B) HCA2 cells were infected with lentiviruses expressing shGFP (control) or shH2AX-A–shH2AX-D, as in A. Infected cells were irradiated with 8 Gy. Two days later, they were stained for the indicated DDR proteins. (C) HCA2 cells were infected with lentiviruses expressing shGFP or shH2AX-A–shH2AX-D. Infected cells were irradiated with 10 Gy. 7 days later, BrdU was added for 24 hours. Top panels: cells were fixed and stained for H2AX (grayscale). Lower panels: cells were fixed and stained for DNA synthesis (BrdU incorporation, red) and DNA (DAPI, blue). (D) Cells in A were treated as in B and analyzed for DNA synthesis (BrdU positive). Shown are the means ± s.d. from three or more independent measurements. (E) HCA2 cells were infected as in A and either untreated or irradiated with 10 Gy. After 2 days, conditioned media were collected over an interval of 24 hours from untreated cells (‘Controls’) and irradiated cells 2 days after irradiation (2–3 days) and 9 days after irradiation (9–10 days). Conditioned media were assessed for IL-6 by ELISA. IL-6 secretion is reported as 10–6 pg per cell per day. Shown are the means ± s.d. from three or more independent measurements.
To determine the functional consequences of H2AX deficiency, we irradiated (10 Gy) control and H2AX-deficient cells. After 7 days, we pulsed the cells for 24 hours with BrdU to assess DNA synthesis. H2AX deficiency increased BrdU incorporation by cells that would normally remain growth arrested (Fig. 5C,D). All four H2AX shRNAs tested gave similar results. We also asked whether H2AX-deficient cells secreted a SASP cytokine in response to senescence-inducing DNA damage (Rodier et al., 2009). H2AX deficiency suppressed IL-6 secretion 2–3 days after IR, when cells just begin secreting, and 9–10 days later, when secretion is more robust (Fig. 5E). Thus, H2AX depletion reduced certain DDR signaling proteins in DNA-SCARS and suppressed the DDR-dependent senescence-associated growth arrest and IL-6 secretion.
DNA-SCARS – multiple senescence inducers, multiple cell types and formation in vivo
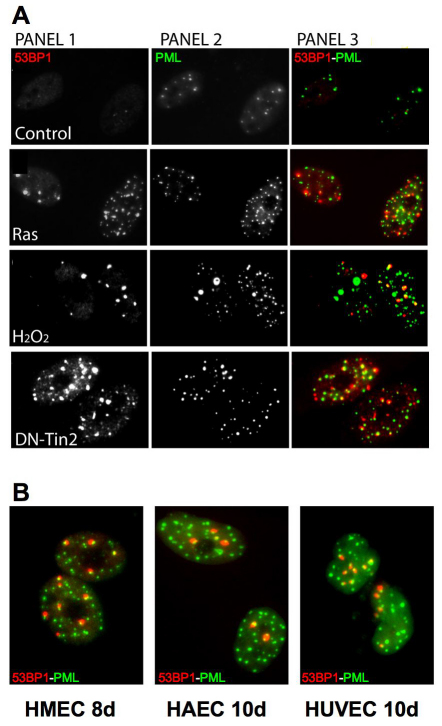
We tested the occurrence of DNA-SCARS, determined by association of 53BP1 foci with PML NBs, in HCA2 cells induced to senesce by means other than IR: expression of an activated oncogene (RAS) (Serrano et al., 1997), expression of a dominant-negative telomeric protein (DN-TIN2) (Kim et al., 2004) or treatment with hydrogen peroxide (H2O2). Each manipulation induced DNA-SCARS within 48 hours (Fig. 6A). Furthermore, we observed DNA-SCARS in four different human fibroblast strains (WI-38, IMR-90, BJ, HCA2) induced to senesce by replicative exhaustion or IR (data not shown) and in IR-induced senescent human mammary epithelial cells (HMECs), human umbilical vein endothelial cells (HUVECs) and human aortic endothelial cells (HAECs) (Fig. 6B). These findings suggest that DNA-SCARS are a general feature of damaged senescent cells.
Fig. 6.
DNA-SCARS are caused by multiple senescence inducers and in several cell types. (A) HCA2 cells were stained for 53BP1 foci (panel 1) and PML NBs (panel 2). Colocalization is shown in yellow (panel 3). Untreated cells were fixed at PD23 (‘Control’), and PD25 cells were infected with a lentivirus encoding either oncogenic Rasv12 (Ras) or a dominant-negative TIN2 protein (DN-Tin2). After a recovery period of 3–4 days, the cells were fixed and stained. Cells were also treated for 1 hour with 100 μM H2O2 (H2O2); 5 days later, they were fixed and stained. (B) Primary HMECs, HUVECs and HAECs were fixed and stained as in A 8 or 10 days, as indicated, after receiving 10 Gy irradiation.
