Abstract
To date, at least 900 different microRNA (miRNA) genes have been discovered in the human genome. These short, single-stranded RNA molecules originate from larger precursor molecules that fold to produce hairpin structures, which are subsequently processed by ribonucleases Drosha/Pasha and Dicer to form mature miRNAs. MiRNAs play role in the posttranscriptional regulation of about one third of human genes, mainly via degradation of target mRNAs. Whereas the target mRNAs are often involved in the regulation of diverse physiological processes ranging from developmental timing to apoptosis, miRNAs have a strong potential to regulate fundamental biological processes also in the lung compartment. However, the knowledge of the role of miRNAs in physiological and pathological conditions in the lung is still limited. This review, therefore, summarizes current knowledge of the mechanism, function of miRNAs and their contribution to lung development and homeostasis. Besides the involvement of miRNAs in pulmonary physiological conditions, there is evidence that abnormal miRNA expression may lead to pathological processes and development of various pulmonary diseases. Next, the review describes current state-of-art on the miRNA expression profiles in smoking-related diseases including lung cancerogenesis, in immune system mediated pulmonary diseases and fibrotic processes in the lung. From the current research it is evident that miRNAs may play role in the posttranscriptional regulation of key genes in human pulmonary diseases. Further studies are, therefore, necessary to explore miRNA expression profiles and their association with target mRNAs in human pulmonary diseases.
A. miRNA definition, biology and function
Discovery of microRNA (miRNA)
lin-4 was the first short non-coding RNA discovered in 1993 as a regulator of developmental timing in Caenorhabditis elegans [1]. The first non-coding RNA identified in humans was let-7, which has been found involved in the control of developmental timing in humans and animals [2,3]. Soon it became evident that these short non-coding RNAs are a part of much larger class of non-coding RNAs and the term microRNA (miRNA) was introduced [4]. To date, more than 900 miRNAs in Homo sapiens have been identified (940 in miRBase v15).
Structure and function of miRNAs
MiRNAs are small non-coding RNAs ~22 nucleotides (nt) long involved in the negative post-transcriptional gene regulation via RNA interference mechanism [5,6]. The sequences of miRNAs are highly conserved among plants-microorganisms-animals, suggesting that miRNAs represent a relatively old and important regulatory pathway [7]. MiRNAs belong to the most abundant class of human gene regulators [8]: up to a third of the human genes are regulated by miRNAs [9]. MiRNAs are, therefore, key regulators of numerous genes in biological processes ranging from developmental timing to apoptosis [e.g. [10-14]]. It has been speculated that miRNAs may be associated with the regulation of almost every aspect of cell physiology [8].
miRNA biogenesis
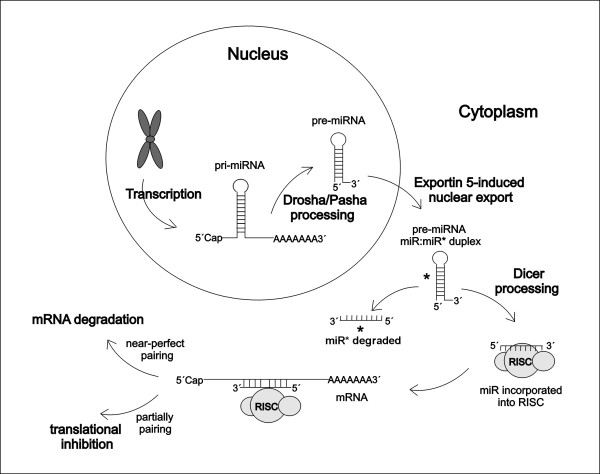
MiRNA genes are localized in the non-coding regions or in the introns of protein-coding genes in the genomic DNA. The miRNA genes are much longer than biologically active, mature miRNAs which originate through a multi-step process [15] (Figure 1). Briefly, transcription by the RNA polymerase II leads to hundred or thousand nucleotides long primary miRNA transcripts (pri-miRNAs) [16]. A local stem-loop structure of pri-miRNAs is then cleaved in the nucleus by the dsRNA-specific ribonuclease Drosha/Pasha to 70 nucleotides long precursor miRNA (pre-miRNA) [17] in a process known as "cropping" [18,19]. Pre-miRNAs are then actively transported from the nucleus to the cytoplasm [20,21]. In the cytoplasm, pre-miRNAs are subsequently cleaved by RNase III Dicer into ~22-nt miRNA duplexes [17,20]. One strand of the short-lived miRNA duplex is degraded ("passenger" strand, miR*), whereas the other ("guide", miR) strand is incorporated into the RNA-induced silencing complex (RISC) and serves as a functional, mature miRNA [8]. Selection of the "guide" strand is based on the base pairing stability of both dsRNA ends [22,23].
