Abstract
We recently identified a new target of microRNA398 (miR398), a conserved miRNA in plants. In Arabidopsis, miR398 targets the mRNAs of two copper/zinc superoxide dismutases (Cu/Zn SODs) by triggering their cleavage or repressing their translation. We analyzed the transcriptomes of mutants impaired in miR398 production, revealing that the mRNAs encoding the chaperone (CCS1), essential for copper delivering to the Cu/Zn SODs of Arabidopsis and to generate the mature proteins, were undiscovered targets of miR398. It is likely that CCS1 was not identified by previous bioinformatic predictions because of the number of mismatches between the mRNA and its target. Since CCS1 has four mismatches and one GU wobble, it would have been excluded by the majority of prediction algorithms. miR398 directs the post-transcriptional regulation of CCS1 mRNAs by cleavage and ARGONAUTE10 (AGO10)-mediated translational repression. Indeed, CCS1 protein accumulate in zwille (ago10) mutants while both miR398 and CCS1 mRNAs levels remain identical to the Landsberg erecta WT plants. Moreover, since AGO10 is a negative regulator of AGO1, the CCS1 protein is more abundant in a double ago1-27 ago10-3 Col mutant compared to the single hypomorphic ago1-27 mutant, as previously shown for CSD2.
Key words: copper chaperone for super-oxide dismutase (CCS1), copper/zinc superoxide dismutase (Cu/Zn SOD), miR398, argonaute (AGO), Arabidopsis
MicroRNAs (miRNAs) are a class of endogenous small RNAs (21 to 24 nucleotides) playing a crucial role in the development of both plant and animals by downregulating gene expression at the post-transcriptional level. In Arabidopsis, hairpin precursors of miRNAs are transcribed from MIR genes by RNA polymerase II, and processed in the nucleus by the ribonuclease III enzyme DICERLIKE1 (DCL1). miRNA duplexes and/or mature miRNAs are methylated by HUA ENHANCER1 (HEN1), exported to the cytoplasm where mature miRNAs bind to ARGONAUTE1 (AGO1) and direct the cleavage and/or translational repression of partially complementary target mRNAs.1
MicroRNA398 (miR398) is a conserved miRNA targeting at least four mRNAs in Arabidopsis: (1) the cytosolic COPPER/ZINC SUPEROXIDE DISMUTASE1 (CSD1), (2) the chloroplastic CSD2, (3) a subunit of the mitochondrial cytochrome c oxidase, COX5b-1,2–4 and (3) the COPPER CHAPERONE FOR SUPEROXIDE DISMUTASE (CCS1)5 that delivers the copper cofactor to the three Cu/Zn SODs (CSD1, 2 and 3). miR398 seems to be involved in responses to biotic and abotic stresses.6–9
miR398 Triggers the Cleavage of a Limited Number of Targets
Our goal was to find targets of miR398 not previously identified by bioinformatic predictions. For this purpose, we isolated T-DNA knockouts corresponding to putative mir398 mutants and we determined, by northern blot analysis, the levels of miR398 in plants homozygous for the mutations. We found two new mutant alleles with T-DNAs inserted in MIR398c, containing lower and higher levels of miR398, compared to their respective WT. We then hypothesised that target transcripts would be upregulated in the mir398c mutant containing less miR398 and downregulated in the mir398c mutant containing higher levels of the miRNA. To find such transcripts, we analyzed and compared the transcriptomes of these mutants and found, in addition to the two previously described targets of miR398 (CSD1 and CSD2,2–4), a new target, CCS1 (At1g12520). CCS1 is the chaperone delivering copper to the Cu/Zn SODs of Arabidopsis.10,11 We have shown that CCS1 mRNAs and protein abundance are determined by the level of miR398. Finally, we further demonstrated that miR398 triggers the cleavage of CCS1 mRNAs by (1) mapping the 5′ end of the cleavage products in the miR398 complementary site of CCS1 and (2) by generating transgenic plants carrying a CCS1 cDNA-form resistant to the cleavage by miR398, finding that CCS1 protein accumulate in these plants even when the miRNA is abundant. Altogether, the number of targets revealed by our transcriptome analyses (designed to identify cleaved targets) is very limited. Indeed, we found only one mRNA target (CCS1) in addition to CSD1 and CSD2 mRNAs.