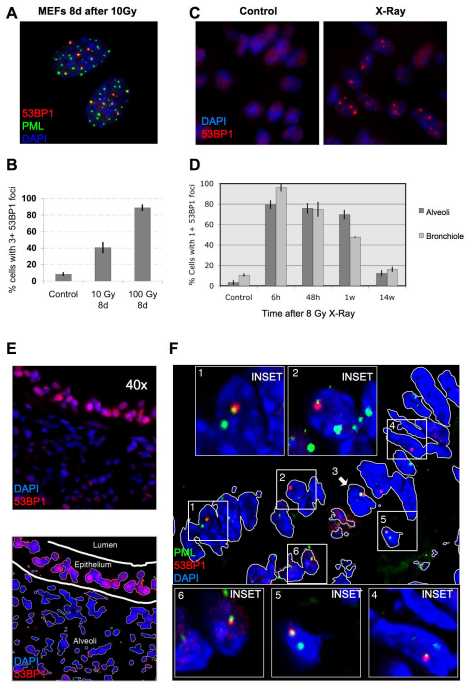
To determine whether DNA-SCARS form in vivo, we examined mouse cells and tissues. First, we exposed primary mouse embryonic fibroblasts (MEFs) to 10–100 Gy IR and, 8 days later, immunostained for 53BP1 and PML. Coincident staining was readily apparent, although higher IR doses were needed relative to those in human cells (Fig. 7A,B). Thus, DNA-SCARS form in both human and mouse cells. We then subjected mice to whole-body non-lethal IR (8 Gy) and prepared frozen tissue sections for immunostaining at varying times thereafter. 53BP1 foci were readily detected in both the alveoli and bronchioles of lungs 6 hours, 48 hours and 7 days after IR (Fig. 7C,D; supplementary material Figs S3, S4). Nearly 100% of cells harbored foci 6 hours after IR. Many foci resolved after 48 hours, especially in the bronchioles, but a fraction of cells (45–70%) maintained at least one 53BP1 focus 1 week after IR (Fig. 7D; see supplementary material Fig. S5 for similar results in liver). After 14 weeks, persistent 53BP1 foci diminished further, but remained two- to four-fold higher than the low level in matched unirradiated controls (Fig. 7D). This delayed decline might reflect damage repair, clearance of damaged cells by the immune system (Ventura et al., 2007; Xue et al., 2007) and/or cell turnover (Le et al., 2010).
Fig. 7.
DNA-SCARS are detected in vivo in irradiated mouse tissues. (A) MEFs (early passage) were irradiated with 10 Gy. Eight days later, they were fixed and stained for 53BP1 (red), PML (green) and DAPI (blue). (B) MEFs were irradiated with 10 or 100 Gy. Eight days later, they were stained as in (A) and analyzed for the percentage of cells with three or more 53BP1 foci associated with PML NBs. Shown are the means ± s.d. from three or more independent measurements. (C) Lungs from three-month-old C57BL6 mice were analyzed by immunofluorescence using fresh frozen sections. Representative 53BP1 staining (red) is shown in a control (left panel) animal or an animal irradiated with 8 Gy and analyzed 7 days later (right panel). Nuclei were counterstained with DAPI (blue). (D) Lung alveoli and bronchiolar epithelial cells from control or irradiated animals that were allowed to recover for the indicated periods after 8 Gy irradiation. The percentage of cells with at least one 53BP1 foci was quantified in three independent images from frozen lung sections similar to that displayed in C. Error bars show the s.d. from three independent measurements (total number of nuclei counted; for each time point, n=600 or more for lung alveolar cells and n=300 or more for bronchiolar epithelial cells). (E) Representative 53BP1 staining (red) of the lung bronchiolar epithelial cells in control mice (nuclei counterstained with DAPI). The lower panel outlines the epithelial cell layer. (F) A lung section from an irradiated animal that was allowed to recover for 7 days after 8 Gy irradiation, stained for 53BP1 (red), PML (green) and DAPI (blue) and analyzed by confocal microscopy. The six cells that displayed 53BP1–PML colocalization are boxed and numbered and magnified in the insets. The thin white lines in the main image outline the periphery of the nuclei.
To determine whether the 53BP1 foci that persisted 1 week after IR were DNA-SCARS, we asked whether they associated with PML NBs. Fig. 7E illustrates how bronchiolar layers were selected and single cells analyzed for coincidental 53BP1 and PML staining. As shown in the Fig. 7F insets, most bronchiolar epithelial cell nuclei showed an association between 53BP1 and PML NBs (cells 1–4). Additionally, non-epithelial cells (underlying the airway epithelial border) also showed associated 53BP1 foci and PML NBs (cells 5–6). Similar results were obtained in liver and lung alveolar cells, although only 30–50% of 53BP1 foci associated with PML NBs in these compartments (supplementary material Figs S4, S5). These results suggest that association between PML NBs and 53BP1 can be used to distinguish early transient DNA damage foci from DNA-SCARS in vivo.