Figure 1.
miRNA biogenesis. MiRNAs are transcribed by RNA polymerase II from the genomic DNA as long (hundred or thousand nucleotides) primary miRNA transcripts (pri-miRNAs). A local stem-loop structure of pri-miRNAs is then cleaved in the nucleus by the dsRNA-specific ribonuclease Drosha/Pasha to produce a 70 nucleotides long precursor miRNA (pre-miRNA). Pre-miRNAs in form of hairpins are then actively transported from the nucleus to the cytoplasm. In the cytoplasm, pre-miRNAs are subsequently cleaved by RNase III Dicer into ~22-nt miRNA duplexes, consisting of the "guide" (miR) strand and the "passenger" (miR*) strand. The "passenger" strand is degraded, the "guide" strand is incorporated into the RNA-induced silencing complex (RISC) and serves as a functional, mature miRNA, acting by two different mechanisms according to the complementarity with the target mRNA. Adopted from Kim [15].
Mechanism of miRNA action
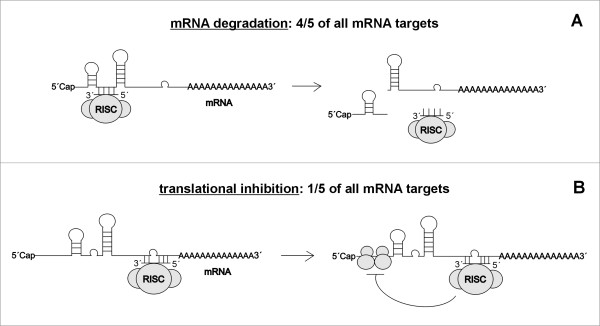
Depending on the complementarity between miRNA and 3' untranslated region (UTR) of target mRNA there are two known mechanisms of miRNAs action on mRNAs: 1) target mRNA degradation and 2) translational inhibition with little or no influence on mRNA levels [24] (Figure 2). Firstly, the deadenylation and subsequent degradation of the target mRNA occurs when miRNA is near-perfectly complementary with target mRNA [25,26]. A recent study proved that mRNA degradation represents the major mechanism of miRNA regulation [27]. The authors showed that about 84% of all protein-coding mRNA targets undergo degradation while recognized by their cognate miRNA [27]. Secondly, the translational inhibition occurs when miRNA is only partially complementary to its target mRNA [28-30]. In light of the recent study by Guo et al [27], this mechanism does not represent a predominant reason for reduced protein output.
Figure 2.
Mechanism of miRNA action on target mRNA. According to the complementarity between miRNA and 3' untranslated region (UTR) of target mRNA, there are two mechanisms of miRNA action: (A) when miRNA is near-perfectly complementary with target mRNA, deadenylation and subsequent degradation of the target mRNA occurs (major mechanism of miRNA action); (B) when miRNA is only partially complementary to its target mRNA, translational inhibition occurs. Adopted from Brodersen et al [24].
Besides the complementarity between miRNA and mRNA, several other factors may influence the miRNA action such as impaired processing, methylation, gene polymorphisms, gene amplification, deletion of Dicer, translocations and others [31].
Targets of miRNAs
It is evident that single miRNAs may regulate translation of numerous downstream mRNAs and each mRNA is likely to be regulated by several miRNAs simultaneously [30,32]. Thus, identification of miRNA target genes has been a great challenge [33]. Numerous computational algorithms [34-43] were established which combined 5' seed matches, thermodynamic stability and conservation analysis in order to maximize specificity when predicting mRNA targets [44] (Table 1). Nevertheless, various algorithms differ in the selection of mRNA targets and simultaneous application of several algorithms is, therefore, highly recommended. Nowadays, many web-based applications [45-52] have been developed by combining existing prediction programs with functional annotations associated to many miRNA, gene, protein or biological pathway resources such as miRBase, Ensembl, Swiss-Prot, UCSC genome browser, KEGG pathway and other databases [44] (Table 2).
Table 1.
Computational algorithms for miRNA target prediction.
| Algorithm | Web link | References |
|---|---|---|
| miRanda | http://www.microrna.org | [34] |
| TargetScan | http://genes.mit.edu/targetscan | [35] |
| TargetScanS | http://genes.mit.edu/targetscan | [36] |
| PicTar | http://pictar.mdc-berlin.de | [37] |
| DIANA-microT | http://diana.pcbi.upenn.edu/cgi-bin/micro_t.cgi | [38] |
| ElMMo | http://www.mirz.unibas.ch/ElMMo2/ | [39] |
| MirTarget2 | http://mirdb.org | [40] |
| miTarget | http://cbit.snu.ac.kr/~miTarget/ | [41] |
| rna22 | http://cbcsrv.watson.ibm.com/rna22.html | [42] |
| RNAhybrid | http://bibiserv.techfak.uni-bielefeld.de/rnahybrid | [43] |
Adopted from Min et al [44].
Table 2.
Methods with extended features for miRNA target prediction.
| Name | Web link | References |
|---|---|---|
| GOmir | http://www.bioacademy.gr/bioinformatics/projects/GOmir | [45] |
| miRDB | http://mirdb.org | [46] |
| miRecords | http://miRecords.umn.edu/miRecords | [47] |
| miRGator | http://genome.ewha.ac.kr/miRGator | [48] |
| miRNAMap | http://miRNAMap.mbc.nctu.edu.tw | [49] |
| mirZ | http://www.mirz.unibas.ch | [50] |
| MMIA | http://156.56.93.156/~MMIA/mmia_main.html | [51] |
| TarBase5.0 | http://diana.cslab.ece.ntua.gr/tarbase | [52] |
Adopted from Min et al [44].
