Abstract
Meiosis is a crucial process for the production of functional gametes. However, the biological significance of many genes expressed during the meiotic phase remains poorly understood, mainly because of the lethal phenotypes of the knockout mice. Functional analysis of such genes using the conditional knockout approach is hindered by the lack of suitable Cre transgenic lines. We describe here the generation of transgenic mice expressing Cre recombinase under the control of the meiotic Spo11 gene. Using LacZ-R26loxP and EYFP-R26loxP reporter mice, we show the specific expression and activity of Cre during meiosis in males and females. Spo11Cre mice were then crossed with floxed Nbs1 and JAM-C mice to produce conditional knockouts. A strong reduction of Nbs1 and JAM-C protein levels was found in the testis. Although Nbs1-deleted mice developed minor gonadal abnormalities, JAM-C-knockout mice showed a spermiogenetic arrest, as previously described for the null mice. These results provide strong evidence that Spo11Cre transgenic mice represent a powerful tool for deleting genes of interest specifically in meiotic and/or in postmeiotic germ cells.
Keywords: Cre recombinase, JAM-C, Meiosis, Nbs1, Spo11
Introduction
Gametogenesis is a maturative process of germ cell formation, in which mitotic, meiotic and differentiative phases orderly occur. In males, meiosis occurs postnatally, when spermatogonia first differentiate into spermatocytes. After two subsequent divisions, spermatocytes give rise to haploid spermatids, which enter into the spermiogenetic process (de Rooij and Mizrak, 2008; Rossi et al., 2003). During this phase, spermatids elongate, condense their chromatin, develop the flagellum and the acrosome, and lose most of their cytoplasm to become spermatozoa (Cheng and Mruk, 2002). In females, meiosis starts during fetal development, when primordial germ cells (PGCs) stop proliferating, and it is eventually completed postnatally, during fertilization.
Although it is well known that meiotic and postmeiotic alterations are critical for the production of functional gametes, the molecular pathways involved in the correct initiation, signaling and resolution of these differentiative processes are poorly understood. This is mainly due to the early embryonic lethality of mice lacking functional copies of candidate genes implicated in meiotic progression. These findings suggest that many of the proteins involved in meiotic progression also have an important role in other cellular processes.
The Nijmegen breakage syndrome (NBS) gene, which encodes nibrin, a protein essential for DNA double-strand breaks (DSB) repair events (Carney et al., 1998; Chapman and Jackson, 2008; Chen et al., 2000; Difilippantonio et al., 2007; Stracker et al., 2007; Zhu et al., 2001), is one such example. Indeed, Nbs1-null mice show early embryonic lethality and have poorly developed embryonic and extra-embryonic tissues (Zhu et al., 2001). However, mice with a hypomorphic mutation in Nbs1 are viable, but females are sterile as result of arrest during meiosis at pachytene stage, whereas males show a delay in the first wave of spermatogenesis (Cherry et al., 2007; Difilippantonio et al., 2005; Morales et al., 2005). Another such example is the junctional adhesion molecule-C (JAM-C), a cell-surface protein of the immunoglobulin superfamily. These proteins colocalize with tight junctions in endothelium and epithelium and they are also found on blood cells, where they are mainly involved in inflammatory events (Cera et al., 2004; Orlova et al., 2006; Santoso et al., 2005; Santoso et al., 2002; Zimmerli et al., 2009). Gliki and co-workers (Gliki et al., 2004) generated JAM-C-null mice by targeted disruption. Although a major proportion of the JAM-C-null offspring die during postnatal development because of infections, about 40% of the mutants are viable. JAM-C-deficient males are infertile and fail to produce mature sperm cells. The testes of JAM-C-null mice were reported to be about 50% smaller than those of littermate controls and lacked differentiated elongated spermatids.
Conditional gene knockout is a commonly used technique to examine the biological function of essential genes, which result in embryonic lethality, in postnatal tissues. This is accomplished by generating conditional alleles of genes by flanking them with loxP sites, so that the gene can be deleted by crossing to Cre expressing transgenic mice. Unfortunately, to date there are no Cre transgenic lines that can be used to delete genes in early stages of meiosis. We describe here the generation of a Cre transgenic line under the genetic control of Spo11.
S. cerevisiae sporulation protein (Spo11) is an evolutionarily conserved topoisomerase-like protein that, in mammals, is functionally expressed in gonads of both male and female during meiosis and is responsible for physiological DNA DSB formation during the early meiotic prophase in spermatocytes and oocytes (Baudat et al., 2000; Keeney et al., 1999; Klein et al., 2002; Romanienko and Camerini-Otero, 1999; Romanienko and Camerini-Otero, 2000). To obtain transgenic mice expressing Cre during early meiosis, we used the bacterial artificial chromosome (BAC) engineering technology in which an IRES-Cre sequence has been inserted into the murine Spo11 locus. After studying the developmental stage of germ cells in which Cre was functional, we tested the efficiency of deletion by breeding the Spo11Cre mice with mice containing conditional alleles of Nbs1 (Reina-San-Martin et al., 2005) or JAM-C (H. F. Langer and T.C., unpublished results). The deletion of the conditional alleles is expected to generate a spermatogenic arrest during the meiotic and postmeiotic phases, respectively.
In Spo11Cre transgenic mice, the Cre recombinase begins to be specifically expressed during meiotic germ cell development. We found that Cre expression driven by Spo11 regulatory regions is able to delete Nbs1loxP and JAM-CloxP alleles, partially displaying the Nbs1 hypomorphic gonadal phenotype and fully recapitulating the JAM-C-null phenotype in male testis. In summary, we describe here the generation of a novel transgenic mouse model in which exogenous Cre is expressed and fully functional during meiosis in both sexes.
Results
Generation of Spo11-IRES-Cre mice
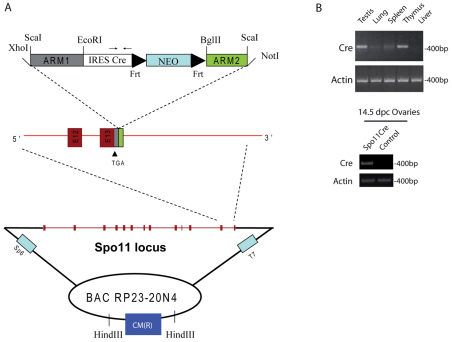
To generate transgenic mice expressing the Cre recombinase in germ cells, presumably in the early stages of meiosis, we inserted the Cre cDNA within the Spo11 genomic locus cloned in a BAC vector. The structure of the targeting construct used to generate transgenic mice expressing Cre during the early meiotic phase of spermatogenesis and oogenesis is depicted in Fig. 1A. The Cre cDNA was inserted immediately downstream from the stop codon present in exon 13 of Spo11 BAC by recombineering as previously described (Liu et al., 2003; Yang and Sharan, 2003), and used for generation of transgenic mice. Two founder mice, D5 and H9, were analyzed in detail. Both founders gave germline transmission and the expression of the transgene was identical to each other in every aspect studied. Furthermore, RT-PCR analysis of Cre mRNA in different tissues from transgenic mice revealed Cre expression in adult testis and thymus, and in ovary from 14.5 days post coitum (d.p.c.) embryos (Fig. 1B, upper and lower panels, respectively), consistent with the reported Spo11 expression pattern (Romanienko and Camerini-Otero, 1999).