Discussion
Constitutive DDR signaling is an important component of the integrated tumor suppressor network that establishes and maintains the senescence phenotypes of growth arrest and the SASP (d'Adda di Fagagna et al., 2003; Herbig et al., 2004; Rodier et al., 2009). Here, we show that senescence-associated DNA damage foci, or DNA-SCARS, are relatively stable structures that are distinct from transient damage foci and functionally important for both the growth arrest and IL-6 secretion, an important SASP feature.
DNA damage foci can be detected by immunofluorescence because chromatin modifications generated by the DDR at DSBs spread over megabases (Meier et al., 2007; Rogakou et al., 1999). Whether these foci form early and transiently after DNA damage or are the persistent foci associated with senescence, they reflect elevated concentrations of modified histones (e.g. γH2AX) and other proteins that are recruited to remodeled chromatin. We identified several molecular events that differentiate transient from persistent foci, which we followed after X-irradiation to generate DSBs (among other lesions) synchronously.
In transient foci, lesions are likely to be repaired by both non-homologous end joining (NHEJ) and HRR, depending on the cell cycle phase in which the damage was inflicted. For cells damaged in S phase, we detected the ssDNA-binding proteins RAD51 and RPA70, which bind the resected DNA that is generated during HRR; these cells also incorporated BrdU, indicative of repair. γH2AX can be found at ssDNA, in addition to its predominant association with DNA DSBs (Lobrich et al., 2010). Nonetheless, both ssDNA-binding proteins and DNA synthesis were absent from persistent foci. Furthermore, individual persistent foci were stable for several hours in living cells, and, in fixed senescent cells, were detected for days and weeks after their formation. Thus, HRR and NHEJ are likely either inactive or incapable of resolving these foci after ~48 hours. Formation of DNA-SCARS was accelerated in cells that are deficient in certain DNA repair proteins (Fig. 4B), supporting the idea that ineffective or defective repair initiates the formation of these structures. We propose that, by ~48 hours after senescence-inducing DNA damage, a general repair phase ends and the remaining foci contain either unresolved lesions or stable chromatin modifications resulting from the damage. Alternatively, repair mechanisms that do not require DNA synthesis, HRR or NHEJ could be actively attempting to repair lesions in DNA-SCARS, but these (presumably abortive) repair processes were not detected in our study. Because these foci persist, and their destabilization relaxed the senescence growth arrest and IL-6 secretion, we term them ‘DNA segments with chromatin alterations reinforcing senescence’ or DNA-SCARS.
DNA-SCARS include TIF, which occur at dysfunctional telomeres (Herbig et al., 2004; Takai et al., 2003). They also include foci generated by senescence-inducing DNA damage inflicted by H2O2 or oncogenic RAS. However, as with other senescence markers, DNA-SCARS are associated with, but not exclusive to, senescent cells. In particular, p53-deficient cells spontaneously develop DNA-SCARS, yet they proliferate. Such cells resemble those in pre-cancerous lesions in which DDR signaling is detected before full transformation (Gorgoulis et al., 2005). Furthermore, PML NBs can harbor damaged telomeres in telomerase-deficient cancer cells (Nabetani et al., 2004). In these ALT (‘alternative lengthening of telomeres’) cells, markers of telomeres, DSBs and PML colocalize in a structure reminiscent of DNA-SCARS. These results suggest that, although DNA-SCARS are novel markers of senescence in normal cells, the association can be uncoupled in preneoplastic or cancer cells. Thus, DNA-SCARS or TIF can be used as senescence markers, but only when combined with other markers, such as senescence-associated beta-galactosidase (SA-Bgal) (Dimri et al., 1995), p16INK4A (Krishnamurthy et al., 2004) or senescence-associated heterochromatic foci (Narita et al., 2003). Notably, only the growth arrest, not inflammatory cytokine secretion, was uncoupled from DNA-SCARS in cells expressing viral oncogenes that inactivate the p53 and/or pRB tumor-suppressor pathways. Therefore, in the context of cancer, DNA-SCARS could mark locally increased inflammatory cytokine secretion, which is associated with persistent DDR signaling (Rodier et al., 2009).
The association between DNA-SCARS and PML NBs, where many repair proteins localize, might occur to allow further processing of lesions. In addition, persistent foci might associate with PML NBs, where many chromatin-modifying proteins also localize, to promote senescence-associated gene expression changes such as the SASP. Alternatively, the presence of repair proteins in PML NBs could simply reflect their normal localization when the immediate repair phase ends. In support of this idea, disruption of PML NB structures did not alter the kinetics of foci formation and resolution. Furthermore, loss of several DDR repair and checkpoint proteins did not prevent, and in some cases accelerated, DNA-SCAR formation. Thus, damage foci might quickly progress to DNA-SCARS in the absence of optimal repair.
