Abstract
Proteins containing bromodomains are capable of binding to acetylated histone tails and have a role in recognizing and deciphering acetylated chromatin. Plant BET proteins contain one bromodomain. Twelve BET-encoding genes have been identified in the Arabidopsis genome. Two of these genes have been functionally characterized, one shows a role in seed germination, the other is involved in the establishment of leaf shape. Recently, we characterized a third AtBET gene, named GTE4. We demonstrated that GTE4 is involved in the activation and maintenance of cell division in the meristems and by this controls cell numbers in differentiated organs. Moreover, the quiescent center (QC) identity is partially lost in the apex of the primary root of gte4 mutant, and there is a premature switch from mitosis to endocycling. Genes involved in the retinoblastoma (RB)-E2F pathway, which is important for coupling cell division and cell differentiation in plants and animals, were either up or downregulated in the gte4 mutant. In this report we also show that the defect in germination observed in gte4 mutant seeds is not rescued by the action of GA3. Further the root pole of the mutant embryo shows irregular cytokinesis in the procambial stem cells, and the QC of the lateral root shows a partial, but not transient, loss of QC identity. These additional results reinforce the importance of GTE4 in the control of cell proliferation.
Key words: arabidopsis, BET bromodomain, cell cycle, E2F, germination
Introduction
The acetylation of histone tails are able to relax the packing of the DNA, facilitating the access to DNA of many regulatory proteins involved in replication, transcription, repair and recombination.1 Proteins containing bromodomains are capable of binding to acetylated histone tails and have therefore a role in recognizing and deciphering acetylated chromatin.2,3 The bromodomain was first discovered in the Drosophila Brahma protein and is present in a broad range of chromatin-modifying proteins.4,5 Some bromodomain proteins exhibit an extra terminal (ET) domain besides the N-terminal bromodomain(s), and are named BET proteins.6 The ET domain functions as an interaction domain to recruit other proteins to acetylated histones.7 BET proteins have been studied in a variety of organisms, where they are implicated in cell cycle progression and contribute to the transmission of the transcriptional memory from one generation of cells to the next.2,8,9 The BET proteins of plants are different from those of yeast and animals, since they have only one bromodomain instead of two.6 Twelve BET-encoding genes have been identified in the Arabidopsis genome, however only two of them, IMBIBITION INDUCIBLE1(IMB1) and GENERAL TRANSCRIPTION FACTOR GROUP E6 (GTE6), were functionally characterized, and showed a role in seed germination and establishment of leaf shape, respectively.10,11 Recently, we characterized GTE4, another gene belonging to a so far uncharacterized group of Arabidopsis BET proteins, and showed that gte4 mutant plants have some characteristic features of cell cycle mutants.12 We demonstrated that GTE4 is involved in the activation and maintenance of cell division in the meristems and by this controls cell numbers in the root, stem and leaf.12 Moreover, we suggest a link between GTE4 and E2F related pathways, which are known to regulate stem cell maintenance in the root meristem and to be involved in cell cycle reactivation in the quiescent center (QC) cells.12–14 Here we present additional data that further support a role of GTE4 in seed germination, the control of cell division and cytokinesis in the root stem cells of the mature embryo, and in the definition of the size of the differentiated leaves. These results reinforce the importance of GTE4 in the control of cell proliferation.
Loss of GTE4 Function Affects Plant Development
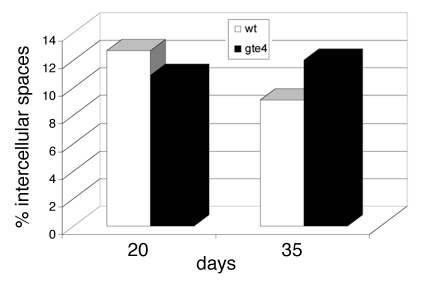
To determine whether the loss of GTE4 function affects Arabidopsis development, we analyzed histologically gte4 mutant during vegetative and reproductive growth. The mutant plants were reduced in size at all stages of development. The leaves were macroscopically smaller than those of the wild type. However, there was no significant change both in the mean cell area,12 and in the mean area of the intercellular spaces, as exemplified for the 6th leaf at days 20 and 35 after germination (Fig. 1A and B and Fig. 2). This shows that in the gte4 leaves the reduced macroscopic size is related to a reduced cell number, because neither the intercellular spaces nor the cell size12 are affected. The thickness of the inflorescence stem was also significantly reduced in the gte4 mutant, and this was due to a reduced cell number. Moreover, a reduction in stamen number occurred in the mutant.12
Figure 1.