However, because of high similarities in miRNA sequences, computational algorithms may predict a large number of putative miRNA binding sites on mRNA targets [33]. Thus, experimental validation in biological system is fundamental to complete the target prediction study [44]; the currently available methods [53-67] are listed in Table 3. Of these, antagomir studies or immunoprecipitation of Ago-bound mRNAs have been specifically developed for miRNA-mRNA studies. Antagomirs represent a novel class of chemically engineered oligonucleotides used to silence endogenous microRNAs [64,65]. Immunoprecipitation is then based on the observation that each member of the Argonaute (Ago) protein family (catalytic components of the RNA-induced silencing complex) can bind to miRNAs and to partially complementary sequences in the 3'-UTR of specific target mRNAs. Thus, using highly specific monoclonal antibodies against members of the Ago protein family, Ago-bound mRNAs can be co-immunoprecipitated [66,67].
Table 3.
Experimental methods to check the functional interaction between miRNA and target mRNA.
| Method | Selected references |
|---|---|
| Luciferase reporter assay | [53] |
| Northern blot analysis | [54] |
| Quantitative real-time PCR | [55] |
| Ribonuclease protection assay | [56] |
| in situ hybridization | [57,58] |
| miRNA mimics | [59] |
| Western blot | [60] |
| Immunocytochemistry | [61] |
| Bead-based flow cytometry method | [62] |
| Suppression of miRNA expression in cells by anti-sense locked-nucleic acid oligonucleotides | [63] |
| Antagomir assays | [64,65] |
| Immunoprecipitation of Ago-bound mRNAs | [66,67] |
B. miRNA in pulmonary physiology and pathology
Role of miRNAs in the lung
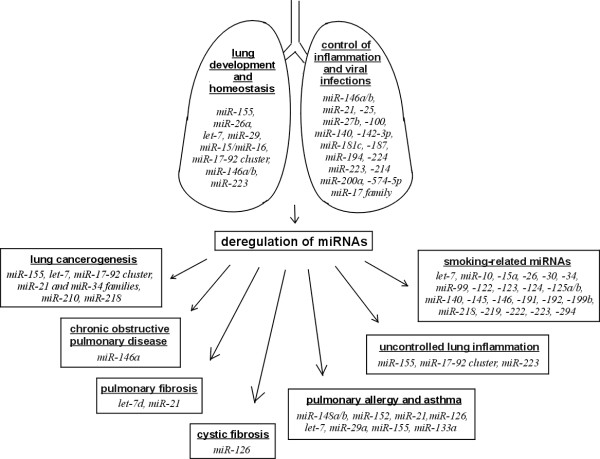
The lung has a very specific miRNA expression profile, highly conserved across mammalian species [68,69]. However, the knowledge of the role of miRNAs in physiological and pathological conditions in the lung compartment is still limited and it is based mainly on the studies in animal models. MiRNAs have been shown to be involved in 1) the lung development and homeostasis, 2) in inflammation and viral infections and 3) miRNA deregulation may contribute to several pulmonary diseases (Figure 3). Hereby, we summarize the knowledge of the involvement of miRNAs in the lung and current information on their posttranscriptional regulation ongoing in the lung compartment. Besides pathology we pay attention also to physiological lung because understanding miRNA function in normal condition is prerequisite to description of its involvement in disease.
Figure 3.
Involvement of miRNAs in physiological and pathological processes in lung. The scheme shows several miRNAs involved in physiological and pathological processes in lung. There has been shown that numerous miRNAs are implicated in maintaining lung homeostasis and development. When deregulation of these miRNAs occurs, pathological processes turn up and may lead to the development of pulmonary diseases.
MiRNAs in the lung in physiological condition
Homeostasis and lung development
Several miRNAs such as miR-155, miR-26a, let-7, miR-29, miR-15/miR-16, miR-223, miR-146a/b and the miR-17-92 cluster have been shown to be involved in homeostasis and in the lung development (Table 4). The pulmonary role of miR-155 was studied in murine lung, where it has been shown that miR-155 is crucially involved in the differentiation of naive T-cells into Th1 and Th2 cells [70,71]. Mice deficient in bic/miR-155 became immunodeficient and displayed increased lung remodelling, higher bronchoalveolar leukocytes and impaired T- and B-cell responses to inflammatory stimuli [70]. Another member of miRNA family, miR-26a, has been shown to be selectively expressed in the bronchial and alveolar epithelial cells in murine lung [72]. Target mRNA of miR-26a is the transcription factor SMAD1, which is involved in the regulation of bone morphogenic protein signalling during lung development and pulmonary vascular remodelling [73,74]. Thus, miR-26a might be important in controlling essential developmental and physiological events in the lung [75]. Also the miR-17-92 cluster is believed to regulate the lung development because its expression is high in embryonic development and steadily declines through development into adulthood [76]. Mice deficient in the miR-17-92 cluster died shortly after birth and lung hypoplasia/ventricular septal defects were demonstrated; moreover the absence of the miR-17-92 cluster let to upregulation of the pro-apoptotic protein Bim and inhibition of B-cell development [77]. On the other side, the overexpression of the miR-17-92 cluster in murine models resulted in an abnormal phenotype manifested by absence of terminal air sacs, which were replaced by highly proliferative, undifferentiated pulmonary epithelium [76]. Other miRNAs found to be involved in the pulmonary homeostasis are members of let-7 family [78], miR-29 [79], miR-15 and miR-16 [80,81], which function as tumor suppressors in lung cells. In addition, another miRNA, miR-223, has been shown to be crucial for normal granulocyte development and function in the lung [82]. MiR-223 mutant mice spontaneously developed neutrophilic lung inflammation with tissue destruction after endotoxin challenge [82].