Senescent cells acquire a complex phenotype, including arrested growth and a multi-factor SASP (Coppe et al., 2008). We propose that DNA-SCARS provide a reservoir for active DDR signaling, which is essential to maintain both the p53-dependent growth arrest and inflammatory cytokine secretion (Beausejour et al., 2003; d'Adda di Fagagna et al., 2003; Gire et al., 2004; Gire and Wynford-Thomas, 1998; Herbig et al., 2004; Rodier et al., 2009). In support of this idea, depletion of the damage-foci-stabilizing core histone H2AX disrupted the DNA-SCARS structure, reducing the levels of SCARS-associated MDC1 and activated CHK2 and relaxed both the senescence growth arrest and IL-6 secretion. Although ATM and CHK2 are essential for the senescence-associated secretion of inflammatory cytokines (Rodier et al., 2009), p53 is not, suggesting that DNA-SCARS could be the reservoir for active p53, CHK2 and other activated DDR proteins. Thus, DNA-SCARS influence two important features of the senescent phenotype – growth arrest and inflammatory cytokine secretion.
DNA-SCARS appeared after a senescence-inducing dose of DNA damage in all primary human and mouse fibroblasts studied, in primary human epithelial and endothelial cells and in mouse tissues. This finding indicates that formation of DNA-SCARS is a robust phenomenon that is conserved between mouse and human, as are many other features of cellular senescence (Coppe et al., 2010), and that it occurs in culture and in vivo. A deeper understanding of how these structures assemble and function will probably enrich our insights into the mechanisms that link DNA damage, inflammation, cancer and aging. Furthermore, it might suggest novel strategies for ameliorating the effects of DNA damage, whether encountered during normal aging, environmental exposure or certain therapies, such as those used to treat many cancers.
Materials and Methods
Cells
In general, cells were cultured at 37°C in 10% CO2, DMEM, 10% fetal bovine serum and 100 U/ml streptomycin–penicillin. Early-passage cells had 24 hours BrdU labeling indices of >75%, and <10% expressed SA-Bgal (Dimri et al., 1995). Cell populations were considered senescent when they had labeling indices of <5% and >75% expressed SA-Bgal. HCA2 cells were from J. Smith (University of Texas, San Antonio, TX). Artemis-deficient cells were from S. Yannone, ATM-deficient cells from E. Blakeley and human mammary epithelial cells from M. Stampfer, all from Lawrence Berkeley National Laboratory. Fibroblasts deficient in ATR (GM09812), NBS1 (GM07166A) or BLM (GM02085) were from the Coriell Institute. Human umbilical vein endothelial cells (HUVECs) and aortic endothelial cells (HAECs) were from CloneTics.
Viruses and infections
Viruses were produced as described previously (Beausejour et al., 2003) and titers adjusted when necessary to achieve ~90% infectivity. Retroviruses encoding eGFP (Rodier et al., 2009), GSE22 (Itahana et al., 2002) or E7 (Itahana et al., 2003) have been described previously. iE1 and PML-RAR were cloned into the pBABE-puro retroviral backbone. Lentiviruses encoding dominant-negative TIN2 (TIN2-15C) (Kim et al., 2004), eGFP (Rodier et al., 2009), SV40LT (Beausejour et al., 2003) and oncogenic RASV12 (Rodier et al., 2009) have been described previously. The full-length coding sequence for human MDC1 was fused with eGFP (entry vector 770-7) and inserted in the lentiviral backbone 775-1 to generate the pLenti CMV/TO GFP–MDC1 expression vector (779-2) (Campeau et al., 2009). Lentiviruses encoding shRNAs against eGFP and H2AX were purchased from Open Biosystems (shGFP3: #RHS4459; shH2AX-A: #TRCN0000073278; shH2AX-B: #TRCN0000073280; shH2AX-C: #TRCN0000073279; and shH2AX-D: #TRCN0000073281).
Irradiation and bleomycin
Cells were X-irradiated at rates equal to or above 0.75 Gy/minute using a Pantak X-ray generator (320 kV/10 mA with 0.5 mm copper filtration). Cells were treated with bleomycin, either 10 μg/ml for 30 minutes with a recovery period of 30 minutes or 20 μg/ml for 2 hours with a recovery period of 10–12 days.
Frozen tissue sections
C57BL6 mice were sacrificed and excised organs were dipped in OCT (Tissue-Tek #4583), flash-frozen in liquid nitrogen and preserved at −80°C. Organs were sectioned (6–10 μm thickness) at −20°C and slices were placed on superfrost plus microslides (VWR #48311-703). Slices were air-dried and stored at −80°C.
Pre-permeabilization extraction
Soluble proteins were extracted as described previously (He et al., 1990). Briefly, cells in chamber-slides were washed on ice with phosphate-buffered saline (PBS) and soluble proteins extracted by an incubation for 10 minutes with ice-cold permeabilization buffer containing inhibitors of RNAses (New England Biolabs), phosphatases (Sigma) and proteases (Complete-Mini Roche). Cells were washed in cold permeabilization buffer and PBS before being fixed in formalin (room temperature) and processed for immunofluorescence (below).