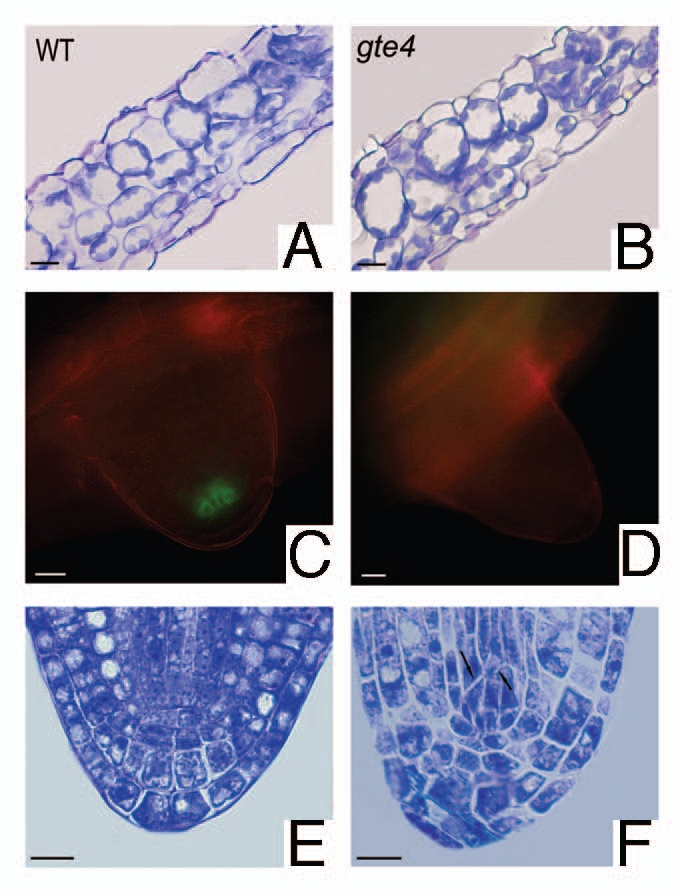
Effects of gte4 mutation on the histological structure of the leaf and the embryonic root pole, and the expression of a QC marker in the lateral root primordium. (A and B) Cross sections of the 6th rosette leaf showing the cell types and the extension of the intercellular spaces in the mesophyll of wild-type (A) and gte4 mutant (B). (C and D) Expression of AGL42:GFP in the apex of a lateral root primordium. A strong GFP signal in the QC cells is present in the wild-type (C), whereas no signal is present in gte4 mutant containing the same construct (D). (E and F) Details of the longitudinal sections of the root pole of the embryo at the cotyledonary stage showing the regularly arranged cell files of the initials in the wild type (E), and the irregularly divided procambial initials (arrows) in the mutant (F). (A, B, E and F) toluidine blue-stained sections observed under light microscopy. (C and D) samples observed under an epifluorescence microscope equipped with a double wavelength filter set (BP 490/20 and BP 575/30) with dichroic filters RKPs 505 and 600 and emission filters BPs 525/20 and 635/40. Bars = 20 µm (A and B), 10 µm (C–F).
Figure 2.
Percentage of intercellular spaces in the mesophyll of rosette leaves of 20- and 35-old gte4 and wild type plants. (5,000 µm2 randomly chosen in the 6th leaf of 10 specimens per genotype).
The primary root of gte4 was macroscopically shorter than that of the wildtype, and the reduction in length was observed in the hairy region, in the elongation region, and in the apical meristem. Cell size in the apical region in the mutant was, instead, higher than in the wild type, resulting in a highly reduced cell number in gte4 apices. They also showed an anomalous organization, and QC defects, as demonstrated by histological and epifluorescence analyses with QC marker lines (two QC-expressed promoter trap GUS lines, i.e., QC25 and QC46, and the AGL42:GFP line). We demonstrated that the loss of GTE4 function causes a partial loss of QC identity, since in the gte4 mutant containing the AGL42:GFP marker, no GFP signal was detected in all stages of root development in which the QC is present, whereas the QC25 and QC46 markers were normally expressed.12 Furthermore, lateral root formation was also reduced in comparison with wild type, and the apex of the lateral root primordium showed an anomalous morphology. To investigate whether the lateral roots exhibited the same features as the primary root apex, we investigated histologically all the stages of lateral root development following QC definition with the same QC markers. We observed that the QC25 and QC46 markers are expressed, the same as in the primary root apex, in both gte4 and wild type lateral root primordia (data not shown). However, in the gte4 mutant containing the AGL42:GFP marker, no GFP signal was detected in comparison with the wild type (Fig. 1C and D) independently on the primordium developmental stage, in accordance with the results obtained for the primary root apex.