Table 4.
MiRNAs involved in physiological processes in the lung.
| miRNA | Function (A animal studies, H human studies) | References |
|---|---|---|
| miR-17-92 cluster | important in lung development and homeostasis (A) | [69,76,77] |
| miR-155 | important for normal lung airway remodelling (A) | [70] |
| alteration of T-cell differentiation (A) | [71] | |
| miR-26a | highly expressed within bronchial and alveolar epithelial cells, important for lung development (H) | [75] |
| let-7 | highly expressed in normal lung tissue, functions as a tumor suppressor in lung cells (H) | [78] |
| miR-29 | functions as tumor suppressor in lung cells (H) | [79] |
| miR-15, miR-16 | function as tumor suppressor genes (H) | [80,81] |
| miR-223 | control of granulocyte development and function (A) | [82] |
| miR-146a/b | central to the negative feedback regulation of IL-1β-induced inflammation (H) | [83,84] |
| miR-200a, miR-223 | contribution to the extreme virulence of the r1918 influenza virus (A) | [85] |
| miR-17 family, miR-574-5p, miR-214 | upregulated at the onset of SARS infection (A, H) | [86] |
Control of pulmonary inflammation and viral infections
Two miRNAs, miR-146a and miR-146b, have been shown to play central role in the negative feedback regulation of IL-1β-induced inflammation; the mechanism is down-regulation of two proteins IRAK1 and TRAF6 involved in Toll/interleukin-1 receptor (TIR) signalling [83,84]. Also other miRNAs have been shown to regulate the inflammation in mouse lung exposed to aerosolized lipopolysaccharide (LPS): miR-21, -25, -27b, -100, -140, -142-3p, -181c, -187, -194, -214, -223 and -224 [72]. Increase in these miRNAs correlated with the downregulation of pro-inflammatory cytokine production such as TNFα [72]. The deregulation of miR-155, the miR-17-92 cluster and miR-223, miRNAs involved in lung development and homeostasis, resulted in the uncontrolled lung inflammation in murine models [70,77,82].
Based on the studies in murine models, there is evidence that miRNA expression may influence also the course of pulmonary viral infections [85,86]. MiR-200a and miR-223 were detected in lethal influenza virus infection presumably contributing to the extreme virulence of the r1918 influenza virus [85]. MiR-17 family, miR-574-5p and miR-214 were upregulated at the onset of SARS infection: these miRNAs may help the virus to evade the host immune system and are responsible for effective transmission at the initial stage of viral infection [86].
Deregulation of miRNAs leads to development of pulmonary diseases
There is evidence that upregulation or downregulation of miRNAs is critical for the lung development/homeostasis and thus may contribute to development of pathological pulmonary conditions, namely to smoking-related diseases including lung cancerogenesis, fibrosis, and other immune-mediated disorders including allergy (Table 5).
Table 5.
MiRNAs involved in pathological processes in the lung.