Immunofluorescence
Frozen tissue-section slides were brought to room temperature for 10 minutes, or cells in four-well chamber-slides were washed in PBS. Frozen sections or cells were fixed in formalin (10%, neutral buffered) for 10 minutes at room temperature, washed in PBS, permeabilized in PBS plus 0.5% Triton X-100 for 10 minutes and washed again. Slides were blocked for 1 hour in PBS plus 1% BSA and 4% serum corresponding to the secondary antibody (goat or donkey) and incubated with primary antibodies diluted in blocking buffer overnight at 4°C. The slides were washed with PBS and incubated with secondary antibodies for 1 hour at room temperature, washed and mounted with slow-fade gold (Molecular Probes). When noted, DAPI was added to secondary antibodies.
BrdU staining
Cells in chamber slides were pulsed with BrdU before fixation and permeabilization, as described above. After two PBS washes and a wash in enzyme reaction buffer [0.75X EXO III reaction buffer (Promega) plus 10 mM MgCl2], DNA was denatured by incubation for 30 minutes at 37°C in enzyme reaction buffer plus 5 U/ml DNASE I (Roche) and 200 U/ml EXOIII (Promega). Cells were washed with PBS and processed for immunofluorescence.
Microscopy
Images were acquired on an Olympus BX60 fluorescence microscope with the spotfire 3.2.4 software (Diagnostics Instruments) or a Nikon PCM-2000 laser confocal scanning microscope and prepared for figure creation using Photoshop CS2 (Adobe).
Western blots
Cells were lysed in 50 mM Tris–HCl pH 6.8, 100 mM DTT, 2% SDS and 10% glycerol. The mixture was sonicated (30 seconds), boiled (10 minutes) and spun. Samples were separated by electrophoresis on 5% or 4–15% gradient Tris–glycine SDS-polyacrylamide gels. Proteins were electroblotted overnight onto PVDF membranes and probed overnight at 4°C in PBS with 5% milk. Secondary antibodies conjugated to horseradish peroxidase were used at 1:15,000 (Jackson ImmunoResearch). Blots were developed using the supersignal pico reagent (PIERCE).
Antibodies
The antibodies used were: 53BP1, Bethyl, BL182 or Upstate, BP13 (against human) or Novus, 100-305 (against mouse); γH2AX, Upstate, JBW301; PML, Santa Cruz Biotechnology, N-19 (against human) or Upstate, 36.1-104 (against mouse); p53, Oncogene Research Products, DO-1; phospho-p53 serine 15, Cell Signaling, #9286; p21, BD Biosciences, 556430; p16INK4A, Neomarkers, JC8; actin, Chemicon, MAB3128; tubulin, Sigma, T5168; phospho-ATM/ATR STK substrates, Cell Signaling #2851; TRF2, Imgenex, IMG-124; CREST, Antibodies Incorporated, 15-235-F; phospho-CHK2 threonine 68, Cell Signaling #2661 (lots #1 and #7) or the rabbit monoclonal #2197; BrdU; Abcam, ab1893 or Roche, BMC 9318; Rad51, Santa Cruz Biotechnology, H-92; RPA70, Calbiochem, NA13; SV40LT, Santa Cruz Biotechnology, Pab 101; E7, Neomarkers, TVG701Y; HSP60, Santa Cruz Biotechnology, N-20; MDC1, rabbit polyclonal (Campeau et al., 2009); donkey secondary antibodies conjugated to Alexa Fluor dyes were from Molecular Probes (Alexa 350, 488 and 594).
ELISA
Conditioned media (CM) were prepared by washing cells three times in PBS and incubating in serum-free DMEM plus antibiotics for 24 hours. CM were filtered and stored at −80°C. Cell number was determined for every sample. ELISAs were performed using kits and procedures from R&D (IL-6 #D06050). The data were normalized to cell number and reported as 10−6 pg secreted protein per cell per day.
