Summary
Macrophages (Mϕ) and dendritic cells (DC) are key cells controlling tissue homeostasis and response to aggressions. In this work, we demonstrate that Mϕ and DC are the major producers of the phenylalanine catabolizing enzyme IL4I1 under inflammatory conditions. IL4I1 was first described in B cells, which indeed can produce IL4I1 in vitro to much lower levels. In vivo, IL4I1 is highly expressed in Mϕ and DC of Th1 granulomas (sarcoidosis, tuberculosis) but poorly detected in Th2 granulomas (schistosomiasis). In vitro, expression of the enzyme is induced in mononuclear phagocytes by various proinflammatory stimuli through the activation of the transcription factors NFκ B and/or STAT1. B cells also express IL4I1 in response to NFκ B activating stimuli such as CD40L. However, in contrast to myeloid cells, they are insensitive to IFNγ but respond to stimulation of the IL4-STAT6 axis. Since we show that expression of IL4I1 by a monocytic cell line inhibits T cell proliferation and production of IFNγ and inflammatory cytokines, we propose that IL4I1 participates to the down regulation of Th1 inflammation in vivo.
Keywords: B-Lymphocytes; drug effects; immunology; metabolism; pathology; CD40 Ligand; pharmacology; Cell Line; Cell Proliferation; Coculture Techniques; Flavoproteins; biosynthesis; genetics; immunology; Humans; Immune Tolerance; Inflammation; Interferon-gamma; pharmacology; Interleukin-4; immunology; metabolism; Mononuclear Phagocyte System; drug effects; immunology; metabolism; pathology; NF-kappa B; metabolism; RNA, Small Interfering; genetics; STAT1 Transcription Factor; metabolism; STAT6 Transcription Factor; metabolism; Th1 Cells; immunology; metabolism; pathology; Th2 Cells; immunology; metabolism; pathology
Keywords: immunosuppressive enzyme, inflammation, granuloma, NFkB, STAT
Introduction
Inflammation is a complex response involving immune and stromal cells, triggered by tissue insults, such as infection, cancer and injuries. This response aims at removing or isolating the source of disturbance to allow the host to restore homeostasis. Central to the initiation and resolution of inflammation are cell populations of the myeloid lineage, particularly macrophages (Mϕ). Mϕ reside in the tissues and can be secondarily differentiated from recruited blood monocytes. They are characterized by a high degree of functional plasticity dictated by a wide range of environmental cytokines and mediators, including pathogen associated molecular patterns [1, 2]. As for Th1 and Th2 lymphocytes, activated Mϕ are often referred to as polarized M1 and M2. Classically activated M1 are induced by IFNγ alone or in concert with microbial stimuli and participate in the anti-bacterial response. Alternatively activated M2 constitute a heterogenic group including cells exposed to IL4 or IL13 and various immunosuppressive stimuli. The latter are induced by helminth infections and participate in the resolution phase of acute inflammation.
When inflammation-inducing conditions are sustained, the resulting chronic state can lead to the formation of a new structure, the granuloma, enriched in Mϕ and other immune cells. In this structure, Mϕ can further differentiate and acquire distinct morphologies [3]. Granulomas aid in the containment of otherwise uncontrollable pathogens and their destruction by T cells could be counter-productive for the host. Therefore, immunosuppressive mechanisms expressed by Mϕ, such as the amino acid degrading enzyme indoleamine 2,3 dioxygenase (IDO), are necessary to prevent excessive T cell activation [4].
We have recently shown that the immunosuppressive enzyme Interleukin-4 Induced Gene 1 (IL4I1) is expressed in various cancers by tumor-associated Mϕ, which are generally considered to belong to the M2 type [5, 6]. Interestingly, Chu et al. first identified IL4I1 as a B cell IL4-inducible gene and we also observed expression of the protein in several types of B-cell lymphomas [5, 7]. Our initial work identified the IL4I1 protein as a secreted L-phenylalanine oxidase that inhibits T cell proliferation through H2O2 production [8].
In this report, we have characterized the conditions of IL4I1 expression in vitro by human myeloid cell populations and B lymphocytes in order to understand which local stimuli may induce IL4I1 expression in inflammatory lesions. We have confirmed that B cells are sensitive to IL4, but surprisingly observed that myeloid cell populations respond to activation of STAT1 by interferons, in accordance with IL4I1 expression in Th1 but not in Th2 granulomas. While STAT1 and STAT6 thus exert dichotomous effects on B lymphoid and myeloid cells, both cell types are sensitive to activation of NFκ B by proinflammatory stimuli.
RESULTS
IL4I1 as a putative anti-inflammatory gene expressed by myeloid cells
We have previously reported that the purified IL4I1 enzyme was able to inhibit both CD4+ and CD8+ T lymphocyte proliferation in vitro [8]. This effect was dependent on the L-phenylalanine oxydase activity, in particular H2O2 production whose toxicity affected preferentially memory cells. In the current study, we have chosen the THP1 monocytic cell line to overexpress human IL4I1 (THP1-IL4I1), as we mainly detected IL4I1 expression in mononuclear phagocytes in vitro and in vivo [5, 8]. IL4I1 activity in the transfected cells was 895±363 pmoles H2O2/h/105 cells (mean from 5 independent tests) and the enzyme was secreted in the culture medium (Fig.1A), in accordance with previous results [8].
Figure 1. IL4I1 expression by monocytes inhibits T cell proliferation and inflammatory cytokine and chemokine secretion.
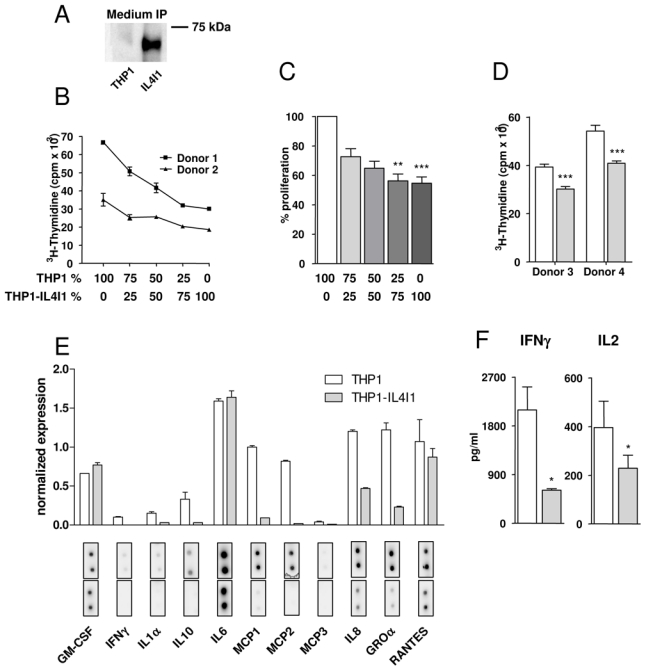
(A) Immunoprecipitation of IL4I1 protein from medium of THP1 and THP1-IL4I1 transfectant cells. (B, C, D) Human PBMC from two different donors co-cultured with irradiated THP1 or THP1-IL4I1 cells were stimulated with an anti-CD3 antibody (B & C) or with PPD (D). Proliferation was measured by 3H-thymidine incorporation during the last 18 hours of a 4-day culture. Results in B are expressed as the average cpm of quadruplicates ± standard deviation (SD) after background proliferation subtraction (see methods) of a representative experiment. Panel C depicts percent proliferation of PBMC cultured with THP1-IL4I1 as compared to PBMC cultured with THP1 cells (mean ± SD of independent experiments). (E) PBMC were co-cultured as in B and culture media harvested at day 3 were analyzed on a Raybiotech cytokine array. (F) PBMC were co-cultured as in B and day 1 and day 3 culture media analyzed respectively for IL2 and IFNγ secretion by ELISA (mean ± SD of 7 and 4 independent experiments, respectively). In D, E and F, white bars and gray bars represent results obtained with THP1 and THP1-IL4I1 cells, respectively. * p < 0.05, ** p < 0.01, *** p < 0.001.