| miRNA | Function (A animal studies, H human studies) | References |
|---|---|---|
| miR-155, miR-17-92 cluster | deregulation results in uncontrolled inflammation (A) | [70,71,77] |
| miR-21, miR-27b, miR-100, miR-181c, miR-223, miR-224 | increased following exposure to LPS (A) | [72] |
| miR-155 | overexpressed in solid tumors, inhibition of tumor suppressor genes (A, H) | [81] |
| miR-223 | impaired granulocyte function, regulator of granulocyte production and inflammatory response (A) | [82] |
| miR-148a/b, miR-152 | allele-specific regulation of asthma susceptibility HLA-G gene (H) | [87] |
| miR-21 | key role in asthma (A) | [88] |
| overexpressed in solid malignancies (A, H) | [103] | |
| up-regulated in bleomycin-induced fibrosis and IPF (A, H) | [110] | |
| miR-126 | suppression of the asthmatic phenotype by blockade of miR-126 (A) | [89] |
| downregulated in cystic fibrosis airway epithelial cells (H) |
[111] | |
| let-7, miR-29a, miR-155 | regulation of allergic inflammation in bronchial epithelial cells (A, H) | [90] |
| let-7 | pro-inflammatory effect in experimental asthma (A) | [91] |
| role in lung cancer progression (H) | [99] | |
| miR-133a | regulator of expression of RhoA, target for asthma therapy (A, H) | [92,93] |
| miR-146a | reduced expression in COPD fibroblasts (H) | [95] |
| miR-218, miR-15a, miR-199b, miR-125a/b, miR-294 | deregulated due to smoking (A, H) | [96,97] |
| miR-218 | tumor suppressor in non-small cell lung cancer (H) | [98] |
| miR-17-92 cluster | overexpressed in lung cancers (H) | [102] |
| miR-34 | regulation of apoptosis in lung cancer cells (H) | [105-107] |
| miR-210 | overexpressed in lung cancer (H) | [108] |
| let-7d | pro-fibrotic effect in pulmonary fibrosis (A, H) | [109] |
Immune-mediated lung diseases
Recent studies have implicated the miRNAs in the pathogenesis of immune system mediated pulmonary diseases. Tan and colleagues [87] described that a single nucleotide polymorphism in the 3'UTR of HLA-G, a known asthma-susceptibility gene, disrupts the binding sites of three miRNAs (miR-148a, miR-148b, miR-152) targeting this gene. Thus, it is likely that the association of the HLA-G gene to asthma-susceptibility may be due to the allele-specific regulation of this gene by miRNAs [87]. MiR-21 is a further miRNA crucially involved in allergic lung inflammation. Its molecular target is IL-12p35, a cytokine contributing to polarization of Th cells toward Th2 cells [88]. MiR-126 is another miRNA found to be involved in the pathogenesis of allergic airways disease [89]. The blockade of miR-126 suppressed the asthmatic phenotype leading to diminished Th2 responses, suppression of inflammation, reduced airways hyperresponsiveness, inhibition of eosinophil recruitment, and lower mucus hypersecretion [89]. In bronchial epithelial cells stimulated with IL-4 and TNFα, let-7, miR-29a and miR-155 have been involved in the regulation of allergic inflammation [90]. Multiple members of let-7 family were also found upregulated in experimental asthma model and the pro-inflammatory role of let-7 miRNAs on the allergic cytokine expression was confirmed [91]. Another study showed that expression of RhoA in bronchial smooth muscle cells (BSMCs), a new target for asthma therapy, is negatively regulated by miR-133a [92]. The same group later revealed that IL-13 is capable of reducing the miR-133a expression in BSMCs and that the miR-133a downregulation causes an upregulation of RhoA, presumably resulting in an augmentation of the contraction [93].
Smoking-related lung diseases
Lung cancer and chronic obstructive pulmonary disease (COPD) share a common environmental risk factor in cigarette smoke exposure [94]. Although extensive studies of the involvement of miRNAs in lung cancer have been performed, there are only few reports focused on the role of miRNAs in COPD. Recent study on fibroblasts from COPD subjects stimulated in vitro with pro-inflammatory cytokines released less miR-146a than smokers without COPD [95]. The reduced miR-146a expression resulted in prolonged mRNA half-life of cyclooxygenase-2, thus increasing prostaglandin E2 in fibroblasts from COPD subjects [95]. There is evidence that smoking has influence also on other miRNAs. Expression profiling study in the rats exposed to environmental cigarette smoke revealed 24 downregulated miRNAs (especially let-7 family, miR-10, -26, -30, -34, -99, -122, -123, -124, -125, -140, -145, -146, -191, -192, -219, -222, and -223) when compared to control group [96]. MiR-294, a known inhibitor of transcriptional repressor genes, was the only miRNA upregulated in smoke-exposed rats [96]. In another study, bronchial airway epithelial cells from current and never smokers differed in the expression of 28 miRNAs (especially miR-218, miR-15a, miR-199b, miR-125a/b, miR-294) in comparison to smokers, whereas the majority of deregulated miRNAs were downregulated in smokers [97]. Similar observation was observed in lung squamous cell carcinoma, where downregulation of miR-218 was associated with a history of cigarette smoking [98].
However, the majority of miRNA studies in smoking-related diseases are focused on the role of miRNAs in lung cancer. Altered expression of miR-155 and let-7 has been reported in lung adenocarcinoma and expression of let-7 related to patient survival [99]. Moreover, it has been shown that let-7 may also play a role in lung cancer progression [99-101]. Further, increased expression of the miR-17-92 cluster has also been detected in lung cancer [102]. Another miRNAs involved in lung cancerogenesis are miR-21 and miR-34 families. MiR-21 was shown to regulate multiple tumor/metastasis suppressor genes in lung solid tumors [103]. MiR-34a/b/c have been identified to be a component of the p53 tumor suppressor network: p53 upregulates in response to DNA damage the members of miR-34 family [104], thus regulating genes involved in the cell cycle and apoptosis [105-107]. Furthermore, miR-210 has been overexpressed in late stages of lung cancer, thus mediated mitochondrial alterations associated with modulation of hypoxia-inducible factor-1 activity [108]. Next, miR-218 was identified as a putative tumor suppressor in non-small cell lung cancer [98].