Fig. 1.
Generation and evaluation of Spo11-IRES-Cre mice. (A) BAC targeting of the murine Spo11 locus after the stop codon of the gene. The DNA fragment containing the homology regions ARM1 and ARM2 for the DNA recombination and the IRES-Cre-Neomycin (Neo) sequences was obtained by ScaI enzymatic digestion from the modified PL459 plasmid. The restriction enzymes used to insert the homology regions into the original PL459 plasmid are indicated. Spo11 exons are shown as red boxes, and exons 12 and 13 are magnified. Frt sites flanking the Neo cassette are shown. In the IRES-Cre sequence the location of primers used for Cre analyses is shown. CM(R) represents the chloramphenicol resistance gene used as a probe to identify founder mice. (B) Representative RT-PCR analysis of testis, lung, liver, spleen and thymus mRNA isolated from a 2-month-old mouse of the D5 founder line (top panel) and ovaries mRNA from 14.5 d.p.c. embryo (bottom panel), showing tissue-specific Cre mRNA expression. Actin PCR was used as a loading control.
Spo11-Cre is expressed in meiotic germ cells
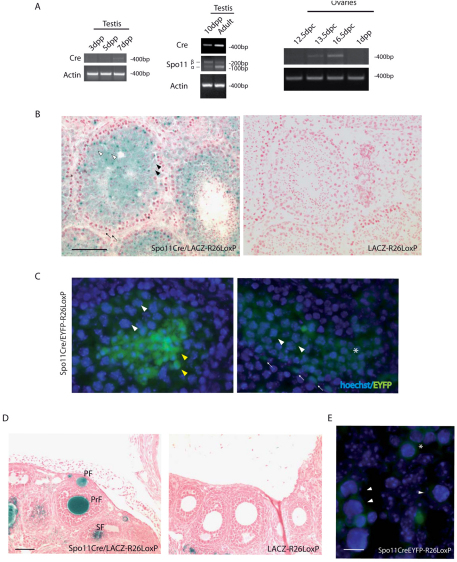
To test the in vivo expression of Cre in germ cells during development, we analyzed Cre mRNA levels by semiquantitative RT-PCR in testes and ovaries of Spo11Cre transgenic mice. Cre mRNA expression started to be evident in testes from 7 days post partum (d.p.p.) mice (Fig. 2A, left panel), when pre-meiotic differentiating spermatogonia are the most abundant germ cells population. Cre mRNA was also evident in testes from 10 d.p.p. mice, which mostly contained spermatocytes at the leptotene stage, but appeared to reach its maximal expression levels in the adult testis (Fig. 2A, middle panel), where pachytene spermatocytes and postmeiotic cells were the prevailing germ cell types.
Fig. 2.
Spo11-Cre is expressed in meiotic germ cells. (A) Semiquantitative PCR of cDNA prepared from 3, 5, 7, 10 d.p.p. and adult testes (left and middle) and from 12.5, 13.5, 16.5 d.p.c. and 1 d.p.p. ovaries (right). Cre and Spo11 alpha and beta isoform mRNA (Bellani et al., 2010; Romanienko and Camerini-Otero, 2000) amplifications are shown. Actin PCR was used as a loading control. (B) β-gal-stained sections of seminiferous tubules of adult testis counterstained with nuclear Fast Red. Black arrows indicate β-gal-negative spermatogonia and somatic Sertoli cells; black and white arrowheads show β-gal-positive pachytene spermatocytes and spermatids, respectively. Asterisk indicates lepto-zygotene spermatocytes. (C) EYFP expression in testis from a 21 d.p.p. male. White and yellow arrowheads indicate pachytene spermatocytes and spermatids, respectively. White asterisk shows early meiotic spermatocytes and white arrows indicate spermatogonia or preleptotene cells. (D) β-gal-stained sections of adult ovaries counterstained with nuclear Fast Red. PF, primordial follicles; PrF, primary follicles; SF, secondary follicles. (E) EYFP expression in fetal ovaries from 14.5 d.p.c. mice. Arrowheads indicate meiotic oocytes and the asterisk, early meiotic preleptotene cells. The cell types were recognized by differential Hoechst 33342 staining of the nuclei. Scale bars: 100 μm in B and D; 10 μm in E.
In fetal ovaries, Cre mRNA was evident at 13.5 d.p.c., coincidently with the beginning of meiosis, it peaked at 16.5 d.p.c., when most of the oocytes are at the pachytene stage, and was downregulated at birth (Fig. 2A, right panel). To verify the specificity of excision achieved by the Spo11Cre transgenic lines, we crossed the two independent founders with ROSA26 loxP-β-galactosidase (lacZ-R26loxP/loxP) mice (Soriano, 1999). These mice express the lacZ transgene only after Cre-mediated excision. The D5 and H9 transgenic Spo11CrelacZ-R26loxP/loxP mice showed a similar intensity of β-galactosidase (β-gal) staining in adult testes and ovaries (Fig. 2B,D).
In Spo11CrelacZ-R26loxP/loxP transgenic testes, Cre activity, as monitored by β-gal staining, was found within the seminiferous tubules as early as at the meiotic stages. Spermatocytes were β-gal-positive cells and corresponded to the germ cell type in which Spo11 is expressed. The basal layer of the tubules, where spermatogonia and somatic cells are located, appeared to be negative for β-gal staining (Fig. 2B, black arrows). A detailed analysis of testis sections revealed an apparent lack of β-gal staining also in germ cells at the lepto-zygotene stage of the first meiotic division, identifiable by their position within tubules and by the typical staining of the condensed chromatin (Fig. 2B, asterisk). On the contrary, a strong and consistent staining was observed in pachytene spermatocytes and in postmeiotic cells (Fig. 2B, black and white arrowheads, respectively), namely round and elongating spermatids. We then bred Spo11Cre transgenic mice with ROSA26 loxP enhanced yellow fluorescent protein (EYFP-R26loxP/loxP) mice (Srinivas et al., 2001). Using this reporter gene, we also observed a consistent EYFP staining of spermatocytes in earlier stages of meiosis and occasionally in some spermatogonia or preleptotene spermatocytes (Fig. 2C).