As THP1 represent immature monocytic cells, they were not able to stimulate efficiently allogeneic PBMC proliferation (less than 5000 cpm, data not shown) unless added together with soluble anti-CD3 antibody, which by binding THP1 Fc receptors may facilitate T cell receptor cross-linking. In these conditions, THP1 represented potent polyclonal stimulators. In contrast, THP1-IL4I1 cells used in the same conditions were significantly less efficient stimulators with 45.2 ± 11.1 % proliferation inhibition in comparison to THP1 (mean of 7 different experiments, p=0.003). Moreover, this inhibitory effect was proportional to the number of THP1-IL4I1 cells added to the culture and detectable for ratios of as little as one THP1-IL4I1 cell per 32 PBMC (Fig. 1B & C and data not shown). To verify this effect in the context of an antigen specific response, we stimulated PBMC with purified protein derivative (PPD) and observed a similar proliferation inhibition, despite the antigen was probably not presented by the allogeneic THP1 cells (Fig. 1D).
Proliferation inhibition did not result from enhanced apoptosis as measured by Annexin V/7AAD or activated caspase 3 labeling (data not shown), but was associated with a decreased IL2 secretion at day 1 (p=0.015, 7 different experiments, Fig. 1F). However, proliferation in the presence of THP1-IL4I1 cells could only be partially restored by adding small amounts of exogenous IL2 (from 40.4 ± 9.1% to 28.3 ± 10.9% inhibition with or without 400 pg/ml IL2 [5 U/ml], respectively). This suggests that the limited IL2 secretion is only one of the mechanisms responsible for the reduced proliferation in the presence of IL4I1. Interestingly, we also measured a decrease in PBMC secretion of IL1α, IFNγ and inflammatory chemokines (IL8/CXCL8, GROα/CXCL1, MCP1/CCL2 and MCP2/CCL8) as shown in Fig. 1E & F. Since T cells were not purified from PBMC in the co-culture, we cannot exclude that some of the modifications in chemokine secretion were due to a cross–talk between the affected T cells and monocytes. Indeed, when LPS-stimulated monocytes purified by elutriation were cultured in conditioned media from THP1 and THP1-IL4I1 cells, no significant difference in their pattern of cytokine and chemokine secretion was observed, suggesting that IL4I1 does not directly regulate monocyte inflammatory properties (data not shown). In conclusion, these results extend our previous data and suggest that IL4I1 may exert an immunoregulatory function in inflammatory conditions.
Granulomas represent chronic inflammatory lesions massively infiltrated by Mϕ-derived cells and DC. While sarcoidosis and tuberculosis are classically associated with Th1 responses, helminth infection, such as schistosomiasis generates Th2 granulomas. In order to confirm that myeloid cells are the main IL4I1 producers in vivo and to explore what kind of cytokine milieu would favor IL4I1 expression in these cells, we selected for immunochemistry analysis, 6 cases of granuloma biopsies from patients with confirmed diagnosis of sarcoidosis, tuberculosis or schistosomiasis. Figure 2 shows the results obtained for two representative cases of Th1 (granulomatous adenitis from a sarcoidosis) and Th2 (bladder schistosomiasis) lesions respectively. The Th1 granulomas are highly organized structures consisting primarily of activated macrophages typically in the form of epithelioid and giant multinucleated cells, mixed with DC and some T lymphocytes [3]. In sarcoidosis and tuberculosis lesions, strong IL4I1 staining was detected in more than 80% of the granuloma cells, particularly in giant and epithelioid cells (Fig. 2A, a case of sarcoidosis). The IL4I1-positive cells displayed a Mϕ or DC morphology, even for the rare cells detected outside the granulomas, in the T cell (CD3-positive) and B cell (CD20-positive) rich zones. Using markers specific for Mϕ (CD68) and DC (S100), we confirmed that these cells were mainly located inside the granulomas, where most of the IL4I1-positive cells were also detected. Moreover, double staining demonstrated that most of the IL4I1-positive cells were also CD68- positive. Both DC-SIGN-positive interstitial DC and CD1a-positive Langerhans cells, which accumulated in some areas between the granulomas, expressed IL4I1. A cell suspension of a sarcoidosis lymph node frozen at diagnosis presented an IL4I1 activity of 70 pmoles/h/106 cells. Flow cytometry analysis of these cells indicated that 26% of CD3 positive and 32 % of CD3 negative cells produced IFNγ, confirming the Th1 context of the inflammatory response (data not shown).
Figure 2. IL4I1 is strongly expressed in Th1 granulomas.
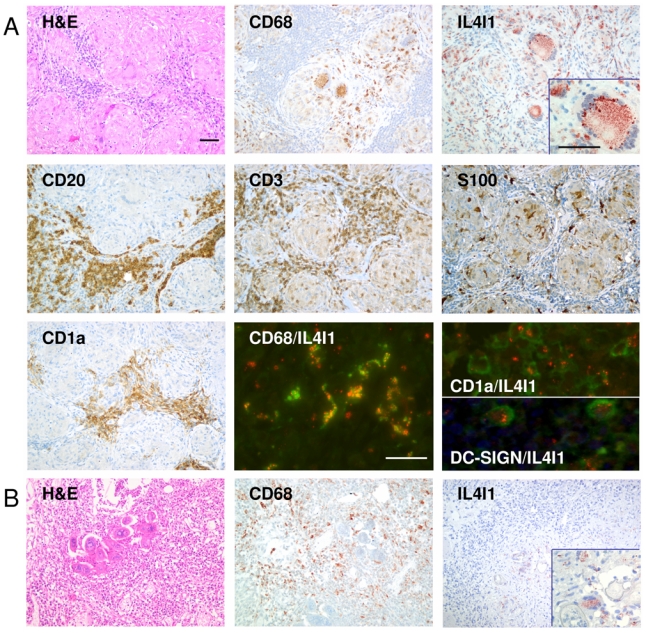
(A) Histomorphology of lymph node sections from a patient with sarcoidosis. H&E staining of a sample slide with prominent granulomas. Magnification ×200. Photographs of immunohistochemical staining with antibodies directed against CD68, IL4I1, CD20, CD3, S100 and CD1a. Magnification ×200, inset ×400. Immunofluorescence double staining of the same patient sample using a combination of anti-CD68 (granular green staining) or anti-CD1a (membrane green staining) or anti-DC-SIGN (membrane green staining) together with anti-IL4I1 (granular red staining) to determine the myeloid origin of IL4I1 expressing cells. CD68-IL4I1 double positive cells are yellow, whereas CD1a-IL4I1 or DC-SIGN-IL4I1 double-positive cells are rimmed with green and contain red granules. Magnification ×400. (B) Histomorphology of lymph node sections from a patient with schistosomiasis. H&E staining of a sample slide with prominent granulomas and parasite eggs. Photographs of immunohistochemical staining with anti-CD68 and anti-IL4I1 antibodies. Magnification ×200, inset ×400.
The structure of the granulomas in schistosomiasis is less organized than in Th1 lesions, with fewer giant cells. In contrast to the results obtained with tuberculosis and sarcoidosis, only very rare (<10%) IL4I1-positive myeloid cells were detected in these granulomas and when such cells were present, the specific staining was significantly less intense (Fig. 2B).
These results suggest that a cytokine milieu rich in IFNγ might favor IL4I1 expression in Mϕ and DC in vivo.