Other lung diseases
Recently, it was reported that miRNAs may play pivotal regulatory role also in the fibrotic processes ongoing in the lung: the downregulation of let-7 d in idiopathic pulmonary fibrosis (IPF) resulted in the pro-fibrotic effects [109]. Also, upregulation of miR-21 was reported in the lungs of IPF patients and in the murine lungs with bleomycin-induced fibrosis, whereas miR-21 expression was enhanced by pro-fibrotic TGF-β1 [110]. Another disease associated with miRNA change was cystic fibrosis. Downregulation of miR-126 was detected in cystic fibrosis bronchial epithelial cells and its expression correlated with upregulation of TOM1 mRNA both in vitro and in vivo [111]. TOM1, a miR-126 target, was reported to be involved in the regulation of innate immune responses through its involvement in the TLR2/4 and IL-1β and TNF-α-induced signalling pathways [111].
Conclusion
Small non-coding RNAs (miRNAs) play pivotal role in the posttranscriptional regulation of numerous human genes, mainly via degradation of target mRNAs. There is evidence that the lung has a very specific miRNA expression profile undergoing changes during the lung development. Studies namely in animal models have provided evidence that miRNAs participate in lung homeostasis and play pivotal role also in the control of pulmonary inflammation and viral infections. Recent studies showed evidence that upregulated or downregulated expression of various miRNAs play an active role in the pathogenesis of pulmonary diseases. Specific miRNA expression profiles were characterized for smoking related-diseases including COPD and lung cancer, immune-mediated pulmonary diseases and pulmonary fibrosis. Moreover, several miRNAs crucial for lung development and homeostasis such as let-7, miR-155 or miR-19-72 cluster have been identified to be deregulated in pulmonary allergy, asthma or lung cancer. The knowledge of altered miRNA expression profiles in diseased lung may thus offer new insights in the biology of pulmonary diseases. Moreover, miRNAs may represent attractive novel diagnostic biomarkers mainly due to their higher stability when compared to mRNAs [112] and could potentially provide possibilities for therapeutic intervention [31,113,114].
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
All authors wrote and revised the manuscript, and approved the final version.
Contributor Information
Tereza Tomankova, Email: tomankot@upol.cz.
Martin Petrek, Email: petrekm@fnol.cz.
Eva Kriegova, Email: kriegova@yahoo.com.
Acknowledgements
Funding was obtained from the Czech Ministry of Health (IGA MZ CR NT/11117-6, IGA MZ CR NS/10267-3, IGA MZ CR NS/10260-3) and in part by the Internal Grant Agency of Palacky University (IGA PU project SV LF_2010_008). The authors declare no conflicting financial interests.
References
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Rougvie AE. Control of developmental timing in animals. Nat Rev Genet. 2001;2:690–701. doi: 10.1038/35088566. [DOI] [PubMed] [Google Scholar]
- Ruvkun G. Molecular biology. Glimpses of a tiny RNA world. Science. 2001;26:797–799. doi: 10.1126/science.1066315. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Grosshans H, Slack FJ. Micro-RNAs: small is plentiful. J Cell Biol. 2002;156:17–21. doi: 10.1083/jcb.200111033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkoly E, Pivarcsi A. Advances in microRNAs: implications for immunity and inflammatory diseases. J Cell Mol Med. 2009;13:24–38. doi: 10.1111/j.1582-4934.2008.00534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes J, Collison A, Foster PS. Emerging role of microRNAs in disease pathogenesis and strategies for therapeutic modulation. Curr Opin Mol Ther. 2008;10:150–157. [PubMed] [Google Scholar]
- Wang Y, Stricker HM, Gou D, Liu L. MicroRNA: past and present. Front Biosci. 2007;12:2316–2329. doi: 10.2741/2234. [DOI] [PubMed] [Google Scholar]
- Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310:1954–1957. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Guo M, Hay BA. MicroRNAs and the regulation of cell death. Trends Genet. 2004;20:617–624. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nature Reviews Molecular Cell Biology. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR. EST analyses predict the existence of a population of chimeric microRNA precursor-mRNA transcripts expressed in normal human and mouse tissues. Genome Biol. 2003;4:403. doi: 10.1186/gb-2003-4-7-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/S0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/S0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nature Reviews Molecular Cell Biology. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nana-Sinkam SP, Hunter MG, Nuovo GJ, Schmittgen TD, Gelinas R, Galas D, Marsh CB. Integrating the MicroRNome into the study of lung disease. Am J Respir Crit Care Med. 2009;179:4–10. doi: 10.1164/rccm.200807-1042PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Kuhn DE, Martin MM, Feldman DS, Terry AV Jr, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. MicroRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidatzis D, van Nimwegen E, Hausser J, Zavolan M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics. 2007;8:69. doi: 10.1186/1471-2105-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- Kim SK, Nam JW, Rhee JK, Lee WJ, Zhang BT. miTarget: microRNA target-gene prediction using a support vector machine. BMC Bioinformatics. 