Supplementary Material
Acknowledgments
We thank Patrick Kaminker for valuable assistance with the confocal microscope, Dolf Beems for comments on mouse histology and Pierre Desprez for critically reading the manuscript. This work was supported by grants from the Canadian Institutes of Health Research MPO79317 to C.B., US NIH AG09909 and AG17242 to J.C. and AG025708 to the Buck Institute, and DOE contract AC03-76SF00098. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/124/1/68/DC1
References
- Acosta J. C., O'Loghlen A., Banito A., Guijarro M. V., Augert A., Raguz S., Furnagalli M., DaCosta M., Brown C., Popov N., et al. (2008). Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 133, 1006-1018 [DOI] [PubMed] [Google Scholar]
- Ahn J. H., Hayward G. S. (1997). The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol. 71, 4599-4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Rashid S. T., Dellaire G., Cuddihy A., Jalali F., Vaid M., Coackley C., Folkard M., Xu Y., Chen B. P., Chen D. J., et al. (2005). Evidence for the direct binding of phosphorylated p53 to sites of DNA breaks in vivo. Cancer Res. 65, 10810-10821 [DOI] [PubMed] [Google Scholar]
- Appella E., Anderson C. W. (2001). Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268, 2764-2772 [DOI] [PubMed] [Google Scholar]
- Bakkenist C. J., Kastan M. B. (2003). DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421, 499-506 [DOI] [PubMed] [Google Scholar]
- Bartek J., Lukas J. (2007). DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19, 238-245 [DOI] [PubMed] [Google Scholar]
- Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L. V., Kolettas E., Niforou K., Zoumpourlis V. C., et al. (2006). Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444, 633-637 [DOI] [PubMed] [Google Scholar]
- Bavik C., Coleman I., Dean J. P., Knudsen B., Plymate S., Nelson P. S. (2006). The gene expression program of prostate fibroblast senescence modulates neoplastic epithelial cell proliferation through paracrine mechanisms. Cancer Res. 66, 794-802 [DOI] [PubMed] [Google Scholar]
- Beausejour C. M., Krtolica A., Galimi F., Narita M., Lowe S. W., Yaswen P., Campisi J. (2003). Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 22, 4212-4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M. B., Bartek J., Lukas J. (2006). Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 173, 195-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliveau A., Bassett E., Lo A. T., Garbe J., Rubio M. A., Bissell M. J., Campisi J., Yaswen P. (2007). p53-dependent integration of telomere and growth factor deprivation signals. Proc. Natl. Acad. Sci. USA 104, 4431-4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I., Weinberg R. A. (2004). When cells get stressed: an integrative view of cellular senescence. J. Clin. Invest. 113, 8-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovich E., Monnat R. J., Kastan M. B. (2007). Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 9, 683-690 [DOI] [PubMed] [Google Scholar]
- Bernardi R., Pandolfi P. P. (2007). Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 8, 1006-1016 [DOI] [PubMed] [Google Scholar]
- Braig M., Schmitt C. A. (2006). Oncogene-induced senescence: putting the brakes on tumor development. Cancer Res. 66, 2881-2884 [DOI] [PubMed] [Google Scholar]
- Campeau E., Ruhl V. E., Rodier F., Smith C. L., Rahmberg B. L., Fuss J. O., Campisi J., Yaswen P., Cooper P. K., Kaufman P. D. (2009). A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS ONE 4, e6529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. (2005). Suppressing cancer: The importance of being senescent. Science 309, 886-887 [DOI] [PubMed] [Google Scholar]
- Campisi J., d'Adda di Fagagna F. (2007). Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8, 729-740 [DOI] [PubMed] [Google Scholar]
- Carbone R., Pearson M., Minucci S., Pelicci P. G. (2002). PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene 21, 1633-1640 [DOI] [PubMed] [Google Scholar]
- Celeste A., Fernandez-Capetillo O., Kruhlak M. J., Pilch D. R., Staudt D. W., Lee A., Bonner R. F., Bonner W. M., Nussenzweig A. (2003). Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5, 675-679 [DOI] [PubMed] [Google Scholar]
- Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A. J., Barradas M., Benguria A., Zaballos A., Flores J. M., Barbacid M., et al. (2005). Tumor biology: senescence in premalignant tumours. Nature 436, 642 [DOI] [PubMed] [Google Scholar]
- Coppe J. P., Patil C. K., Rodier F., Sun Y., Munoz D., Goldstein J., Nelson P. S., Desprez P. Y., Campisi J. (2008). Senescence-associated secretory phenotypes reveal cell non-automous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6, 2853-2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppe J. P., Patil C. K., Rodier F., Krtolica A., Beausejour C. M., Parrinello S., Hodgson J. G., Chin K., Desprez P. Y., Campisi J. (2010). A human-like senescence-associated secretory phenotype is conserved in mouse cells dependent on physiological oxygen. PLoS ONE 5, e9188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna F. (2008). Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer 8, 512-522 [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F., Reaper P. M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N. P., Jackson S. P. (2003). A DNA damage checkpoint response in telomere-initiated senescence. Nature 426, 194-198 [DOI] [PubMed] [Google Scholar]
- Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre M., Nuciforo P. G., Bensimon A., et al. (2006). Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444, 638-642 [DOI] [PubMed] [Google Scholar]