Differential role of IL4 and IFNγ in IL4I1 induction in B lymphocytes and myeloid cells
Our results were unexpected since IL4I1 has been originally described in B cells as an IL4-inducible gene [7]. This led us to explore in vitro the level of IL4I1 activity in myeloid cells, i.e. monocytes, monocyte-derived DC and Mϕ, versus B cells after stimulation with IL4 and IFNγ (Fig. 3A). As already described, IL4 was able to induce a two-fold increase in IL4I1 activity in B lymphocytes, whereas IFNγ did not have any effect. Conversely, IFNγ was a strong inducer of IL4I1 activity in all the myeloid populations, particularly in monocytes, which have a low basal activity (5 folds). No increase of IL4I1 activity in myeloid cells was observed in the presence of IL4, although we were not able to evaluate the IL4 effect on DC since this cytokine is necessary for their differentiation. However, only a minor induction of IL4I1 was observed after IL4 stimulation of purified circulating myeloid DC, whereas IFNγ confirmed its strong inducing effect on this population (supplementary table S1). Thus, IL4I1 can be induced both in B lymphocytes and myeloid cells, with IL4 and IFNγ exerting dichotomous roles in these populations. However, Mϕ and DC display much higher levels of IL4I1 activity than B lymphocytes (at least 50 folds more), with activities in a similar range of those measured in the THP1-IL4I1 cell line. As IL4I1 mRNA is known to be induced from the first hours post-IL4 stimulation in B cells, we determined the kinetics of IL4I1 production in myeloid cells. IL4I1 activity continued to increase after 48h of stimulation of DC, Mϕ or B cells with their respective stimulating cytokine (Fig. 3B), indicating that IL4I1 protein progressively accumulated over time. This suggests that threshold levels necessary to T cell inhibitory functions may be reached in terminally differentiated mononuclear phagocytic cells.
Figure 3. Dichotomy of IL4I1 activity induction by IFNγ and IL4.
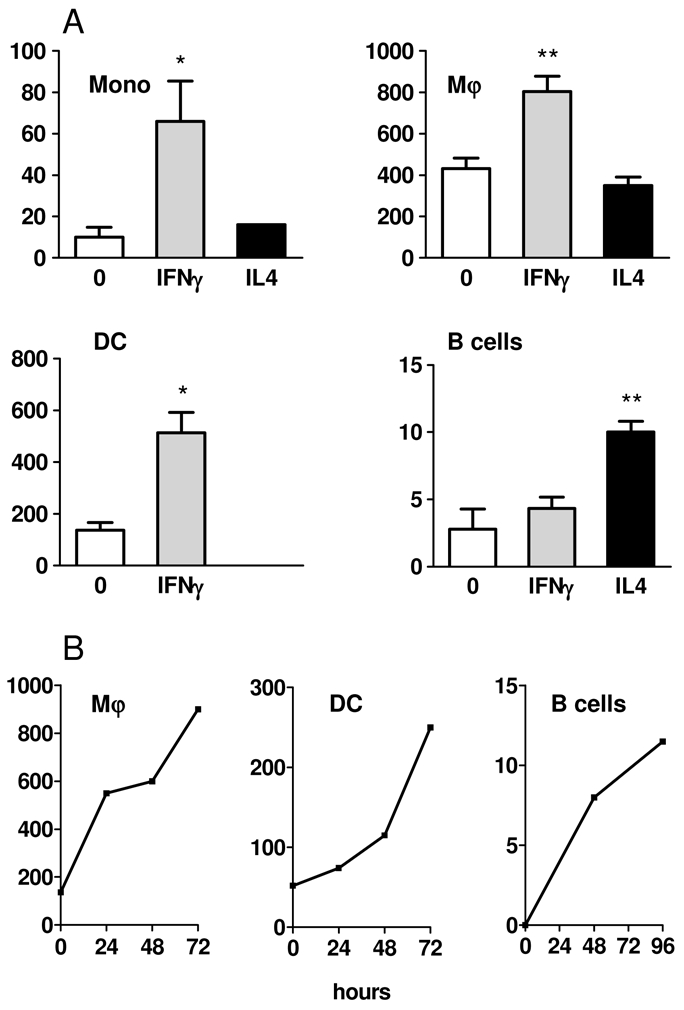
(A) 105 monocytes, monocyte-derived Mϕ, monocyte-derived DC and B cells were stimulated with IFNγ or IL4 and IL4I1 enzymatic activity against phenylalanine measured after 48h. The mean of 3 experiments ± SD is shown. (B) Kinetics of IL4I1 enzymatic activity of IFNγ-treated Mϕ and DC (left and center) and IL4-treated B lymphocytes (right). Representative experiments are shown. Enzymatic activity is expressed as pmoles H2O2 produced by 105 cells per hour using phenylalanine as substrate. * p < 0.05; ** p < 0.01.
Cytokinic and non-cytokinic stimuli associated with IL4I1 induction
The delayed kinetics of expression of this immunosuppressive enzyme in myeloid cells may point to a regulatory role in T cell activation during antigen presentation. We thus examined whether other stimuli associated with Mϕ and DC activation could induce IL4I1 activity. Mϕ were left unstimulated or stimulated with IFNγ, LPS, IL4, IL13 or different combinations of these agents (Fig. 4A). LPS induced similar levels of IL4I1 activity as IFNγ and the combination of both produced an additional increase. In contrast, neither IL4 nor IL13 modified the basal level of activity in unstimulated Mϕ or influenced the IL4I1 inducing effect of IFNγ. Similar results were observed for monocytes and THP1 cells (data not shown). We next tested activation of DC with different agents associated with acquisition of a mature or tolerogenic phenotype (Fig. 4B). DC were left untreated (iDC) or stimulated with Toll-like receptor (TLR) ligands (LPS, poly-IC and zymosan), CD40 ligand (CD40L), pro-inflammatory cytokines (IL1β, TNFα, IL6), interferons (IFNα, IFNβ, IFNγ) and tolerogenic stimuli (PGE2, TGFβ, IL10). All the stimuli which activate STAT1 – i.e type I IFN and INFγ – and/or NFκ B – i.e. TLR-ligands, CD40L, IL1β and TNFα – were able to strongly induce IL4I1 activity, while the tolerogenic cytokines TGFβ and IL10 seemed to repress the basal activity of iDC. The minimal doses of LPS and IFNγ able to induce IL4I1 activity in DC and THP1 cells were in the range of 0.5 to 2 ng/ml for IFNγ and 0.2 to 2 ng/ml for LPS. Results obtained for IFNγ in DC are shown in Fig. 4C. Combinations of minimal doses of INFγ, LPS and/or a standard dose of IL4 were tested on THP1 cells (Fig. 4D). As expected, IL4 did not enhance IL4I1 activity induced by IFNγ or LPS, while the combination of IFNγ and LPS was additive, suggesting that signalization through STAT1 and NFκ B participates independently to IL4I1 induction. In B cells, NFκ B involvement in the control of IL4I1 expression was suggested by the effect of CD40L, which seemed able to induce slightly IL4I1 as single agent but significantly enhanced the effect of IL4 (Fig. 4E). Accordingly, EBVinfected lymphoblastoid cell lines (LCL), which are known to express the NFκ B activating protein LMP1 [9], spontaneously displayed IL4I1 activity and were not further stimulated by CD40L, despite still being responsive to IL4 (Fig. 4F). Overall, these results suggest that IL4I1 transcription may be induced by STAT1 and STAT6 in myeloid cells and B cells, respectively, while NFκ B may represent a common regulator in both cell types.
Figure 4. IL4I1 activity in different myeloid cell populations and B cells.