2006;7:411. doi: 10.1186/1471-2105-7-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H, Yoon S. Got target? Computational methods for microRNA target prediction and their extension. Exp Mol Med. 2010;42:233–244. doi: 10.3858/emm.2010.42.4.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubelakis MG, Zotos P, Papachristoudis G, Michalopoulos I, Pappa KI, Anagnou NP, Kossida S. Human microRNA target analysis and gene ontology clustering by GOmir, a novel stand-alone application. BMC Bioinformatics. 2009;10:S20. doi: 10.1186/1471-2105-10-S6-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. miRDB: A microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. MiRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S, Kim B, Shin S, Lee S. miRGator: an integrated system for functional annotation of microRNAs. Nucleic Acid Res. 2008;36:D159–164. doi: 10.1093/nar/gkm829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC, Hsu PW, Wong YH, Chen YH, Chen GH, Huang HD. miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res. 2008;36:D165–169. doi: 10.1093/nar/gkm1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser J, Berninger P, Rodak C, Jantscher Y, Wirth S, Zavolan M. MirZ: an integrated microRNA expression atlas and target prediction resource. Nucleic Acids Res. 2009;36:W266–272. doi: 10.1093/nar/gkp412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S, Li M, Choi K, Balch C, Kim S, Nephew KP. MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Res. 2009;37:W356–362. doi: 10.1093/nar/gkp294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37:D155–158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MM, Lee EJ, Buckenberger JA, Schmittgen TD, Elton TS. MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblasts. J Biol Chem. 2006;281:18277–18284. doi: 10.1074/jbc.M601496200. [DOI] [PubMed] [Google Scholar]
- Norden-Krichmar TM, Holtz J, Pasquinelli AE, Gaasterland T. Computational prediction and experimental validation of Ciona intestinalis microRNA genes. BMC Genomics. 2007;8:445. doi: 10.1186/1471-2164-8-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurteau GJ, Spivack SD, Brock GJ. Potential mRNA degradation targets of hsa-miR-200c, identified using informatics and qRT-PCR. Cell Cycle. 2006;5:1951–1956. doi: 10.4161/cc.5.17.3133. [DOI] [PubMed] [Google Scholar]
- Gu Z, Eleswarapu S, Jiang H. Identification and characterization of microRNAs from the bovine adipose tissue and mammary gland. FEBS Lett. 2007;581:981–988. doi: 10.1016/j.febslet.2007.01.081. [DOI] [PubMed] [Google Scholar]
- Martin MM, Buckenberger JA, Jiang J, Malana GE, Nuovo GJ, Chotani M, Feldman DS, Schmittgen TD, Elton TS. The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates microrna-155 binding. J Biol Chem. 2007;282:24262–24269. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wheeler G, Valoczi A, Havelda Z, Dalmay T. In situ detection of animal and plant microRNAs. DNA Cell Biol. 2007;26:251–255. doi: 10.1089/dna.2006.0538. [DOI] [PubMed] [Google Scholar]
- Wang F, Xue X, Wei J, An Y, Yao J, Cai H, Wu J, Dai C, Qian Z, Xu Z, Miao Y. hsa-miR-520 h downregulates ABCG2 in pancreatic cancer cells to inhibit migration, invasion, and side populations. Br J Cancer. 2010;103:567–574. doi: 10.1038/sj.bjc.6605724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Xia W, Huang B, Chen L, Su X, Li S, Wang F, Ding H, Shao N. A strategy to rapidly identify the functional targets of microRNAs by combining bioinformatics and mRNA cytoplasmic/nucleic ratios in culture cells. FEBS Lett. 2010;584:3198–3202. doi: 10.1016/j.febslet.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saïb A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, Boldrini R, Donfrancesco A, Federici V, Giacomini P, Peschle C, Fruci D. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AE, Moschos SA, Perry MM, Barnes PJ, Lindsay MA. Maternally imprinted microRNAs are differentially expressed during mouse and human lung development. Dev Dyn. 2007;236:572–580. doi: 10.1002/dvdy.21047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AE, Perry MM, Moschos SA, Lindsay MA. microRNA expression in the aging mouse lung. BMC Genomics. 2007;8:172. doi: 10.1186/1471-2164-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Schambach F, DeJong CS, Hammond SM, Reiner SL. Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. Eur J Immunol. 2010;40:225–231. doi: 10.1002/eji.200939381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics. 2007;8:240. doi: 10.1186/1471-2164-8-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Sun J, Bringas P Jr, Chen Y, Warburton D, Shi W. Smad1 expression and function during mouse embryonic lung branching morphogenesis. Am J Physiol Lung Cell Mol Physiol. 2005;288:L1033–9. doi: 10.1152/ajplung.00277.2004. [DOI] [PubMed] [Google Scholar]