In adult female gonads, we detected a strong signal exclusively in oocytes from primordial, primary and secondary follicles (Fig. 2D), indicating that Cre deletion occurred during the early phases of embryogenesis when the meiotic process takes place. To verify the Cre-mediated deletion event during the meiotic phase, we analyzed fetal ovaries from 14.5 d.p.c. embryos born from mating Spo11Cre and EYFP-R26loxP/loxP mice. The EYFP signal was observed specifically both in early and late meiotic germ cells, identified according to previous reports (Dolci and De Felici, 1990), but not in somatic cells (Fig. 2E). Together, these results demonstrate that the Cre transgene driven by Spo11 regulatory elements is functionally expressed in vivo, in both male and female mice starting from the meiotic stages of gametogenesis.
Nbs1Δ/− mice partially recapitulate the meiotic phenotype of hypomorphic Nbs1 mice
It is known that Nbs1 hypomorphic females lack oocytes and that spermatogenesis is delayed in the male mice (Difilippantonio et al., 2005). To investigate the role of Nbs1 during the meiotic phase of oogenesis and spermatogenesis in a conditional null background, we crossed Spo11Cre mice with the Nbs1loxP/− mice, which we have previously used to investigate the role of Nbs1 protein in B cells (Reina-San-Martin et al., 2005).
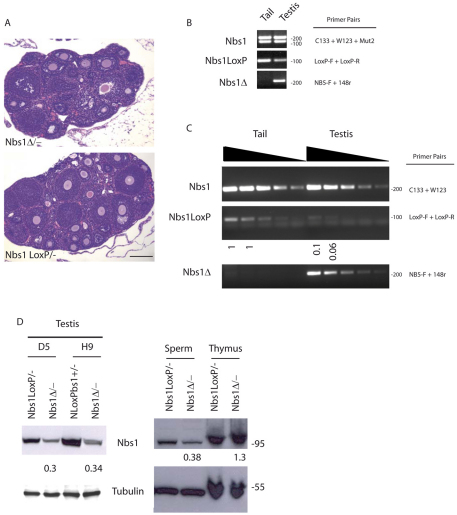
Transgenic Spo11CreNbs1loxP/− (Nbs1Δ/−) males and females were both fertile. Indeed, when females were euthanized at 5 weeks of age, they did not show the canonical Nbs1 hypomorphic phenotype (Difilippantonio et al., 2005), even though we noticed that about 50% of the analyzed animals showed a reduction of the ovary size and oocyte number (Fig. 3A and supplementary material Fig. S1A). To investigate the efficiency of Cre deletion in oocytes, we crossed two Nbs1Δ/− females with wild-type males. One hundred pups were screened by PCR to verify the presence of loxP sites or the deleted band (see below) in their tail DNA. We estimated that 52% of the newborns had undergone the deletion as expected for mendelian inheritance and only 6% were loxP positive (supplementary material Fig. S1B).
Fig. 3.
Nbs1Δ/− mice do not show the canonical meiotic phenotype of hypomorphic Nbs1 mice. (A) Hematoxylin and eosin stained sections of ovaries from 5-week-old females. Scale bar: 100 μm. (B) Identification of Cre-mediated recombination by semiquantitative PCR of genomic DNA obtained from tail and testis of 2-week-old Nbs1loxP/− and Nbs1Δ/− mice. Nbs1 genotype was determined by C133, W123 and Mut2 primers, as previously described (Zhu et al., 2001). Primer pairs LoxP-F and LoxP-R show the extent of Cre-mediated deletion of loxP sites, whereas NB5F and 148-R were used for detection of Cre-mediated band (Reina-San-Martin et al., 2005). (C) Identification of Cre-mediated recombination. For PCRs we used sequential tenfold dilutions of genomic DNA (indicated by wedges) obtained from tail and testis of 3-week-old mice. As determined by semiquantitative PCR, 90–94% of the loxP-flanked Nbs1 alleles were deleted from the genome in Nbs1Δ/− testis cells (middle). NBS primers C133 and W123 in the NBS 59 region were used as control to normalize for the amount of input DNA (top). Primer pairs NB5F and 148-R detected the Cre-mediated recombination band (bottom). Data are representative of three independent experiments. (D) Western blot analysis of murine Nbs1 in whole-cell extracts prepared from total testis (left panel), sperm and thymocytes (right) of Nbs1Δ/− mice relative to Nbs1loxP/− normalized to tubulin. Numbers reflect the intensity of bands representing the amount of Nbs1 protein. No reduction of the Nbs1 protein was evident in thymocytes. Data shown are representative of two independent experiments.
Conditionally deleted males did not present the strong delay of spermatogenesis at different ages that was instead described for the hypomorphic Nbs1 mice (Difilippantonio et al., 2005) (data not shown). The extent of Cre-mediated excision was also evaluated in males. Using PCR analysis, we monitored the disappearance of the loxP sites and the appearance of a band that is amplified as a result of the deletion event in testis DNA of Nbs1Δ/− males (Fig. 3B,C, respectively). Undiluted or diluted DNA extracts, obtained from tails and germ cells isolated from testis of 2 and 3 weeks old animals were analyzed by semiquantitative PCR. As expected, we did not observe the deleted band in genomic DNA obtained from tails but a newly formed strong band was amplified in all DNA dilutions prepared from testicular cells, indicating that a deletion event occurred following Cre activity (Fig. 3B,C, bottom panels). On the contrary, loxP sites were present in tail DNA in all dilutions tested and at extremely low levels in testis samples, confirming again that a dramatic deletion event occurred. We estimated a 90–94% reduction for the loxP sites, as calculated by using Nbs1 PCR product as a loading control (Fig. 3C, top and middle panels).
Because we did not observe any spermatogenetic delay or fertility problems, despite an almost 100% efficiency of deletion, we evaluated Nbs1 protein levels in transgenic Nbs1Δ/− testicular cells at 3 weeks of age, when spermatocytes are the predominant germ cell type in the testis (Fig. 3D, left). Densitometric analysis revealed a residual Nbs1 band corresponding to 30% of the wild-type band in both D5 and H9 transgenic founder lines. To rule out the possibility that Nbs1 is expressed in contaminant spermatogonia or Sertoli cells, we prepared protein lysates from spermatozoa, isolated from the cauda of Nbs1Δ/− and Nbs1loxP/− mice, and we compared the relative Nbs1 protein level in the two samples (Fig. 3D, right). A similar 70% reduction in Nbs1 was observed in spermatozoa from transgenic mice. However, we cannot exclude the possibility that the residual Nbs1 protein arises from somatic epididymal cells that contaminate these sperm preparations. Nbs1Δ/− thymus, which was positive for Cre mRNA expression (Fig. 1C), did not show decreased levels of Nbs1 protein compared with levels in Nbs1loxP/− mice, indicating the absence of Cre activity in this organ.