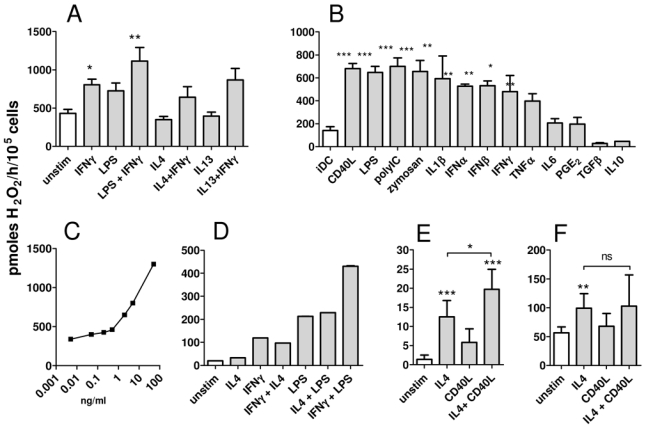
(A) IL4I1 activity was measured on 105 monocyte-derived Mϕ wich had been left unstimulated or activated for 48h with different M1 type stimuli (IFNγ, LPS or the combination of both) or M2 type stimuli (IL4, IL13) or a combination of these (IL4 and IFNγ or IL13 and IFNγ). (B) IL4I1 activity was measured on monocyte-derived DC which were left immature or stimulated 48h with maturation factors, as indicated on the graph (C) IL4I1 activity was measured on DC treated with graded doses of IFNγ. (D) IL4I1 activity of THP1 cells was measured after 48h stimulation with minimal doses of IFNγ (5 ng/ml), IL4 (5 ng/ml), LPS (2 ng/ml) or with combinations of these stimuli. IL4I1 activity was measured on primary tonsil B cells (E) or LCL cells (F) treated or not with IL4 and/or CD40L. Enzymatic activity is expressed as pmoles H2O2 produced by 105 cells per hour using phenylalanine as substrate. The results presented are the mean ± SD from at least 3 independent experiments, except for C and D, which show representative ones. * p < 0.05; ** p < 0.01, *** p < 0.001
Role of STAT proteins and NFκ B in IL4I1 gene expression
We therefore analyzed in silico the 1 kB region upstream of the IL4I1 coding sequence using the Genomatix software to identify consensus sequences for STAT and NFκ B transcription factors. Seven and 4 sequences were identified with putative affinity scores of 0.78 to 0.92 and of 0.92 to 1 for STAT and NFκ B, respectively (Fig. 5A). Since THP1 cells or B cell lines proved to be resistant to retroviral or plasmidic transfection, we carried on the transcriptional studies in a more classical system, i.e the HEK293 cell line. The 1kB hypothetical promoter region was cloned in a reporter vector and cells were transfected with this construct. HEK293 cells do not express TLR4 but are responsive to TNFα, which is an inducer of NFκ B activity. After stimulation with TNFα, a 2 to 4 fold induction of the Renilla luciferase reporter gene was measured, indicating that NFκ B can directly modulate IL4I1 expression via binding to its promoter sequence (Fig. 5B, p = 0.0005). Surprisingly, no significant expression of Renilla luciferase was induced by treatment of HEK293 cells with IFNγ, type I IFN or IL4 nor the combined treatment of the reporter cells with IL4 or IFNγ and TNFα increased the expression of the reporter gene compared to TNFα alone (data not shown). To improve the sensitivity of the assay and evaluate its performance, we transfected STAT1 and STAT6 expressing vectors and compared the activation of the IL4I1-promoter region to the activation of reference promoters, TAP-1 and Bcl-xL respectively. After treatment with the corresponding cytokine and despite phosphorylation of the overexpressed STAT factors (data not shown), a minor induction of transcription from TAP-1 and Bcl-xL promoter could be induced. In these conditions, we could also observe a small induction of transcription from the IL4I1- promoter region when STAT1 was activated. However, no consistent induction of the IL4I1-promoter could be measured after STAT6 activation (Fig. 5C & D). These data indicate that the IL4I1 promoter contains functional binding sequences for NFκ B. However, the low or absent reporter gene activity into HEK293 cells after stimulation with IL4 or IFNγ suggests that STAT proteins alone may not be sufficient to drive expression of IL4I1 in non-hematopoietic cells.
Figure 5. Transcriptional analysis of IL4I1 putative promoter region.
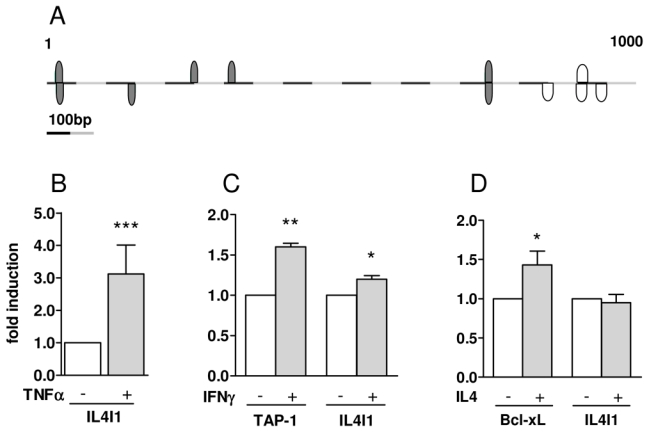
(A) Genomatix analysis of the 1000 bases upstream of transcription initiation of IL4I1 gene showing NFκ B putative binding sites (gray) and putative STAT binding sites (white). (B) HEK293 cells were cotransfected with a control firefly luciferase vector together with the 1000 bp sequence upstream of IL4I1 transcription initiation site inserted in pRL Null Renilla luciferase vector and stimulated or not with 25 ng/ml of TNFα. After an additional 24 hours, the cultures were harvested for determination of luciferase activity and Renilla luminescence reported to firefly luciferase luminescence as transfection normalisation. Results are expressed as the fold increase of specific luminescence compared to untreated controls. Data are the mean of 9 independent experiments ±SD. (C & D) HEK293 cells were co-transfected with 500 ng plasmid vectors coding for STAT1α(C) or STAT6 (D) either together with the IL4I1-promoter vector or the control TAP-1- or Bcl-xL-promoter vectors and the appropriate transfection controls as in B. Data are the mean of 5 independent experiments ±SD. * p < 0.05; ** p < 0.01, *** p < 0.001.
We thus evaluated the role of STAT1, STAT6 and NFκ B in our B and monocyte model cell lines, i. e. LCL and THP1 cells. The purine analog, fludarabine has been shown to inhibit specifically STAT1 expression at doses that are not associated with significant cell apoptosis [10, 11]. In THP1 cells pretreated with fludarabine, addition of IFNγ did not stimulate STAT1 expression (Fig. 6A) and had a minor effect on cell viability (respective cell recovery for the IFNγ-stimulated and unstimulated condition was 67% and 71% of the cells before fludarabine treatment). In accordance with our hypothesis, induction of IL4I1 expression by IFNγ was significantly inhibited by fludarabine (Fig. 6B), independently of its modest antiproliferative effect (all enzymatic activity measurements were calculated as a function of the number of viable cells). These results demonstrate the role of STAT1 in the induction of IL4I1 expression after IFNγ stimulation of monocytic cells.
Figure 6. STAT1, STAT6 and NFκB control of IL4I1 expression.