- Frank DB, Abtahi A, Yamaguchi DJ, Manning S, Shyr Y, Pozzi A, Baldwin HS, Johnson JE, de Caestecker MP. Bone morphogenetic protein 4 promotes pulmonary vascular remodeling in hypoxic pulmonary hypertension. Circ Res. 2005;97:496–504. doi: 10.1161/01.RES.0000181152.65534.07. [DOI] [PubMed] [Google Scholar]
- Williams AE, Larner-Svensson H, Perry MM, Campbell GA, Herrick SE, Adcock IM, Erjefalt JS, Chung KF, Lindsay MA. MicroRNA expression profiling in mild asthmatic human airways and effect of corticosteroid therapy. PloS One. 2009;4:e5889. doi: 10.1371/journal.pone.0005889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MM, Moschos SA, Williams AE, Shepherd NJ, Larner-Svensson HM, Lindsay MA. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol. 2008;180:5689–5698. doi: 10.4049/jimmunol.180.8.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chan EY, Li J, Ni C, Peng X, Rosenzweig E, Tumpey TM, Katze MG. MicroRNA expression and virulence in pandemic influenza virus-infected mice. J Virol. 2010;84:3023–3032. doi: 10.1128/JVI.02203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick B, Ghosh Z, Chakrabarti J. MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PLoS One. 2009;4:e7837. doi: 10.1371/journal.pone.0007837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, Solway J, Gern JE, Lemanske RF, Nicolae D, Ober C. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci USA. 2009;106:18704–18709. doi: 10.1073/pnas.0905063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Zhong Z, Chen CY, Xia Z, Song L, Blackburn MR, Shyu AB. Coordinated changes in mRNA turnover, translation, and RNA processing bodies in bronchial epithelial cells following inflammatory stimulation. Mol Cell Biol. 2008;28:7414–7426. doi: 10.1128/MCB.01237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikepahad S, Knight JM, Naghavi AO, Oplt T, Creighton CJ, Shaw C, Benham AL, Kim J, Soibam B, Harris RA, Coarfa C, Zariff A, Milosavljevic A, Batts LM, Kheradmand F, Gunaratne PH, Corry DB. Pro-inflammatory role for let-7 microRNAs in experimental asthma. J Biol Chem. 2010;285:30139–30149. doi: 10.1074/jbc.M110.145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y, Tanabe M, Goto K, Sakai H, Misawa M. Down-regulation of miR-133a contributes to up-regulation of Rhoa in bronchial smooth muscle cells. Am J Respir Crit Care Med. 2009;180:713–719. doi: 10.1164/rccm.200903-0325OC. [DOI] [PubMed] [Google Scholar]
- Chiba Y, Misawa M. MicroRNAs and Their Therapeutic Potential for Human Diseases: MiR-133a and Bronchial Smooth Muscle Hyperresponsiveness in Asthma. J Pharmacol Sci. 2010;114:264–268. doi: 10.1254/jphs.10R10FM. [DOI] [PubMed] [Google Scholar]
- Caramori G, Casolari P, Cavallesco GN, Giuffrè S, Adcock I, Papi A. Mechanisms involved in lung cancer development in COPD. Int J Biochem Cell Biol. in press . [DOI] [PubMed]
- Sato T, Liu X, Nelson A, Nakanishi M, Kanaji N, Wang X, Kim M, Li Y, Sun J, Michalski J, Patil A, Basma H, Holz O, Magnussen H, Rennard SI. Reduced MiR-146a Increases Prostaglandin E2 in Chronic Obstructive Pulmonary Disease Fibroblasts. Am J Respir Crit Care Med. 2010;182:1020–1029. doi: 10.1164/rccm.201001-0055OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23:806–812. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schembri F, Sridhar S, Perdomo C, Gustafson AM, Zhang X, Ergun A, Lu J, Liu G, Zhang X, Bowers J, Vaziri C, Ott K, Sensinger K, Collins JJ, Brody JS, Getts R, Lenburg ME, Spira A. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci USA. 2009;106:2319–2324. doi: 10.1073/pnas.0806383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson MR, Larsen JE, Yang IA, Hayward NK, Clarke BE, Duhig EE, Passmore LH, Bowman RV, Fong KM. MicroRNA-218 is deleted and downregulated in lung squamous cell carcinoma. PLoS One. 2010;5:e12560. doi: 10.1371/journal.pone.0012560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- Du L, Pertsemlidis A. microRNAs and lung cancer: tumors and 22-mers. Cancer Metastasis Rev. 2010;29:109–122. doi: 10.1007/s10555-010-9204-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puisségur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, Maurin T, Lebrigand K, Cardinaud B, Hofman V, Fourre S, Magnone V, Ricci JE, Pouysségur J, Gounon P, Hofman P, Barbry P, Mari B. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. in press . [DOI] [PMC free article] [PubMed]
- Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, Richards T, Selman M, Watkins SC, Pardo A, Ben-Yehudah A, Bouros D, Eickelberg O, Ray P, Benos PV, Kaminski N. Inhibition and Role of let-7 d in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. 2010;182:220–229. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oglesby IK, Bray IM, Chotirmall SH, Stallings RL, O'Neill SJ, McElvaney NG, Greene CM. miR-126 is downregulated in cystic fibrosis airway epithelial cells and regulates TOM1 expression. J Immunol. 2010;184:1702–1709. doi: 10.4049/jimmunol.0902669. [DOI] [PubMed] [Google Scholar]
- Giglio S, Vecchione A. Role of microRNAs in the molecular diagnosis of cancer. Journal of Nucleic Acids Investigation. 2010;1:e4. [Google Scholar]
- Nana-Sinkam SP, Karsies T, Riscili B, Ezzie M, Piper M. Lung microRNA: from development to disease. Expert Rev Respir Med. 2009;3:373–385. doi: 10.1586/ers.09.30. [DOI] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]