JAM-CΔ/− mice display the postmeiotic phenotype of JAM-C-null mice
Since Spo11Cre mice showed the maximal Cre activity in pachytene spermatocytes, we thought that the Spo11-Cre protein would be useful to reveal a phenotype in postmeiotic germ cells. To test this hypothesis, we bred the Spo11Cre mice with a conditional knockout mouse model of JAM-C (H. F. Langer and T.C., unpublished results), whose homozygous deletion causes defects during spermiogenesis (Gliki et al., 2004). By breeding the Spo11CreJAM-C+/− mice with JAM-CloxP/− mice, we generated Spo11CreJAM-CloxP/− (JAM-CΔ/−) mice, which were analyzed at different stages of development. JAM-CΔ/− mice showed a reduction in testis size (collected at 7 months of age) compared with JAM-CloxP/− mice (supplementary material Fig. S2A). Starting from 2 months of age, we observed a strong decrease in sperm number ranging from 90% to 95% relative to control mice.
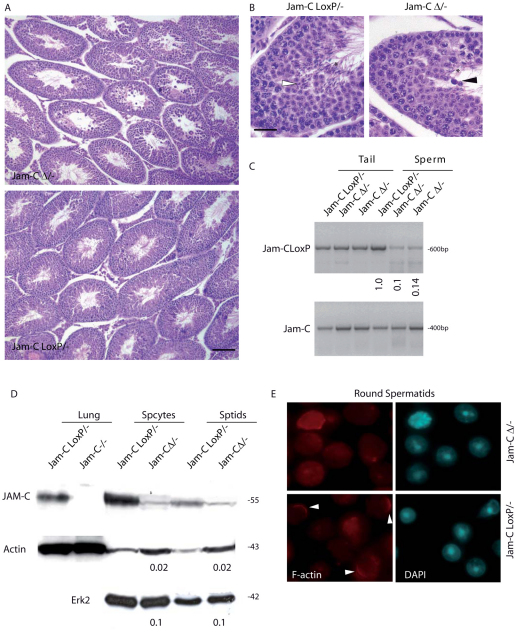
JAM-C-null mice are infertile as a result of a maturation arrest from round spermatids to elongated spermatids. This phenotype is related to the role of JAM-C in the assembly of a cell polarity complex (Gliki et al., 2004). Histological analysis of JAM-CΔ/− testes showed that more than 80–90% of the tubules were arrested at the round stage of spermatid differentiation and rare elongating spermatids were found (Fig. 4A). Within the JAM-CΔ/− tubules, several degenerating spermatids and spermatocytes were also observed (Fig. 4A,B). TUNEL assay confirmed a widespread apoptosis in JAM-CΔ/− testes but not in mice expressing Spo11CreJAM-C+/−, which were used as control (supplementary material Fig. S2B), indicating that the germ cell degeneration is effectively due to the deletion of JAM-C allele and not to the presence of the transgene per se.
Fig. 4.
JAM-CΔ/− mice display the postmeiotic phenotype of JAM-C-null mice. (A) Hematoxylin and eosin stained sections of testis from 10-week-old littermates showing the depletion of elongated spermatids in JAM-CΔ/− relative to control JAM-CloxP/− mice and the consequent spermatogenetic arrest. Scale bar: 100 μm. (B) Higher magnification of sections in A of seminiferous tubules from JAM-CΔ/− and JAM-CloxP/− mice. The white arrowhead indicates elongated spermatids in control littermate and the black arrowhead shows degenerating spermatids in JAM-CΔ/− mice. Asterisks indicate spermatozoa in control and JAM-CΔ/− mice. Scale bar: 10 μm. (C) Identification of Cre-mediated recombination by semiquantitative PCR of genomic DNA obtained from tails and sperm of JAM-CloxP/− and JAM-CΔ/− mice. JAM-C F1 and JAM-C R1 primer pairs show the extent of Cre-mediated deletion of loxP sites, whereas JAM-C F1 and JAM-C R4 were used for detection of the null allele, used as control to normalize for the amount of input DNA. The ratio between the intensity of the JAM-CloxP/− bands in sperm and tail was arbitrarily considered 1. Data shown are representative of three independent experiments. (D) Western blot analysis of JAM-C in whole-cell extracts prepared from lung, spermatocytes (Spcytes) and spermatids (Sptids) of JAM-C−/−, JAM-CΔ/− mice relative to JAM-CloxP/−. Numbers represents the ratio of residual JAM-C protein in JAM-CΔ/− germ cells with respect to JAM-CloxP/− germ cells, using either actin or Erk2 as loading controls. (E) Immunofluorescence in freshly isolated round spermatids showing polarized F-actin (phalloidin, red), in most of the control JAM-CloxP/− cells (arrowheads), but disorganized in JAM-C-deleted spermatids.
To monitor the Cre-mediated recombination event, we performed a semiquantitative PCR analysis of loxP sites on DNA extracted from tails and spermatozoa of adult JAM-CloxP/− and JAM-CΔ/− mice. Whereas the tail genomic DNA showed a similar loxP content, the JAM-CΔ/− sperm DNA revealed only 10–20% of the original loxP sites compared with JAM-CloxP/− sperm DNA (Fig. 4C), providing an explanation for the previous observed phenotype. We then analyzed the protein levels of JAM-C in isolated spermatocytes and spermatids that were conditionally deleted. As shown in Fig. 4D, JAM-C was expressed in normal JAM-CloxP/− spermatocytes and its levels were higher than those in spermatids. However, in JAM-CΔ/− mice, we found a dramatic reduction of the protein levels in isolated spermatocytes and spermatids, indicating that the deletion occurred efficiently. The reduction ranged between 90% and 98%, according to whether Erk2 or actin were used as loading controls. It is known that F-actin is abnormally distributed in JAM-C-null spermatids, and that bundles fail to form at Sertoli–spermatid junctions. We observed similar cytoskeletal defects in most JAM-CΔ/− isolated round spermatids. As previously reported in JAM-CloxP/− mice (Gliki et al., 2004), F-actin appeared uniformly distributed within the cytoplasm in spermatids from JAM-CΔ/− mice, rather than being localized in restricted zones of the cells (Fig. 4E). We detected the F-actin polarization only in a subpopulation of control spermatids, probably because we disrupted the Sertoli–spermatid connection during germ cell purification, but this polarization was preserved extremely rarely in deleted cells.