One of the intriguing points about CCS1 was to understand why the miR398 target site remained undiscovered until now and why CCS1 mRNA has escaped all bioinformatic predictions, including the ones that are taking into account the minimum free energy of the miRNA/mRNA duplexes. In order to minimize the number of false positives and to find real targets, most prediction algorithms limit the number of allowable authentic mismatches. Since CCS1 has four mismatches and one GU wobble (Fig. 1), it has been excluded from the majority of predictions. We found about 120 duplexes in Arabidopsis that were validated by 5′-RACE PCR since the discovery of miRNAs in plants and out of this, we found only five duplexes that exceed four mismatches (if we consider GU as a mismatch): miR-JAW and TCP3, 4 or 1012, the one formed by miR159a and CSD3,13 and finally miR398 with CCS1 mRNA. All of them are corresponding to target genes identified by experimental evidences. We concluded that CCS1 was not identified before because of the number of mismatches between the mRNA and its target. It is likely that new algorithms, allowing more than four mismatches, and applying more flexible rules to the pairing in the seed region and the GU content, combined to the transcriptomic analyses of mutants impaired in the production of miRNAs, are likely to open the way to new target discoveries.
Figure 1.
miR398 targets in Arabidopsis. The sequence (Col-0) of the miR398 complementary sites in the target mRNA s is aligned with the sequences of miR398a and miR398b/c. In Arabidopsis, miR398 is encoded by three genes, MIR398a, MIR398b and MIR398c.2–4 The miRNAs produced from MIR398b and c are identical and differ by one nucleotide from the one produced by MIR398a. The arrows indicate the cleavage sites experimentally validated by a modified version of the 5′-RACE PCR18 and localized between the nucleotides 10 and 11 of the miRNA , like for many other targets of miRNAs in Arabidopsis.1 The region corresponding to the seed is in bold.
Mutations in AGO Genes Affect the Regulation of CCS1 mRNA by miR398
Arabidopsis contains ten AGO proteins involved in miRNA- and siRNA-mediated regulations.14 Among them, AGO1 plays an essential role by associating with most miRNAs. AGO10 (i.e., ZWILLE (ZLL) or PINHEAD (PNH)) is the closest paralogue of AGO1. Recently, Brodersen et al.15 demonstrated that some miRNAs can trigger both the cleavage and the translational repression of their target mRNAs, through the action of AGO1 and/or AGO10. In the Landsberg erecta (Ler) accession, the CSD2 protein accumulate in zll (ago10) mutants whereas both the levels of the corresponding mRNA and the level of miR398 remain almost unchanged.5,15 This is suggesting that AGO10 is probably involved in miR398-mediated translational inhibition of CSD2 mRNA. We have shown that CCS1 protein accumulate in zll mutants in a similar way, suggesting that AGO10 is also involved in miR398-directed post-transcriptional regulation of CCS1.5 Thus, miR398 acts through both translational repression and cleavage of at least two of its mRNA targets identified so far: CSD2 and CCS1.
The ago10 mutation have different effects in the Ler or the Columbia (Col) accession backgrounds. Ler zll mutants are impaired in shoot apical meristem development16 while Col ago10 mutants (i.e., ago10-1, SALK_000457 and ago10-3, SALK_519738) present only subtle morphological changes of certain flowers (also observed in the Wassileskija accession background; Bouché N, unpublished data). To study the specific and overlapping functions of AGO1 and AGO10, ago1 ago10 double mutants were generated.17 Since the combination of null ago1 alleles with ago10 alleles are embryo lethal in both Col and Ler, the hypomorphic Col ago1-27 mutant (with an AGO1 function partially compromised) was combined with ago10-1 or ago10-3 Col mutants. This revealed that an ago10 mutation partially restores leaf development as well as siRNA and miRNA pathways of ago1-27.17 Indeed, in the ago1-27 ago10-3 double mutant, miR398 levels were elevated compared to single ago1-27 mutant (although not restored to the WT level), and consequently, both CSD2 mRNAs and protein levels were reduced (Fig. 2 and see ref. 17 for miR398 and CSD2 mRNA levels). This was also observed for CCS1 as shown in Figure 2. In fact, AGO10 seems to be involved in the translational repression of the AGO1 mRNA itself possibly through the action of miR168 that targets AGO1 mRNAs. Thus, the lack of AGO10 activity increases the accumulation of AGO1 protein in the double ago1-27 ago10-3 mutant.17 This ultimately results in the accumulation (although not to WT levels) of miRNAs and their targets such as CSD2 and CCS1. Whether AGO10 could bind miR168, miR398 or any other miRNAs remains to be determined.