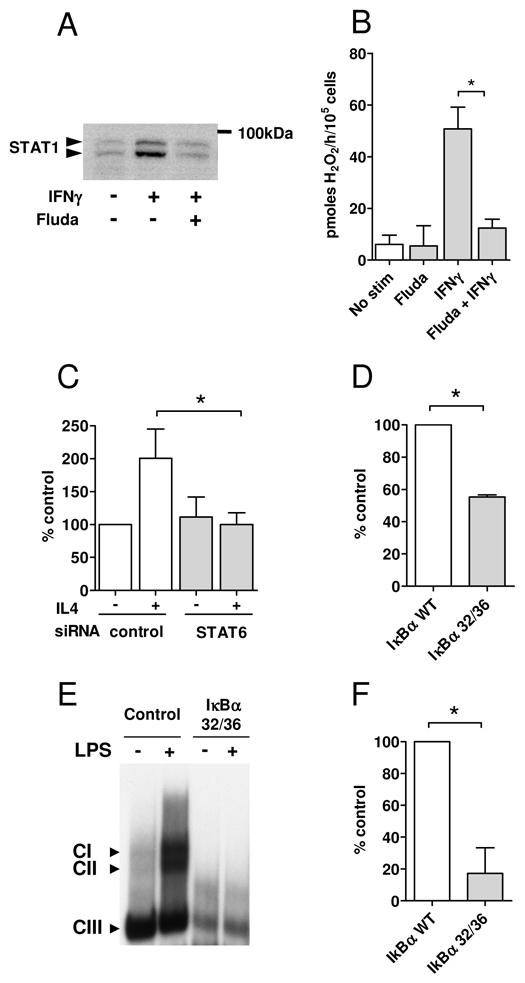
(A & B) IL4I1 activity in monocytic cells after inhibition of STAT1 expression with fludarabine. THP1 cells were pre-treated or not with 50μM of fludarabine 24h before stimulation with IFNγ. Total STAT1 protein expression (STAT1α and STAT1β) was analyzed by Western blot with untreated THP1 cells as control (A) and IL4I1 activity was measured as in Fig. 3 (B). (C) Role of STAT6 in IL4I1 activity in B cells. LCL cells were transfected with 3 μg control siRNA or STAT6 siRNA 72h prior stimulation with IL4. IL4I1 activity was measured 48h later and reported to control transfected cells left untreated. (D, E & F) Role of NFκ B in IL4I1 activity in monocytic and B cells. LCL cells were transiently transfected with a plasmid vector coding for the dominant IκBα S32A-S36A mutant (IκBα 32/36) or wild type (WT) IκBα as a control. IL4I1 activity was measured at day 3 after the transfection (D). THP1 cell line stably expressing IκBα 32/36 was established by infecting THP1 cells with pTRIP-ΔU3-EF1α-IRES GFP lentiviral vector and sorting of GFP+ cells. Iκ Bα 32/36 THP1 cells and control THP1 cells were stimulated by LPS (20 ng/ml for 90 min). Nuclear extracts were prepared and analyzed by EMSA using a 32P-labeled HIV-LTR tandem κ B oligonucleotide as a probe, to verify that NF-κB DNA binding activity was blocked in IκBα 32/36 THP1 cells (E). IL4I1 activity was measured 48h post-stimulation by LPS Data are presented as percent activity of mutant IκBα-expressing cells in comparison to their respective controls (F). All the IL4I1 activity results are the mean of 3 independent experiments ± SD. * p < 0.05. Note that the cell lysate samples used for all the measures are prepared with a fixed quantity of viable cells to avoid a bias due to variable cell viabilities in the different test conditions.
The role of STAT6 was explored by transfecting LCL cells with a STAT6- specific siRNA before IL4 treatment. As our two LCL did not display the same basal level of IL4I1 activity, the IL4I1 activity measured in control siRNA transfected cells not treated with IL4 was used as reference. As expected, IL4 significantly induced IL4I1 expression in control cells, while it did not have any effect on STAT6 siRNA transfected cells. These results demonstrate the role of STAT6 in IL4I1 induction after IL4 stimulation of B cells.
Finally, to confirm the role of NFκ B, we transfected LCL and LPS-activated THP1 cells with a plasmid coding for the dominant-negative Iκ Bα S32A-S36A mutant (Iκ Bα 32/36), which inhibits NFκ B activity. Despite the poor transfection efficacy (below 20% of the cells), there was a significant inhibition of IL4I1 activity in LCL cells in comparison to control cells transfected with WT Iκ Bα (Fig. 6D). A similar tendency was observed in THP1 cells. To improve the efficacy of NFκ B blocking, THP1 cells were transduced with a lentivirus encoding Iκ Bα 32/36 mutant, and then GFPpositive cells were sorted to generate a stable NFκ B-repressed cell line. A similar process was attempted on LCL cells, but due to their dependence on NFκ B activation for survival, the rare selected cells rapidly died in culture. As expected, nuclear translocation of p50/p65 complexes was abolished in Iκ Bα 32/36 THP1 transfectants stimulated with LPS (Fig. 6E and S2). Importantly, IL4I1 activity was profoundly depressed in these cells (Fig. 6F). We thus demonstrated that NFκ B, STAT1 and STAT6 participate to the induction of the active IL4I1 enzyme. However and much surprisingly, STAT1 is operative only in monocytic cells, while STAT6 induces IL4I1 in B lymphocytes. The results obtained with the reporter system suggest that cofactors may be necessary to full activity of the STAT factors in each cell type.
DISCUSSION
In this work, we demonstrate that, under inflammatory conditions, DC and Mϕ – and not B cells, as could have been expected from the literature [7, 12] – are the major IL4I1 producers in vitro and in vivo. Among proinflammatory stimuli, NFκ Bactivating microbial products and cytokines as well as type I IFN and IFNγ potently induce IL4I1 production by Mϕ and DC. This is illustrated by the major IL4I1 expression in granulomas formed in the context of a Th1 immune response in comparison to Th2-associated schistosomiasis granulomas. Furthermore, IL4I1 production by myeloid cells downregulates the Th1 response by limiting T cell proliferation and secretion of IFNγ and proinflammatory chemokines. This study highlights a second fundamental difference in IL4I1 expression by myeloid cells and B cells: while the former are sensitive to STAT1 activation by interferons, the latter respond to the IL4-STAT6 axis.
We had previously demonstrated that recombinant human IL4I1 coated on beads in a transwell system was able to inhibit CD3ζ expression leading to decreased CD4+ and CD8+ T cell proliferation. In this work, we have confirmed and extended these results by using a monocytic cell line constitutively expressing IL4I1 at similar levels to activated DC or Mϕ in order to mimic IL4I1 expression during an inflammatory reaction. In both polyclonal and antigen-specific conditions, we measured a significant decrease of T cell functions. However, we could still measure residual T cell proliferation. This might be due to the presence of 1 mg/L of the oxygen radical scavenger glutathione in the RPMI 1640 culture medium, which could limit the toxic effect of H2O2 produced by IL4I1 enzymatic activity. The effect of IL4I1 activity would have probably been greater in the absence of this molecule, but was impossible to measure since T lymphocytes did not proliferate in glutathione-free classical medium (DMEM, data not shown).
In addition, the inhibition of IL2 production and T cell proliferation was accompanied by diminished secretion of IFNγ, IL1α and proinflammatory chemokines. Since we did not observe a direct effect of IL4I1 on monocytes, the T cells which produce less IFNγ may instruct the monocytes present in the PBMC population to limit their chemokine production. Moreover, IL4I1 reaches its maximal expression in terminally differentiated Mϕ and DC, as has also been reported for the IDO enzyme [4, 13, 14], supporting a regulatory function. Thus, the role of IL4I1 – and therefore of its catabolite H2O2 – may be to control the proinflammatory environment via diminished T cell proliferation and reduced recruitment and activation of monocytic populations. In line with this, it has been suggested that the production of reactive oxygen species is involved in limiting the inflammatory response [15].
In vivo, a particularly high expression of IL4I1 was observed in giant and epithelioid cells from chronic Th1 lesions, such as tuberculosis and sarcoidosis. This is in accordance with microarray data showing the induction of IL4I1 transcription in mouse lung by aerogenic infection with Mycobacterium tuberculosis [16, 17]. Interestingly, another immunosuppressive enzyme, IDO, has been detected in listeriosis and to a lesser extent in other granulomatous diseases, including sarcoidoisis and tuberculosis [4]. Thus, expression of immunosuppressive enzymes in chronic Th1 lesions could be a common regulatory mechanism induced at variable levels by different pathogens. Granulomas are known to assume two complementary functions: they limit the proliferation of bacteria and prevent their systemic spread. Therefore, two independent roles might be attributable to immunosuppressive enzymes in these structures. (1) As suggested by Popov et al [4], local inhibition of destructive T cells could allow maintenance of the bacterial containment function. (2) At the same time, the enzymatic activity could have an antibacterial role via local consumption of essential amino acids and production of toxic catabolites (H2O2 for IL4I1 and kynurenine for IDO). We present evidence here that IL4I1 exert an inhibitory effect on T cell functions. Preliminary experiments indicate that IL4I1 also inhibits bacterial growth (VMF and FC unpublished data).