Discussion
Generation of Spo11-IRES-Cre mice and Cre expression
Conditional gene targeting has proved to be a powerful approach to study the function of genes in particular cell lineages without compromising gene expression in other tissue and cell compartments (Lobe and Nagy, 1998). The success of this approach clearly relies on the availability of mice that express Cre in the cell or tissue of interest. Several mouse models of Cre recombinase, conditionally expressed in germ cells during different developmental stages, have been reported: PGK-1-Cre, TNAP-Cre and Nanos3-Cre mice express Cre in primordial germ cells (Lallemand et al., 1998; Lomeli et al., 2000; Suzuki et al., 2008); Zp3-Cre mice in oocytes (Lewandoski et al., 1997); c-Kit-Cre in germ cells and Stra8-Cre in male premeiotic germ cells (Bergqvist et al., 1998; Sadate-Ngatchou et al., 2008); Pgk2-Cre and Sycp1-Cre in spermatocytes (Ando et al., 2000; Bhullar et al., 2001; Chung et al., 2004; Vidal et al., 1998); and Prm1-Cre in haploid spermatid (O'Gorman et al., 1997; Schmidt et al., 2000). However, several of these transgenic and knock-in mice show ectopic or mosaic expression of Cre (Bergqvist et al., 1998; Bhullar et al., 2001; Lallemand et al., 1998; Lomeli et al., 2000; Suzuki et al., 2008), and inefficient or abnormal Cre activity (Schmidt et al., 2000; Suzuki et al., 2008). Furthermore, even though the Cre-expressing mice have been proposed to be a valuable tool for studying genes potentially essential for spermatogenesis or oogenesis, very few have been crossed to test the actual Cre-mediated deletion of meiotic or postmeiotic genes targeted with loxP sequences.
Sycp1-Cre is a current mouse model in which Cre is restricted to the zygotene through the pachytene and later stages during male meiosis, but it is not expressed during oogenesis (Chung et al., 2004; Sage et al., 1999; Vidal et al., 1998). These mice have been successfully used to delete the androgen receptor gene in spermatids from young animals, but the Cre-induced recombination was completely lost in older mice (Tsai et al., 2006). The same failure of loxP sites recombination over several generations was described for germ-cell-specific nuclear-factor-κB knockdown mice obtained through a floxed IkB kinase β gene (Rasoulpour and Boekelheide, 2006).
In this study, we report a new meiotic deleter strain, Spo11Cre mice, which show specific Cre activity during meiosis both in male and female germ cells. Since the Spo11 promoter region has not been fully characterized and it is well known that other regulatory elements, such as enhancer sequences, are important for proper spatial and temporal regulation of transgenes, we inserted the Cre cDNA sequence within a BAC containing the entire genomic Spo11 locus, to correctly drive Cre mRNA expression.
In Spo11CreLacZ-R26loxP/loxP males, we observed that Cre activity was apparently not detectable during the lepto-zygotene stage, even though Cre is expressed at 7 d.p.p., but it was clearly evident at the pachytene stage of meiosis, as judged by β-gal staining of the testes. This result is in apparent contrast with the expression pattern and activity of Spo11 gene, which was found in all the stages of the meiotic prophase up to pachytene cells (Baudat et al., 2000; Romanienko and Camerini-Otero, 2000; Shannon et al., 1999). This discrepancy can be explained by a difference in the timing of Cre translation with respect to Spo11, and/or by a Cre recombinase activity below the threshold detectable by β-gal staining at the beginning of male meiosis, arguing for the requirement of an accumulation of Cre to efficiently delete loxP sites. This latter hypothesis is supported by the evidence that Cre is expressed at lower levels in 10 d.p.p. versus adult testis, when leptotene and lepto-zygotene cells are first observed. Moreover, it is known that Spo11 gene expression actually increases in pachytene spermatocytes, where an alternative splicing product accumulates (Baudat et al., 2000; Bellani et al., 2010). However, by using EYFP as a reporter gene, whose detection assay is more sensitive compared with β-gal staining, we also found Cre activity in spermatogonia committed to meiotic entry and early meiotic cells.
In female gonads, positive β-gal staining was evident in diplotene oocytes within primordial follicles, suggesting that Cre was expressed at earlier meiotic stages. This hypothesis was confirmed by revealing Cre in fetal ovaries through semiquantitative RT-PCR analysis of Cre expression and EYFP staining. Notably, we found that Cre is expressed at the beginning of meiosis, it increases at the pachytene stage and it is subsequently downregulated at birth. In 14.5 d.p.c. ovaries, preleptotene and early meiotic oocytes expressed EYFP, suggesting that the timing of Spo11Cre transgene expression overlapped that of the endogenous Spo11 gene.
To further validate actual gene deletions in meiosis, we used Spo11Cre to delete floxed Nbs1 and JAM-C alleles, which have been described to have a role in meiotic and postmeiotic cells, respectively (Difilippantonio et al., 2005; Gliki et al., 2004).
Nbs1 and JAM-C gene deletions mediated by Spo11-Cre
The role of Nbs1 in gametogenesis has been previously studied in hypomorphic animals (Difilippantonio et al., 2005) because null mice are embryonic lethal (Zhu et al., 2001). Although females are completely sterile, the onset of spermatogenesis is delayed in Nbs1 hypomorphic males. Our Nbs1Δ/− females were fertile despite an efficient deletion occurring, as evaluated in the progeny of these mice. These results might suggest a potential crucial role of Nbs1 before meiosis, during the mitotic phases of oogenesis. However, we observed a reduction of ovarian size and oocyte number in about 50% of Nbs1Δ/− females. Because Cre activity, as monitored by EYFP, was found in preleptotene oocytes, we hypothesize that a portion of the preleptotene oocytes that underwent deletion are also sensitive to Nbs1 levels. Nbs1Δ/− males were also fertile and we did not observe any gross spermatogenetic defect by morphological analysis. Although Cre-mediated deletion was almost complete in Nbs1Δ/− mice, western blot analysis indicated that a small amount of Nbs1 protein was still detectable in Nbs1Δ/− isolated spermatozoa. If this residual protein is not due to somatic contaminants, this discrepancy could be due to the stability of the protein already present before the Cre-mediated deletion of the conditional allele, and, as in females, Nbs1 could be required also before meiosis. However, we cannot completely exclude a detrimental effect of the deleted form of Nbs1 protein that is produced in hypomorphic Nbs1 mice (Difilippantonio et al., 2005), which is not present in our conditionally deleted mice.
Proper gene ablation in meiotic cells by Spo11-Cre was further confirmed when we specifically targeted JAM-C, a gene involved in cytoskeletal protein polarization during spermatid differentiation (Gliki et al., 2004). In JAM-CΔ/− mice, we noticed an apparent increase in actin protein levels in germ cells, which might be correlated to the dramatic alteration of F-actin polarization that we observed in mutant spermatids.
Similarly to what observed in JAM-C-knockout mice (Gliki et al., 2004), adult JAM-CΔ/− mice displayed a consistent reduction of testis size and a strong decrease in sperm number. A detailed analysis showed an arrest of round spermatid differentiation and few elongating spermatids were found within the affected tubules.
Our conditional mouse model provides a clear evidence of a direct role played by JAM-C during spermiogenesis, as previously suggested by the JAM-C-null phenotype (Gliki et al., 2004). Furthermore, with the use of this conditional knockout strategy we demonstrated that the testicular phenotype of JAM-C-null mice is germ cell autonomous and we could exclude defects during the mitotic phase of spermatogenesis. In all the different breeding that we have generated, we did not observe a decrease in the efficiency of deletion in aged mice, as described for Sycp1-Cre mice (Rasoulpour and Boekelheide, 2006; Tsai et al., 2006). Moreover, we did not observe the illegitimate chromosome rearrangements in transgenic spermatids that were reported in mice carrying a promoter fragment of Prm1 driving Cre in spermatids (Schmidt et al., 2000).