We also analyzed the formation of the embryo during seed development to find whether the gte4 mutant has defects in the definition of the root pole at the embryonic level. The root pole was already defined in the wild type embryo at the cotyledonary stage, whereas it showed irregularly shaped QC cells and irregularly divided columella initials in the mutant at the same embryonic stage.12 Moreover, we presently show that also the division planes in the procambial initials were anomalous in position (Fig. 1E and F), demonstrating that cytokinesis is affected in various types of stem cells in the mutant. The anomalous positioning of cell walls continued to be observed during post-embryonic development, still occurring in the procambial initials, in particular, in the apex of the primary root at day 4 after germination.12
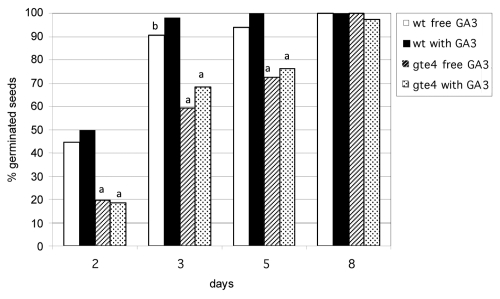
Previously, we have shown that there is a delay in germination in gte4 mutant seeds in comparison with the wildtype.12 However, there is the possibility that an exogenous hormonal treatment may counteract GTE4-induced delay in germination. To investigate whether GA3, i.e., the hormone which promotes germination in Arabidopsis,15 is able to overcome the delay in germination, gte4 and wild type seeds were sown on solid half strength MS medium,16 with 1 µM GA3 under a 16 h light/day photoperiod. As expected, the treatment with GA3 enhanced seed germination in the wild type whereas no stimulation of germination was observed for gte4 mutant seeds (Fig. 3). Moreover, the delay in germination of the mutant was observed up to day 5, both in the absence and in the presence of GA3, and without significant differences in the percentages of germinated seeds between the treatments (Fig. 3). Thus, the mutation causes a transient defect in germination which is not rescued by the action of GA3. It is interesting to note that IMB1, another BET-encoding gene of Arabidopsis, belonging to a different subfamily in comparison with GTE4,12 similarly does not control GA3-regulated germination.10
Figure 3.
Seed germination in the presence and absence of GA3, in gte4 in comparison with wild type. The percentages of germinated seeds on the total of sowed seeds are shown at different days in culture. aValues of gte4 statistically different at P < 0.01 level with respect to the corresponding values of wild type grown under the same conditions. bValue statistically different at P < 0.05 level with respect to wild type grown with GA3 at the same day. Values of the same genotype and day followed by no symbol or the same symbol are not significantly different (N = 90).
Is GTE4 Involved in the E2F Related Cell Cycle Pathway?
To investigate the onset of cell proliferation during seed germination, wild type and gte4 mutant seeds were imbibited and grown in the presence of bromodeoxyuridine (BrdU), which allows the detection of cells entering the S phase.12 We observed that the delay in root emergence from the seed coat (i.e., germination) was related to the delay in cell cycle activation. Moreover, applying the BrdU pulse/chase method along with the determination of mitotic index and flow cytometry,17 we showed that a substantial number of gte4 cells exit from the mitotic cell cycle earlier than the corresponding wild-type cells, and switch from mitosis to endoreduplication. This results into an increase in ploidy levels in the differentiated organs of gte4 plants.12 The retinoblastoma (RB)-E2F pathway is one of the most important pathways involved in the control and coupling of cell division and cell differentiation in animals and plants.18 We tested by quantitative RT-PCR the expression of a few genes important for a correct cell proliferation/endoreduplication and related to the RB-E2F pathway (i.e., E2Fc, E2Fe/DEL1, CDC6 and CCS52A2). We found that, except for E2Fe/DEL1, whose expression did not significantly change, E2Fc and CCS52A2 expression was upregulated, while that of CDC6 was downregulated in 4-days-old gte4 plantlets. These results show a possible involvement of GTE4 in the RB-E2F pathway. According to the ability of animal BRD2 (RING3) BET protein to bind E2F factor,19 we can speculate that GTE4 interacts with DNA, deciphering the acetylation-based epigenetic code, and recruits E2F transcription factors by its ET domain modulating target gene expression.