- Dimri G. P. (2005). What has senescence got to do with cancer? Cancer Cell 7, 505-512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri G. P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E. E., Linskens M., Rubelj I., Pereira-Smith O. M., et al. (1995). A novel biomarker identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92, 9363-9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyck J. A., Maul G. G., Miller W. H., Chen J. D., Kakizuka A., Evans R. M. (1994). A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell 76, 333-343 [DOI] [PubMed] [Google Scholar]
- Erusalimsky J. D., Kurz D. J. (2005). Cellular senescence in vivo: its relevance in ageing and cardiovascular disease. Exp. Gerontol. 40, 634-642 [DOI] [PubMed] [Google Scholar]
- Ferbeyre G., de Stanchina E., Querido E., Baptiste N., Prives C., Lowe S. W. (2000). PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 14, 2015-2027 [PMC free article] [PubMed] [Google Scholar]
- Gire V., Wynford-Thomas D. (1998). Reinitiation of DNA synthesis and cell division in senescent human fibroblasts by microinjection of anti-p53 antibodies. Mol. Cell. Biol. 18, 1611-1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gire V., Roux P., Wynford-Thomas D., Brondello J. M., Dulic V. (2004). DNA damage checkpoint kinase Chk2 triggers replicative senescence. EMBO J. 23, 2554-2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V., Seluanov A., Mao Z., Hine C. (2007). Changes in DNA repair during aging. Nucleic Acids Res. 35, 7466-7474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis V. G., Vassiliou L. V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R. A., Kastrinakis N. G., Levy B., et al. (2005). Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907-913 [DOI] [PubMed] [Google Scholar]
- He D. C., Nickerson J. A., Penman S. (1990). Core filaments of the nuclear matrix. J. Cell Biol. 110, 569-580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig U., Jobling W. A., Chen B. P., Chen D. J., Sedivy J. (2004). Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol. Cell 14, 501-513 [DOI] [PubMed] [Google Scholar]
- Herbig U., Ferreira M., Condel L., Carey D., Sedivy J. M. (2006). Cellular senescence in aging primates. Science 311, 1257 [DOI] [PubMed] [Google Scholar]
- Huyen Y., Zgheib O., Ditullio R. A., Gorgoulis V. G., Zacharatos P., Petty T. J., Sheston E. A., Mellert H. S., Stavridi E. S., Halazonetis T. D. (2004). Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432, 406-411 [DOI] [PubMed] [Google Scholar]
- Itahana K., Dimri G. P., Hara E., Itahana Y., Zou Y., Desprez P. Y., Campisi J. (2002). A role for p53 in maintaining and establishing the quiescence growth arrest in human cells. J. Biol. Chem. 277, 18206-18214 [DOI] [PubMed] [Google Scholar]
- Itahana K., Zou Y., Itahana Y., Martinez J. L., Beausejour C., Jacobs J. J., Van Lohuizen M., Band V., Campisi J., Dimri G. P. (2003). Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol. Cell. Biol. 23, 389-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli P. V., Lengauer C., Vogelstein B., Bunz F. (2003). The Chk2 tumor suppressor is not required for p53 responses in human cancer cells. J. Biol. Chem. 278, 20475–20479 [DOI] [PubMed] [Google Scholar]
- Jeyapalan J. C., Ferreira M., Sedivy J. M., Herbig U. (2007). Accumulation of senescent cells in mitotic tissue of aging primates. Mech. Ageing Dev. 128, 36-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Beausejour C., Davalos A. R., Kaminker P., Heo S. J., Campisi J. (2004). TIN2 mediates the functions of TRF2 at telomeres. J. Biol. Chem. 279, 43799-43804 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J., Torrice C., Ramsey M. R., Kovalev G. I., Al-Regaiey K., Su L., Sharpless N. E. (2004). Ink4a/Arf expression is a biomarker of aging. J. Clin. Invest. 114, 1299-1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krtolica A., Parrinello S., Lockett S., Desprez P., Campisi J. (2001). Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc. Natl. Acad. Sci. USA 98, 12072-12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T., Peeper D. S. (2009). Senescence-messaging secretome: SMS-ing cellular stress. Nat. Rev. Cancer 9, 81-94 [DOI] [PubMed] [Google Scholar]
- Kuilman T., Michaloglou C., Vredeveld L. C. W., Douma S., van Doorn R., Desmet C. J., Aarden A. L., Mooi W. J., Peeper D. S. (2008). Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133, 1019-1031 [DOI] [PubMed] [Google Scholar]
- Le O. N., Rodier F., Fontaine F., Coppe J. P., Campisi J., DeGregori J., Laverdiere C., Kokta V., Haddad E., Beausejour C. M. (2010). Ionizing radiation-induced long-term expression of senescence markers in mice is independent of p53 and immune status. Aging Cell 9, 398-409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Barlow J. H., Burgess R. C., Rothstein R. (2004). Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699-713 [DOI] [PubMed] [Google Scholar]
- Liu D., Hornsby P. J. (2007). Senescent human fibroblasts increase the early growth of xenograft tumors via matrix metalloproteinase secretion. Cancer Res. 67, 3117-3126 [DOI] [PubMed] [Google Scholar]
- Lobrich M., Shibata A., Beucher A., Fisher A., Ensminger M., Goodarzi A. A., Barton O., Jeggo P. A. (2010). gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle 9, 662-669 [DOI] [PubMed] [Google Scholar]
- Lukas C., Falck J., Bartkova J., Bartek J., Lukas J. (2003). Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat. Cell Biol. 5, 255-260 [DOI] [PubMed] [Google Scholar]