Collectively, our results demonstrate that the expression pattern of Cre recombinase overlaps with that of the endogenous Spo11 gene during the meiotic prophase of both oogenesis and spermatogenesis. Its expression, which reaches greatest levels at the pachytene stage, is able to efficiently recombine the loxP sites in the Nbs1 and JAM-C loci specifically in meiotic germ cells. Thus, Spo11Cre mice might be useful to investigate whether a gene required for fertility has an essential role in the meiotic or post-meiotic phases of gametogenesis.
Materials and Methods
Generation of mice
To generate transgenic mice, the murine BAC RP23-20N4, which consists of a 260-kilobase (kb) insert, including 188 kb of sequence upstream and 57 kb of sequence downstream of the Spo11 initiation and stop codons, respectively, was engineered by homologous recombination as previously described (Liu et al., 2003; Yang and Sharan, 2003). Briefly, we electroporated the IRES-Cre-Neo fragment of the PL459 plasmid (Liu et al., 2003), flanked by two homology regions (ARM1 and ARM2) of the Spo11 gene immediately downstream from the stop codon site, into bacteria containing the Spo11 BAC. XhoI-ScaI-Spo11 for EcoRI-Spo11 rev and BglII-Spo11 for ScaI-Not1-Spo11 rev pair primers were used to amplify ARM1 (295 bp) and ARM2 (135 bp) fragments (supplementary material Table S1).
The IRES sequence was inserted upstream of Cre to allow proper translation of the Cre protein. The Neomycin (Neo) resistance was used for the screening of the positive recombined bacteria containing the Spo11-IRES-Cre-Neo BAC. Subsequently, the recombined BAC was electroporated into (L)-arabinose inducible EL250 330G11 bacteria for flp-mediated Neo deletion. Spo11-Ires-Cre BAC was purified and microinjected into the pronuclei of fertilized Balb C57 eggs by standard methods. Founder lines were identified by southern blotting analysis using a probe amplified in the chloramphenicol resistance cassette with CMfor and CMrev primers. All experiments were performed with D5 and H9 founder lines, unless otherwise noted.
Transgenic founders were crossed to homozygous LacZ-R26loxP (a generous gift from Richard J. Hodes, EIB_NCI_NIH, USA), homozygous EYFP-R26loxP (kindly provided by GianGiacomo Consalez, University of San Raffaele, Milan, Italy), Nbs1loxP/− (Zhu et al., 2001) and JAM-CloxP/− mice. Briefly, the ploxPFRTPGKneoFRT (kindly provided by Günther Schütz, German Cancer Research Center, Heidelberg, Germany) containing exon 1 and 560 bp of the promoter region of JAM-C gene flanked by loxP sites, as well as the 6.4 kb 5′ long and 3 kb 3′ short arms for homologous recombination, was used as targeting vector. The targeted ES cells were injected into blastocysts and chimeric mice were crossed with C57BL/6 to select for germline transmission. Positive mice were crossed with an flp-deleter mouse line, resulting in a wt/flox mouse. JAM-CloxP/− mice were propagated in a mixed 129:B6 background. For Nbs1 genotyping C133, W123 and Mut2 primers were used as previously described (Zhu et al., 2001). JAM-C F1, JAM-C R1 and JAM-C F1 and JAM-C R4 were used for tail DNA screening of JAM-C mice. JAM-C F1 and JAM-C R1 primers amplify a floxed band of 640 bp and a wild-type band of 460 bp, whereas JAM-C F1 and JAM-C R4 primers amplify a floxed band of 1.3 kb, a wild-type band of 1 kb and a knockout band of 400 bp. All experiments were performed in compliance with the Tor Vergata University Institutional Animal Care and National Institutes of Health Intramural Animal Care and Use program. All procedures adhered to the standards published in Guide for the Care and Use of Laboratory Animals.
Germ cell isolation and sperm count
Testicular cells were obtained at 10, 12 d.p.p. or from adult mice by sequential digestions in 0.5 mg/ml type II collagenase (Gibco) and saline solution (Cellgro) and used for DNA and protein extraction. Spermatocytes and spermatids were isolated by elutriation as previously described (Barchi et al., 2009; Meistrich, 1977; Pellegrini et al., 2003). Briefly, testes were sequentially digested in 0.25 mg/ml collagenase type IX (Sigma) (dissolved in EKRB), trypsin 0.5 mg/ml (Sigma) and 0.05 mg/ml DNase type I (Sigma). The cell suspension was loaded into the elutriator (JE-6B Beckman). Spermatozoa were squeezed with fine needles in 1 ml PBS from caudae epididymi. Cells were counted on a Thoma chamber and used for DNA and protein extraction.
Semiquantitative PCR
For semiquantitative RT-PCR (sqRT-PCR), testis RNA was extracted using Trizol reagent (Invitrogen) and contaminating genomic DNA was removed using DNA-Free RNA Kit (Zymo Research). cDNA synthesis was subsequently performed with the SuperScript™ III First-Strand Synthesis System (Invitrogen). Spo11-2-F and Spo11-lex-R primers amplify the mRNA for alpha (134 bp) and beta (248 bp) isoforms of Spo11. Actin mRNA was amplified by β-actin F and R primers (400 bp). For semiquantitative genomic PCR, tail and testis DNA was extracted following standard procedures and serial dilutions were prepared. Spo11Cre transgene was detected with Crefor and Crerev pair primers (490 bp). LoxP-F, LoxP-R, NB5F and 148-r amplified the loxP band or the deleted band of Nbs1 as previously described (Reina-San-Martin et al., 2005). According to the different primer sets, samples were amplified at 55–60°C for 25–30 cycles. Primer sequences for sqRT-PCRs are listed in supplementary material Table S1. Densitometry was performed using a Molecular Dynamics Densitometer and ImageQuant software.
Western blotting and immunofluorescence
For western blotting analysis, cells were lysed in 1% Triton X-100 lysis or in RIPA buffers. Sperm protein extraction was performed using Laemmli 1 × buffer. Proteins were separated by 10% SDS-PAGE and transferred to nitrocellulose membrane (Amersham). Membranes were incubated overnight at 4°C with rabbit polyclonal anti-Nbs1 (Chen et al., 2000), goat polyclonal anti-JAM-C (1:500, R&D AF1213), rabbit polyclonal anti-Erk2 (sc-154), rabbit polyclonal anti-actin (sc-7210) and mouse monoclonal anti-tubulin (Sigma). The horseradish-peroxidase-conjugated secondary antibodies were revealed by chemiluminescence. Densitometry was performed using a Molecular Dynamics Densitometer and ImageQuant software. Protein values were normalized against tubulin or actin levels. For immunofluorescence analysis, isolated round spermatids were fixed for 10 minutes in 2% paraformaldehyde (PFA), permeabilized with 0.1% Triton X-100 and processed with α-phalloidin-TRITC antibody (1:200, Sigma 77418) and Hoechst 33342 for DNA staining.