In marked contrast to the observations made in Th1 granulomas, only faint and rare expression of IL4I1 was detected in Th2 lesions. Indeed, we clearly demonstrated in vitro that IL4I1 expression in myeloid cell populations is insensitive to IL4, whether alone or in association with other stimuli. Only B cells produced IL4I1 in response to IL4, albeit to a dramatically lesser extent (more than 50 fold lower) than Mϕ or DC stimulated by interferons. This could explain why IL4I1 expression is not revealed by immunohistochemistry in the B cells present in Th2 lesions. We currently lack an explanation for this striking difference in cytokine sensitivity between B lymphocytes and myeloid cells. Both types of cells are equipped with STAT1 and STAT6, which can modulate some of their functions in vitro and in vivo [1, 18, 19]. Moreover, our bioinformatic analysis of the promoter region of IL4I1 confirmed the presence of STAT-binding consensus sites. In line with this, fludarabine, an inhibitor of STAT1 transcription, and a STAT6-specific siRNA inhibited IL4I1 production, in monocytic and B cell lines respectively. In mouse splenocytes, STAT6 has previously been shown to control IL4I1 transcription [19]. However, we detected only small STAT1- and no STAT6-transcriptional activity on the putative IL4I1 promoter region in the HEK293 cell gene reporter system. The minor induction of transcription from bona fide STAT1 and STAT6 promoters indicates that HEK293 cells are not optimal for the analysis of STAT promoter regions. Factors absent in epithelial cells may be necessary for the optimal transcriptional activation of IL4I1 by STAT proteins. These factors might be specific to mononuclear phagocytes and B lymphocytes, and regulate STAT signalling or associate with the STATs on the IL4I1 promoter.
In addition to STAT1 and STAT6, we identified NFκB as a key factor in IL4I1 expression that operates in both myeloid and B cells. Accordingly, IL4I1 transcription has been shown to be induced by both NFκB family members c-Rel and p65 in a murine macrophage model [20]. As for the STATs, the presence of additional factors specifically expressed by haematopoietic cells may be required to obtain an optimal transcriptional effect. This hypothesis is supported by the observation that cancerous epithelial cells only very rarely express IL4I1 despite frequent association with an inflammatory context [5, 21].
Numerous pathogen-associated molecular patterns (PAMPs) activate the NFκ B pathway and in our hands, LPS, zymosan and polyIC were able to induce in vitro a similar level of IL4I1 expression in myeloid cells. However, in vivo, in the context of Th2 granulomas, IL4I1 was only weakly detected. Thus, functional conditioning of the Mϕ in a Th2 environment may render them less sensitive to IL4I1 induction by NFκB, although this effect was not detectable in vitro (no influence of IL4 on LPS induction of IL4I1 in Mϕ, no reduction of the reporter gene expression when combining TNFα with IL4). An alternative explanation would be that stimulation with the multiple schistosmiasis PAMPs results in a modulation of NFκ B associated with a decrease in IL4I1 induction. Indeed, it has been recently shown that stimulation of DC-SIGN with mannose or fucose containing pathogens results in induction of IL12, IL6 and IL10 or IL10 only, respectively, via differential modulation of the TLR4-NFκ B axis [22].
In conclusion, our data demonstrate that myeloid cells from the mononuclear phagocyte lineage represent the major IL4I1 producing population in vitro and in vivo. In contrast to the B lymphocytes, IL4I1 activity in these cells is controlled by the interferon family of cytokines. By producing IL4I1 in response to STAT1 and NFκ B inducing stimuli, myeloid cells may play a major role in the regulation of chronic Th1 inflammation.
MATERIALS AND METHODS
Antibodies, cytokines and chemical inhibitors
Human recombinant IL4 (50ng/ml), IL6 (10ng/ml), IL10 (10ng/ml) and IL13 (50ng/ml) were purchased from R&D systems (Lille, France), LPS (20ng/ml), PGE2 (1μg/ml), polyIC (10μg/ml), zymosan (50μg/ml) and fludarabine (50μM) from Sigma Aldrich, human recombinant IFNα and IFNβ (10ng/ml), IFNγ (50ng/ml except otherwise stated), TNFα (10ng/ml), IL1β (10ng/ml), GM-CSF (50 mg/ml), M-CSF (100ng/ml), soluble CD40L (1μg/ml), TGFβ (10ng/ml) from Preprotech France (Levallois Perret, France). Monoclonal anti-CD20 (clone L26), monoclonal anti-CD68 (clone KP1) were purchased from DAKO (Glostrup, Denmark); monoclonal anti-S100 (clone S100A2/1) from Labvision (Interchim, Montluçon, France), monoclonal anti-CD1a (clone 010) from Beckman Coulter (Marseille, France). Monoclonal anti-DC-SIGN (clone 5D7) and polyclonal anti-IL4I1 were from AbCam (Cambridge, Great Britain). Monoclonal antibody blocking IFNγ clone MMHG-1 from Calbiochem was used at 4μg/ml. Monoclonal functional grade anti-CD3 (OKT3) was purchased from eBioscience (San Diego, USA). Anti-RelA, RelB, cRel, p105/p50 and p100/p52 supershifting antibodies were purchased from Santa Cruz Biotechnology.
Cell culture and transduction
The Human Embryonic Kidney (HEK) 293 cell line and the monocytic THP1 cell line were cultivated respectively in DMEM and RPMI 1640 (Invitrogen, Cergy Pontoise France) containing 10% foetal calf serum (FCS). Human lymphoblastoid cell lines (LCL) were cultivated in RPMI 1640 containing 10% heat-inactivated human AB serum (PAA, France). All media were supplemented with 2mM L-glutamine, 100 UI/ml penicillin and 100 μg/ml streptomycin (all from Invitrogen). Human peripheral blood mononuclear cells (PBMC) from healthy subjects were obtained after informed consent from the French Blood Bank (EFS) and purified on a lymphocyte separation gradient (Eurobio, France). Human monocytes were enriched by adherence from PBMC. DC were differentiated in AIM-V serum free medium (Invitrogen) supplemented with glutamine and 50ng/ml IL4 and 50ng/ml GM-CSF. Cells obtained after 5–6 days in culture were designated as immature DC (iDC). Mature DC (mDC) were obtained by culturing 1×106 iDC with different maturation stimuli in AIM-V serum-free medium in 6-well plates for up to 48–72 h. Adherent mDC were removed by gentle scraping. Monocyte-derived macrophages were seeded in sealed gaspermeable bags (Baxter Healthcare corporation, Deerfield, USA) under non adherent conditions at a density of 1×106 cells/ml containing 100ng/ml of M-CSF. After 6 days of culture, macrophages were recovered, counted and seeded in 6 well plates at 1×106 cells/ml and activated with different stimuli. B lymphocytes were obtained from human tonsils obtained from tonsillectomies performed at the Intercommunal Hospital of Créteil with ethical approval. Tissue was cut into small pieces in RPMI 1640 containing 10% FCS and forced through a 100μm mesh nylon screen to disrupt the pieces into singles cells. After two washes in RPMI/FCS, the cells were counted. The viability of all cells or cell lines was established by Trypan blue exclusion. Production of infectious recombinant lentiviruses was performed by transient transfection of HEK293 cells as described by Kieusseian et al [23]. For infections, 1×105 cells in 35- mm dishes were transduced with 5000 ng/ml of p24 (HIV-1 capsid protein). An identical amount of lentiviral vector particles was added 24 hours later, and the incubation was prolonged for a total of 72 hours. Cells were then washed, and fresh medium was added. The culture was then continued as described above.