- Meier A., Fiegler H., Muñoz P., Ellis P., Rigler D., Langford C., Blasco M. A., Carter N., Jackson S. P. (2007). Spreading of mammalian DNA-damage response factors studied by ChIP-chip at damaged telomeres. EMBO J. 26, 2707-2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani A., Yokoyama O., Ishikawa F. (2004). Localization of hRad9, hHus1, hRad1, and hRad17 and caffeine-sensitive DNA replication at the alternative lengthening of telomeres-associated promyelocytic leukemia body. J. Biol. Chem. 279, 25849-25857 [DOI] [PubMed] [Google Scholar]
- Nakamura A. J., Chiang Y. J., Hathcock K. S., Horikawa I., Sedelnikova O. A., Hodes R. J., Bonner W. M. (2008). Both telomeric and non-telomeric DNA damage are determinants of mammalian cellular senescence. Epigenetics Chromatin 1, 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M., Nunez S., Heard E., Narita M., Lin A. W., Hearn S. A., Spector D. L., Hannon G. J., Lowe S. W. (2003). Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703-716 [DOI] [PubMed] [Google Scholar]
- Ossovskaya V. S., Mazo I. A., Chernov M. V., Chernova O. B., Strezoska Z., Kondratov R., Stark G. R., Chumakov P. M., Gudkov A. V. (1996). Use of genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc. Natl. Acad. Sci. USA 93, 10309-10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello S., Coppe J. P., Krtolica A., Campisi J. (2005). Stromal-epithelial interactions in aging and cancer: senescent fibroblasts alter epithelial cell differentiation. J. Cell Sci. 118, 485-496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos J. F., Nelson G., Wang C., Richter T., Simillion C., Proctor C. J., Miwa S., Olijslagers S., Hallinan J., Wipat A., et al. (2010). Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol. Syst. Biol. 6, e347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull T. T., Rogakou E. P., Yamazaki V., Kirchgessner C. U., Gellert M., Bonner W. M. (2000). A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10, 886-895 [DOI] [PubMed] [Google Scholar]
- Pearson M., Carbone R., Sebastiani C., Cioce M., Fagioli M., Saito S., Higashimoto Y., Appella E., Minucci S., Pandolfi P. P., et al. (2000). PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406, 207-210 [DOI] [PubMed] [Google Scholar]
- Price J. S., Waters J., Darrah C., Pennington C., Edwards D. R., Donell S. T., Clark I. M. (2002). The role of chondrocyte senescence in osteoarthritis. Aging Cell 1, 57-65 [DOI] [PubMed] [Google Scholar]
- Prieur A., Peeper D. S. (2008). Cellular senescence in vivo: a barrier to tumorigenesis. Curr. Opin. Cell Biol. 20, 150-155 [DOI] [PubMed] [Google Scholar]
- Regulus P., Duroux B., Bayle P. A., Favier A., Cadet J., Ravanat J. L. (2007). Oxidation of the sugar moiety of DNA by ionizing radiation or bleomycin could induce the formation of a cluster DNA lesion. Proc. Natl. Acad. Sci. USA 104, 14032-14037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F., Campisi J., Bhaumik D. (2007). Two faces of p53: aging and tumor suppression. Nucleic Acids Res. 35, 7475-7584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F., Coppe J. P., Patil C. K., Hoeijmakers W. A. M., Munoz D., Raza S. R., Freund A., Campeau E., Davalos A. R., Campisi J. (2009). Persistent DNA damage signaling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 11, 973-979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E. P., Boon C., Redon C., Bonner W. M. (1999). Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146, 905-916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W. (1997). Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593-602 [DOI] [PubMed] [Google Scholar]
- Takai H., Smogorzewska A., de Lange T. (2003). DNA damage foci at dysfunctional telomeres. Curr. Biol. 13, 1549-1556 [DOI] [PubMed] [Google Scholar]
- Varadaraj A., Dovey C. L., Laredj L., Ferguson B., Alexander C. E., Lubben N., Wyllie A. H., Rich T. (2007). Evidence for the receipt of DNA damage stimuli by PML nuclear domains. J. Pathol. 211, 471-480 [DOI] [PubMed] [Google Scholar]
- Ventura A., Kirsch D. G., McLaughlin M. E., Tuveson D. A., Grimm J., Lintault L., Newman J., Reczek E. E., Weissleder R., Jacks T. (2007). Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661-665 [DOI] [PubMed] [Google Scholar]
- Verdun R. E., Crabbe L., Haggblom C., Karlseder J. (2005). Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol. Cell 20, 551-561 [DOI] [PubMed] [Google Scholar]
- Wajapeyee N., Serra R. W., Zhu X., Mahalingam M., Green M. R. (2008). Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell 132, 363-374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Jurk D., Maddick M., Nelson G., Martin-Ruiz C., von Zglinicki T. (2009). DNA damage response and cellular senescence in tissues of aging mice. Aging Cell 8, 311-323 [DOI] [PubMed] [Google Scholar]
- Wang Y., Blandino G., Givol D. (1999). Induced p21waf expression in H1299 cell line promotes cell senescence and protects against cytotoxic effect of radiation and doxorubicin. Oncogene 18, 2643–2649 [DOI] [PubMed] [Google Scholar]
- Xu Z. X., Timanova-Atanasova A., Zhao R. X., Chang K. S. (2003). PML colocalizes with and stabilizes the DNA damage response protein TopBP1. Mol. Cell. Biol. 23, 4247-4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Zender L., Miething C., Dickins R. A., Hernando E., Krizhanovsky V., Cordon-Cardo C., Lowe S. W. (2007). Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656-660 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.