Histology
Testes and ovaries were fixed in 10% formalin and paraffin embedded. Sections were stained with hematoxylin and eosin. TUNEL analysis was performed using the In Situ Cell Death Detection kit according to manufacturer's instructions (Roche). For β-galactosidase staining, testes and ovaries were processed as previously described (Sanes et al., 1986). Paraffin sections were counterstained with Nuclear Fast Red (Sigma). For EYFP detection, ovaries were fixed in 4% PFA and then immediately frozen in OCT compound (Tissue-Tek). 5-μm-thick cryosections were counterstained with Hoechst 33342 before observation by epifluorescence using Pan-Neofluar 10 ×/0.30, 20 ×/0.50 or 100 ×/1.3 objective lenses onto an Axioskop microscope (Carl Zeiss). Images were acquired using an RT slider camera (Diagnostic Instruments) and IAS2000 software (Biosystem 82).
Supplementary Material
Acknowledgments
We thank Steve Jay, Fabio Lancia, Simonetta Pucci, Graziano Bonelli and Angela Catizone for technical assistance, Dirk G. de Rooij for helpful suggestions on the manuscript. We are greatful to Richard Hodes for providing the LacZ-R26loxP mice and to Giacomo Consalez for the EYFP-R26loxP mice. This work was supported by grants from the Italian Association for Cancer Research (AIRC), the Italian Ministry of University (Grant Prin 2007 200788TPYE_002) and by the Intramural Research Program of the National Institutes of Health (NIH_NCI). Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/124/1/91/DC1
References
- Ando H., Haruna Y., Miyazaki J., Okabe M., Nakanishi Y. (2000). Spermatocyte-specific gene excision by targeted expression of Cre recombinase. Biochem. Biophys. Res. Commun. 272, 125-128 [DOI] [PubMed] [Google Scholar]
- Barchi M., Geremia R., Magliozzi R., Bianchi E. (2009). Isolation and analyses of enriched populations of male mouse germ cells by sedimentation velocity: the centrifugal elutriation. Methods Mol. Biol. 558, 299-321 [DOI] [PubMed] [Google Scholar]
- Baudat F., Manova K., Yuen J. P., Jasin M., Keeney S. (2000). Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6, 989-998 [DOI] [PubMed] [Google Scholar]
- Bellani M. A., Boateng K. A., Mc Leod D., Camerini-Otero R. D. (2010). The expression profile of the major mouse SPO11 isoforms indicates that SPO11β introduces the double strand breaks and suggests that SPO11α has an additional role in prophase both in spermatocytes and oocytes. Mol. Cell. Biol. 30, 4391-4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergqvist I., Eriksson B., Eriksson M., Holmberg D. (1998). Transgenic Cre recombinase expression in germ cells and early embryogenesis directs homogeneous and ubiquitous deletion of loxP-flanked gene segments. FEBS Lett. 438, 76-80 [DOI] [PubMed] [Google Scholar]
- Bhullar B., Schmidt J. V., Truong T., Rancourt D., van der Hoorn F. A. (2001). Germ cell specific promoter drives ectopic transgene expression during embryogenesis. Mol. Reprod. Dev. 59, 25-32 [DOI] [PubMed] [Google Scholar]
- Carney J. P., Maser R. S., Olivares H., Davis E. M., Le Beau M., Yates J. R., 3rd, Hays L., Morgan W. F., Petrini J. H. (1998). The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93, 477-486 [DOI] [PubMed] [Google Scholar]
- Cera M. R., Del Prete A., Vecchi A., Corada M., Martin-Padura I., Motoike T., Tonetti P., Bazzoni G., Vermi W., Gentili F., et al. (2004). Increased DC trafficking to lymph nodes and contact hypersensitivity in junctional adhesion molecule-A-deficient mice. J. Clin. Invest. 114, 729-738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J. R., Jackson S. P. (2008). Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 9, 795-801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. T., Bhandoola A., Difilippantonio M. J., Zhu J., Brown M. J., Tai X., Rogakou E. P., Brotz T. M., Bonner W. M., Ried T., et al. (2000). Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science 290, 1962-1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C. Y., Mruk D. D. (2002). Cell junction dynamics in the testis: Sertoli-germ cell interactions and male contraceptive development. Physiol. Rev. 82, 825-874 [DOI] [PubMed] [Google Scholar]
- Cherry S. M., Adelman C. A., Theunissen J. W., Hassold T. J., Hunt P. A., Petrini J. H. (2007). The Mre11 complex influences DNA repair, synapsis, and crossing over in murine meiosis. Curr. Biol. 17, 373-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S. S., Cuzin F., Rassoulzadegan M., Wolgemuth D. J. (2004). Primary spermatocyte-specific Cre recombinase activity in transgenic mice. Transgenic Res. 13, 289-294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij D. G., Mizrak S. C. (2008). Deriving multipotent stem cells from mouse spermatogonial stem cells: a new tool for developmental and clinical research. Development 135, 2207-2213 [DOI] [PubMed] [Google Scholar]
- Difilippantonio S., Celeste A., Fernandez-Capetillo O., Chen H. T., Reina San Martin B., Van Laethem F., Yang Y. P., Petukhova G. V., Eckhaus M., Feigenbaum L., et al. (2005). Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat. Cell Biol. 7, 675-685 [DOI] [PubMed] [Google Scholar]
- Difilippantonio S., Celeste A., Kruhlak M. J., Lee Y., Difilippantonio M. J., Feigenbaum L., Jackson S. P., McKinnon P. J., Nussenzweig A. (2007). Distinct domains in Nbs1 regulate irradiation-induced checkpoints and apoptosis. J. Exp. Med. 204, 1003-1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolci S., De Felici M. (1990). A study of meiosis in chimeric mouse fetal gonads. Development 109, 37-40 [DOI] [PubMed] [Google Scholar]
- Gliki G., Ebnet K., Aurrand-Lions M., Imhof B. A., Adams R. H. (2004). Spermatid differentiation requires the assembly of a cell polarity complex downstream of junctional adhesion molecule-C. Nature 431, 320-324 [DOI] [PubMed] [Google Scholar]
- Keeney S., Baudat F., Angeles M., Zhou Z. H., Copeland N. G., Jenkins N. A., Manova K., Jasin M. (1999). A mouse homolog of the Saccharomyces cerevisiae meiotic recombination DNA transesterase Spo11p. Genomics 61, 170-182 [DOI] [PubMed] [Google Scholar]
- Klein U., Esposito G., Baudat F., Keeney S., Jasin M. (2002). Mice deficient for the type II topoisomerase-like DNA transesterase Spo11 show normal immunoglobulin somatic hypermutation and class switching. Eur. J. Immunol. 32, 316-321 [DOI] [PubMed] [Google Scholar]
- Lallemand Y., Luria V., Haffner-Krausz R., Lonai P. (1998). Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res. 7, 105-112 [DOI] [PubMed] [Google Scholar]