Conclusions
Our results point to a role of GTE4 in cell cycle regulation, possibly through the E2F-pathway. GTE4 is specifically involved in the maintenance of the mitotic cell cycle in the meristems and thereby controls the number of cells in the differentiated organs. A role of GTE4 on cytokinesis is also suggested by our results. Future studies directed to identify target genes that are under the control of GTE4 will be needed to clarify through which genes this BET protein controls these processes.
Acknowledgements
This work was supported by PRIN 2008 and Progetti Ateneo of Sapienza Università di Roma (to M.M.A.).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11571
References
- 1.de la Cruz X, Lois S, Sànchez-Molina S, Martìnez-Balbàs MA. Do protein motifs read the histone code? BioEssay. 2005;27:164–175. doi: 10.1002/bies.20176. [DOI] [PubMed] [Google Scholar]
- 2.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Wang X, Zhang J, Huang H, Ding B, Wu J, et al. Structural basis and binding properties of the second bromodomain of Brd4 with acetylated histone tails. Biochemistry. 2008;47:6403–6417. doi: 10.1021/bi8001659. [DOI] [PubMed] [Google Scholar]
- 4.Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, et al. Brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 5.Yang XJ. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessay. 2004;26:1076–1087. doi: 10.1002/bies.20104. [DOI] [PubMed] [Google Scholar]
- 6.Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, et al. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Platt GM, Simpson GR, Mittnacht S, Schulz TF. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J Virol. 1999;73:9789–9795. doi: 10.1128/jvi.73.12.9789-9795.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mochizuki K, Nishiyama A, Jang MK, Dey A, Ghosh A, Tamura T, et al. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J Biol Chem. 2008;283:9040–9048. doi: 10.1074/jbc.M707603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunque P, Chua NH. IMB1, a bromodomain protein induced during seed imbibition, regulates ABA- and phyA-mediated responses of germination in Arabidopsis. Plant J. 2003;35:787–799. doi: 10.1046/j.1365-313x.2003.01848.x. [DOI] [PubMed] [Google Scholar]
- 11.Chua YL, Channeliere S, Mott E, Gray JC. The bromodomain protein GTE6 controls leaf development in Arabidopsis by histone acetylation at ASYMMETRIC LEAVES1. Genes Dev. 2005;19:2245–2254. doi: 10.1101/gad.352005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Airoldi AA, Della Rovere F, Falasca G, Marino G, Kooiker M, Altamura MM, et al. The Arabidopsis BET bromodomain factor GTE4 is involved in maintenance of the mitotic cell cycle during plant development. Plant Physiol. 2010;152:1–15. doi: 10.1104/pp.109.150631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korth-out H, et al. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell. 2005;123:1337–1349. doi: 10.1016/j.cell.2005.09.042. [DOI] [PubMed] [Google Scholar]
- 14.Lammens T, Boudolf V, Kheibarshekan L, Zalmas LP, Gaamouche T, Maes S, et al. Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc Natl Acad Sci USA. 2008;105:14721–14726. doi: 10.1073/pnas.0806510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Debeaujon I, Koornneef M. Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristic and embryonic abscisic acid. Plant Physiol. 2000;122:415–424. doi: 10.1104/pp.122.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murashige T, Skoog F. A revised medium for rapid growth and bioassay with tobacco tissue coltures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 17.Galbraith W, Wagner MC, Chao J, Abaza M, Ernst LA, Nederlof MA, et al. Imaging cytometry by multiparameter fluorescence. Cytometry. 1991;12:579–596. doi: 10.1002/cyto.990120702. [DOI] [PubMed] [Google Scholar]
- 18.van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol. 2008;9:713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- 19.Denis GV, Vaziri C, Guo N, Faller DV. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ. 2000;11:417–424. [PMC free article] [PubMed] [Google Scholar]