Plasmids, siRNA and transfection conditions
Human IL4I1 cDNA was cloned in pCDN3.1 myc-his (Invitrogen) as described in Boulland et al. [8]. Wild-type (WT) Iκ Bα and S32A-S36A dominant negative mutants in a pCMV vector were purchased from Clontech Laboratories (Mountain view, CA, USA). pTRIP-ΔU3-EF1α-IRES GFP lentiviral vector [23] for IκBα S32A-S36A mutant was a kind gift from M. Fontenay (Institut Cochin, Paris, France). STAT6 cDNA cloned in pcDNA was described by Ritz et al [24] and STAT1α cDNA cloned in pRC/CMV vector was purchased from Addgene (Addgene inc, Cambridge, MA, USA). Control (AAUUCUCCGUUCGUGUCACGU) or STAT6 (ACGGAUAGGCAGGAACAUACA)-targeting small interfering RNA (siRNA) were obtained from Qiagen (Courtaboeuf, France). TAP-1 and Bcl-xL luciferase reporter vectors were a kind gift from I. Dusanter, Institut Cochin, Paris [25]. The THP1 monocytic cell line and lymphoblastoid cell lines were nucleofected (Lonza, Germany) using the nucleofection kit V and the program L13 and O17 respectively. Stable THP1 cell lines expressing hIL4I1 (THP1-IL4I1) were selected with 0.75mg/ml of G418 (Invitrogen). IL4I1 activity of the THP1-IL4I1 cell lines was 895±363 pmoles H2O2/h/105 cells and was in the range of the activities measured on IFNγ stimulated Mϕ or DC.
Enzymatic assay
IL4I1 enzymatic assay was performed as described in Carbonnelle-Puscian et al. [5] using Amplex Ultra Red (Invitrogen, France) and expressed as pmoles H2O2 produced in the presence of phenylalanine per hour per 105 viable cells unless otherwise specified.
Proliferation assays
All functional assays were performed in complete RPMI 1640 supplemented with 10% heat-inactivated human AB-serum. PBMC (2×105 cells/well) stimulated with anti- CD3 (20ng/ml) were co-cultured 4 days in 96 well round bottom plates with 2.5×104 to 105 irradiated (150 Gy) THP1 and THP1-IL4I1 cells in various proportions (as indicated in figure 1A). Supernatants were collected for cytokines and chemokines measurements (IL2, day1, IFNγ and multiple cytokine detection, day3). Alternatively, the PBMC were incubated with 100U/ml of purified protein derivative (PPD, Aventis Pasteur) and irradiated THP1 or THP1-IL41 cells for 5 days. 3H-thymidine (1 μCi/well, Amersham, Saclay, France) was added for the final 18 h of coculture. T cell proliferation is expressed as cpm 3H-thymidine incorporated. Background proliferation of PBMC or cell lines alone never exceded 1000 cpm and was subtracted from final results.
Cytokine measurements
Cytokine and chemokine secretion in proliferation experiments was detected using the human cytokine antibody array 1 from RayBiotech according to manufacturer’s instructions. Dot blots were quantified using the Labwork software (UVP, United Kingdom). No production of cytokines or chemokines was detected in supernatants of irradiated THP1 or THP1-IL4I1 cells alone. Human IL2 and IFNγ were measured with OptEIA ELISA kits from BD Biosciences (Le Pont de Claix, France) according to manufacturer’s instructions.
Immunohistochemistry and immunofluorescence
IL4I1 immunohistochemistry on formol-fixed paraffin embedded thin sections was performed as described by Carbonnelle-Puscian et al. [5]. Images were captured with an Olympus BX40 microscope (Olympus, Tokyo, Japan). Photographs were taken with an Olympus C-5060 camera. Images were processed with Adobe Photoshop v7.0 (Adobe Systems, San Jose, CA). Double immunofluorescence labeling of deparaffinated 3–5μm tissue slices was performed after antigen retrieval in citrate buffer pH=6 at 98°C for 30min. Mouse primary antibodies were revealed using Alexa Fluor 488 conjugated anti-mouse antibodies (Molecular Probes, Invitrogen). IL4I1 antibodies were revealed using an anti-rabbit biotinylated antibody followed by a streptavidin-Texas Red. Photographs acquisition was performed with a Hamamatsu C8484 digital camera and Fluovision IMSTAR software. This study was approved by the French ethical committee “Comité de Protection des Personnes” Ile-de-France IX.
Reporter assay
The 1Kb non-transcribed fragment in 5′ of the IL4I1 coding sequence was analysed for putative transcription factor binding sequences using Genomatix MatInspector (Genomatix Software GmbH, Munchen, Germany). The 5′ region of IL4I1 was amplified from genomic DNA extracted from PBMC by PCR reaction using the following primers (forward 5′-agagagctcgagtctttgatttactgcgggagtt’-3; reverse 5′-agagaggaattctctgctgtggccctttct-3′) and High Fidelity Taq polymerase (Invitrogen, France) according to the following program: 94°C 2min, 5 cycles 94°C 30sec, 55°C 30 sec, 68°C 1min, 25 cycles 94°C 30sec, 65°C 30sec, 68°C 1min with a final elongation at 68°C for 10 min. The PCR product digested with XhoI and EcoRI was cloned in pRL-Null reporter plasmid (Promega, Charbonnieres, France) (IL4I1-prom). All cloned DNA was confirmed by sequencing. Reporter assays were performed in HEK 293 cells seeded in 24w plates and transiently transfected using Transfast reagent (Promega) according to the manufacturer’s instructions with 500 ng of IL4I1- prom together with 50 ng of pGL3 basic plasmid. In co-transfection experiments, 500 ng of STAT1α or STAT6 DNA were added. Twenty-four hours after the transfection cells were stimulated with 25 ng/ml of TNFα, IFNγ or IL4 for an additional 24 hours and fluorescence analysis performed using the Promega Dual-luciferase reporter assay system and a Fluostar Optima plate reader (BMG Labtech, Champigny sur Marne, France). Results are expressed as the fold increase of fluorescence of treated cells compared to untreated cells.
Immunoprecipitation and Western blot
One ml of conditioned medium from THP1 or THP1-IL4I1 was immunoprecipitated by addition of 4.5μg of rabbit anti-IL4I1 antibodies [8] overnight at 4°C followed by protein A/G sepharose (Santa Cruz, USA). After extensive washes, bound proteins were eluted in SDS-PAGE loading buffer and separated by SDS-PAGE followed by transfer onto polyvinylidene difluoride membranes (Millipore, St Quentin en Yvelines, France). Revelation was performed using the same specific primary antibody, followed by revelation using anti-rabbit HRP-conjugates and Enhanced Chemi Luminescence (GE ECL plus®) detection with the Autochemi System and Labworks Analysis Software (UVP, Cambridge, UK). For STAT1 Western blot, extracts containing similar amounts of proteins were migrated by SDS-PAGE as above and revealed after transfer by a rabbit polyclonal anti-STAT1 antibody (Cell signaling).
Electrophoretic Mobility Shift Assays (EMSA)
Nuclear extracts were prepared as in Pallard et al [26] and analyzed for DNA binding activity using the HIV-LTR tandem κB oligonucleotide as κB probe [27]. For supershift assays, nuclear extracts were incubated with specific antibodies for 30 min on ice, before incubation with the labeled probe.
Statistical analysis
Statistical analyses were performed using one-way Anova test followed by Bonferroni post-test or Wilcoxon matched paired t test.
Supplementary Material
Acknowledgments
This work was supported by ARC subvention fixe 4883 to FC. VB is supported by ANR, ARC, Belgian InterUniversity Attraction Pole, Cancéropole Ile-de-France and Institut National du Cancer.
We thank Michaela Fontenay for providing the lentiviral construct and Isabelle Dusanter for providing TAP1 and Bcl-xL reporter constructs. We are grateful to Amélie Carbonnelle-Puscian and Yves Allory for providing precious human samples and help in the interpretation of immunohistochemistry results. We also thank William Hempel for a critical reading of the manuscript. CC was supported by ARTGIL (granted by Amgen and Roche France).
Abbreviations
- IL4I1
IL4-induced gene 1
- LCL
lymphoblastoid cell line
- HEK
Human Embryonic Kidney 293 cell line
- PPD
purified protein derivative
Footnotes
Conflict-of-interest disclosure: The authors declare no competing financial interests.