- Lewandoski M., Wassarman K. M., Martin G. R. (1997). Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr. Biol. 7, 148-151 [DOI] [PubMed] [Google Scholar]
- Liu P., Jenkins N. A., Copeland N. G. (2003). A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13, 476-484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobe C. G., Nagy A. (1998). Conditional genome alteration in mice. BioEssays 20, 200-208 [DOI] [PubMed] [Google Scholar]
- Lomeli H., Ramos-Mejia V., Gertsenstein M., Lobe C. G., Nagy A. (2000). Targeted insertion of Cre recombinase into the TNAP gene: excision in primordial germ cells. Genesis 26, 116-117 [PubMed] [Google Scholar]
- Meistrich M. L. (1977). Separation of spermatogenic cells and nuclei from rodent testes. Methods Cell Biol. 15, 15-54 [DOI] [PubMed] [Google Scholar]
- Morales M., Theunissen J. W., Kim C. F., Kitagawa R., Kastan M. B., Petrini J. H. (2005). The Rad50S allele promotes ATM-dependent DNA damage responses and suppresses ATM deficiency: implications for the Mre11 complex as a DNA damage sensor. Genes Dev. 19, 3043-3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Gorman S., Dagenais N. A., Qian M., Marchuk Y. (1997). Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl. Acad. Sci. USA 94, 14602-14607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova V. V., Economopoulou M., Lupu F., Santoso S., Chavakis T. (2006). Junctional adhesion molecule-C regulates vascular endothelial permeability by modulating VE-cadherin-mediated cell-cell contacts. J. Exp. Med. 203, 2703-2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M., Grimaldi P., Rossi P., Geremia R., Dolci S. (2003). Developmental expression of BMP4/ALK3/SMAD5 signaling pathway in the mouse testis: a potential role of BMP4 in spermatogonia differentiation. J. Cell Sci. 116, 3363-3372 [DOI] [PubMed] [Google Scholar]
- Rasoulpour R. J., Boekelheide K. (2006). The Sycp1-Cre transgenic mouse and male germ cell inhibition of NF-κB. J. Androl. 27, 729-733 [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B., Nussenzweig M. C., Nussenzweig A., Difilippantonio S. (2005). Genomic instability, endoreduplication, and diminished Ig class-switch recombination in B cells lacking Nbs1. Proc. Natl. Acad. Sci. USA 102, 1590-1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko P. J., Camerini-Otero R. D. (1999). Cloning, characterization, and localization of mouse and human SPO11. Genomics 61, 156-169 [DOI] [PubMed] [Google Scholar]
- Romanienko P. J., Camerini-Otero R. D. (2000). The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6, 975-987 [DOI] [PubMed] [Google Scholar]
- Rossi P., Dolci S., Sette C., Geremia R. (2003). Molecular mechanisms utilized by alternative c-kit gene products in the control of spermatogonial proliferation and sperm-mediated egg activation. Andrologia 35, 71-78 [DOI] [PubMed] [Google Scholar]
- Sadate-Ngatchou P. I., Payne C. J., Dearth A. T., Braun R. E. (2008). Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 46, 738-742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage J., Martin L., Meuwissen R., Heyting C., Cuzin F., Rassoulzadegan M. (1999). Temporal and spatial control of the Sycp1 gene transcription in the mouse meiosis: regulatory elements active in the male are not sufficient for expression in the female gonad. Mech. Dev. 80, 29-39 [DOI] [PubMed] [Google Scholar]
- Sanes J. R., Rubenstein J. L., Nicolas J. F. (1986). Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 5, 3133-3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoso S., Sachs U. J., Kroll H., Linder M., Ruf A., Preissner K. T., Chavakis T. (2002). The junctional adhesion molecule 3 (JAM-3) on human platelets is a counterreceptor for the leukocyte integrin Mac-1. J. Exp. Med. 196, 679-691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoso S., Orlova V. V., Song K., Sachs U. J., Andrei-Selmer C. L., Chavakis T. (2005). The homophilic binding of junctional adhesion molecule-C mediates tumor cell-endothelial cell interactions. J. Biol. Chem. 280, 36326-36333 [DOI] [PubMed] [Google Scholar]
- Schmidt E. E., Taylor D. S., Prigge J. R., Barnett S., Capecchi M. R. (2000). Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc. Natl. Acad. Sci. USA 97, 13702-13707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon M., Richardson L., Christian A., Handel M. A., Thelen M. P. (1999). Differential gene expression of mammalian SPO11/TOP6A homologs during meiosis. FEBS Lett. 462, 329-334 [DOI] [PubMed] [Google Scholar]
- Soriano P. (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70-71 [DOI] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C. S., William C. M., Tanabe Y., Jessell T. M., Costantini F. (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker T. H., Morales M., Couto S. S., Hussein H., Petrini J. H. (2007). The carboxy terminus of NBS1 is required for induction of apoptosis by the MRE11 complex. Nature 447, 218-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Tsuda M., Kiso M., Saga Y. (2008). Nanos3 maintains the germ cell lineage in the mouse by suppressing both Bax-dependent and -independent apoptotic pathways. Dev. Biol. 318, 133-142 [DOI] [PubMed] [Google Scholar]
- Tsai M. Y., Yeh S. D., Wang R. S., Yeh S., Zhang C., Lin H. Y., Tzeng C. R., Chang C. (2006). Differential effects of spermatogenesis and fertility in mice lacking androgen receptor in individual testis cells. Proc. Natl. Acad. Sci. USA 103, 18975-18980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal F., Sage J., Cuzin F., Rassoulzadegan M. (1998). Cre expression in primary spermatocytes: a tool for genetic engineering of the germ line. Mol. Reprod. Dev. 51, 274-280 [DOI] [PubMed] [Google Scholar]
- Yang Y., Sharan S. K. (2003). A simple two-step, ‘hit and fix’ method to generate subtle mutations in BACs using short denatured PCR fragments. Nucleic Acids Res. 31, e80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Petersen S., Tessarollo L., Nussenzweig A. (2001). Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 11, 105-109 [DOI] [PubMed] [Google Scholar]
- Zimmerli C., Lee B. P., Palmer G., Gabay C., Adams R., Aurrand-Lions M., Imhof B. A. (2009). Adaptive immune response in JAM-C-deficient mice: normal initiation but reduced IgG memory. J. Immunol. 182, 4728-4736 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.