References
- 1.Mantovani A, Sica A, Locati M. Macrophage polarization comes of age. Immunity. 2005;23:344–346. doi: 10.1016/j.immuni.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 3.Emile JF. Systemic granulomatosis. Anatomy/pathology of granuloma. Rev Med Interne. 2005;26 (Suppl 1):S4–5. doi: 10.1016/j.revmed.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Popov A, Abdullah Z, Wickenhauser C, Saric T, Driesen J, Hanisch FG, Domann E, Raven EL, Dehus O, Hermann C, Eggle D, Debey S, Chakraborty T, Kronke M, Utermohlen O, Schultze JL. Indoleamine 2,3-dioxygenase-expressing dendritic cells form suppurative granulomas following Listeria monocytogenes infection. J Clin Invest. 2006;116:3160–3170. doi: 10.1172/JCI28996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carbonnelle-Puscian A, Copie-Bergman C, Baia M, Martin-Garcia N, Allory Y, Haioun C, Cremades A, Abd-Alsamad I, Farcet JP, Gaulard P, Castellano F, Molinier-Frenkel V. The novel immunosuppressive enzyme IL4I1 is expressed by neoplastic cells of several B-cell lymphomas and by tumor-associated macrophages. Leukemia. 2009;23:952–960. doi: 10.1038/leu.2008.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 7.Chu CC, Paul WE. Fig1, an interleukin 4-induced mouse B cell gene isolated by cDNA representational difference analysis. Proc Natl Acad Sci U S A. 1997;94:2507–2512. doi: 10.1073/pnas.94.6.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulland ML, Marquet J, Molinier-Frenkel V, Moller P, Guiter C, Lasoudris F, Copie-Bergman C, Baia M, Gaulard P, Leroy K, Castellano F. Human IL4I1 is a secreted L-phenylalanine oxidase expressed by mature dendritic cells that inhibits T-lymphocyte proliferation. Blood. 2007;110:220–227. doi: 10.1182/blood-2006-07-036210. [DOI] [PubMed] [Google Scholar]
- 9.Kuppers R. The biology of Hodgkin’s lymphoma. Nat Rev Cancer. 2009;9:15–27. doi: 10.1038/nrc2542. [DOI] [PubMed] [Google Scholar]
- 10.Frank DA, Mahajan S, Ritz J. Fludarabine-induced immunosuppression is associated with inhibition of STAT1 signaling. Nat Med. 1999;5:444–447. doi: 10.1038/7445. [DOI] [PubMed] [Google Scholar]
- 11.Huang Y, Walstrom A, Zhang L, Zhao Y, Cui M, Ye L, Zheng JC. Type I interferons and interferon regulatory factors regulate TNF-related apoptosis-inducing ligand (TRAIL) in HIV-1-infected macrophages. PLoS One. 2009;4:e5397. doi: 10.1371/journal.pone.0005397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chavan SS, Tian W, Hsueh K, Jawaheer D, Gregersen PK, Chu CC. Characterization of the human homolog of the IL-4 induced gene-1 (Fig1) Biochim Biophys Acta. 2002;1576:70–80. doi: 10.1016/s0167-4781(02)00295-6. [DOI] [PubMed] [Google Scholar]
- 13.Jurgens B, Hainz U, Fuchs D, Felzmann T, Heitger A. Interferongamma- triggered indoleamine 2,3-dioxygenase competence in human monocyte-derived dendritic cells induces regulatory activity in allogeneic T cells. Blood. 2009;114:3235–3243. doi: 10.1182/blood-2008-12-195073. [DOI] [PubMed] [Google Scholar]
- 14.Chung DJ, Rossi M, Romano E, Ghith J, Yuan J, Munn DH, Young JW. Indoleamine 2,3-dioxygenase-expressing mature human monocyte-derived dendritic cells expand potent autologous regulatory T cells. Blood. 2009;114:555–563. doi: 10.1182/blood-2008-11-191197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hultqvist M, Olsson LM, Gelderman KA, Holmdahl R. The protective role of ROS in autoimmune disease. Trends Immunol. 2009;30:201–208. doi: 10.1016/j.it.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez-Juarrero M, Kingry LC, Ordway DJ, Henao-Tamayo M, Harton M, Basaraba RJ, Hanneman WH, Orme IM, Slayden RA. Immune response to Mycobacterium tuberculosis and identification of molecular markers of disease. Am J Respir Cell Mol Biol. 2009;40:398–409. doi: 10.1165/rcmb.2008-0248OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller C, Hoffmann R, Lang R, Brandau S, Hermann C, Ehlers S. Genetically determined susceptibility to tuberculosis in mice causally involves accelerated and enhanced recruitment of granulocytes. Infect Immun. 2006;74:4295–4309. doi: 10.1128/IAI.00057-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baran-Marszak F, Feuillard J, Najjar I, Le Clorennec C, Bechet JM, Dusanter-Fourt I, Bornkamm GW, Raphael M, Fagard R. Differential roles of STAT1alpha and STAT1beta in fludarabine-induced cell cycle arrest and apoptosis in human B cells. Blood. 2004;104:2475–2483. doi: 10.1182/blood-2003-10-3508. [DOI] [PubMed] [Google Scholar]
- 19.Schroder AJ, Pavlidis P, Arimura A, Capece D, Rothman PB. Cutting edge: STAT6 serves as a positive and negative regulator of gene expression in IL-4-stimulated B lymphocytes. J Immunol. 2002;168:996–1000. doi: 10.4049/jimmunol.168.3.996. [DOI] [PubMed] [Google Scholar]
- 20.Sanjabi S, Williams KJ, Saccani S, Zhou L, Hoffmann A, Ghosh G, Gerondakis S, Natoli G, Smale ST. A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes Dev. 2005;19:2138–2151. doi: 10.1101/gad.1329805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 22.Gringhuis SI, den Dunnen J, Litjens M, van der Vlist M, Geijtenbeek TB. Carbohydrate-specific signaling through the DC-SIGN signalosome tailors immunity to Mycobacterium tuberculosis, HIV-1 and Helicobacter pylori. Nat Immunol. 2009;10:1081–1088. doi: 10.1038/ni.1778. [DOI] [PubMed] [Google Scholar]
- 23.Kieusseian A, Chagraoui J, Kerdudo C, Mangeot PE, Gage PJ, Navarro N, Izac B, Uzan G, Forget BG, Dubart-Kupperschmitt A. Expression of Pitx2 in stromal cells is required for normal hematopoiesis. Blood. 2006;107:492–500. doi: 10.1182/blood-2005-02-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritz O, Guiter C, Castellano F, Dorsch K, Melzner J, Jais JP, Dubois G, Gaulard P, Moller P, Leroy K. Recurrent mutations of the STAT6 DNA binding domain in primary mediastinal B-cell lymphoma. Blood. 2009 doi: 10.1182/blood-2009-03-209759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouyez MC, Lestingi M, Charon M, Fichelson S, Buzyn A, Dusanter-Fourt I. IFN regulatory factor-2 cooperates with STAT1 to regulate transporter associated with antigen processing-1 promoter activity. J Immunol. 2005;174:3948–3958. doi: 10.4049/jimmunol.174.7.3948. [DOI] [PubMed] [Google Scholar]
- 26.Pallard C, Gouilleux F, Benit L, Cocault L, Souyri M, Levy D, Groner B, Gisselbrecht S, Dusanter-Fourt I. Thrombopoietin activates a STAT5-like factor in hematopoietic cells. Embo J. 1995;14:2847–2856. doi: 10.1002/j.1460-2075.1995.tb07284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacque E, Tchenio T, Piton G, Romeo PH, Baud V. RelA repression of RelB activity induces selective gene activation downstream of TNF receptors. Proc Natl Acad Sci U S A. 2005;102:14635–14640. doi: 10.1073/pnas.0507342102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