Figure 2.
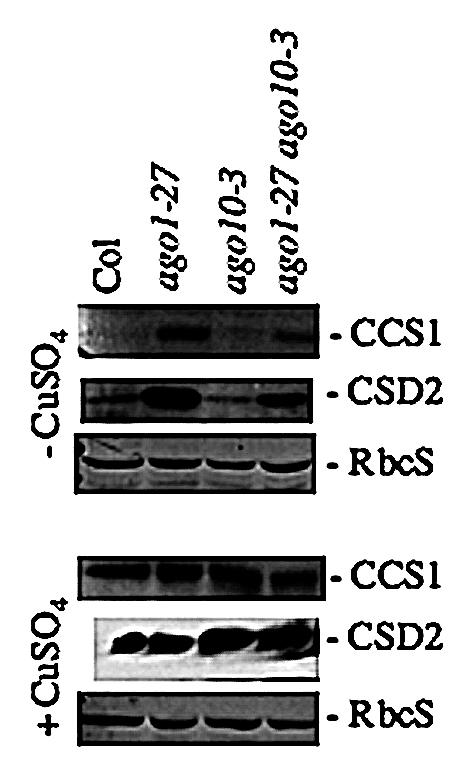
CCS1 protein levels in ago1, ago10 and ago1 ago10 Col mutants. Immunodetection of CCS1 and CSD2 proteins from Col, ago1-27 hypomorphic mutant,19 ago10-3,17 (SALK_519738) and the double ago1-27 ago10-3,17 mutant. Plants were grown for 12 days in vitro, with (0.5 µM of CuSO4) or without copper added to the medium. In limiting copper conditions, miR398 levels are kept high, and as a consequence, CSD1, CSD2,20 and CCS1,5 transcripts are cleaved resulting in low amounts of proteins. Thus, the changes of Cu contents in the medium are directly modulating the levels of miR398. Total proteins (10 µg) were extracted in bulks (10 to 15 plants) and CCS1 or CSD2 proteins were detected with anti-CCS1 or anti-CSD1/2 polyclonal antibodies (Agrisera AB, Sweden) after SDS-PAGE. Staining of Rubisco small subunit (RbcS) by Coomassie blue served as control of equal loading. mRNA and miRNA levels are described in reference 17.
Abbreviations
- AGO
argonaute
- CCS
copper chaperone for superoxide dismutase
- Col
columbia arabidopsis accession
- Ler
Landsberg erecta arabidopsis accession
- miRNA
microRNA
- SOD
superoxide dismutase
- ZLL
ZWILLE
Addendum to: Beauclair L, Yu A, Bouché N. MicroRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J. 2010 doi: 10.1111/j.1365-313X.2010.04162.x. In press.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11573
References
- 1.Mallory AC, Bouché N. MicroRNA-directed regulation: to cleave or not to cleave. Trends Plant Sci. 2008;13:359–367. doi: 10.1016/j.tplants.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Bonnet E, Wuyts J, Rouze P, Van de Peer Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc Natl Acad Sci USA. 2004;101:11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 4.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beauclair L, Yu A, Bouché N. MicroRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J. 2010;62:454–462. doi: 10.1111/j.1365-313X.2010.04162.x. [DOI] [PubMed] [Google Scholar]
- 6.Jia X, Wang WX, Ren L, Chen QJ, Mendu V, Willcut B, et al. Differential and dynamic regulation of miR398 in response to ABA and salt stress in Populus tremula and Arabidopsis thaliana. Plant Mol Biol. 2009;71:51–59. doi: 10.1007/s11103-009-9508-8. [DOI] [PubMed] [Google Scholar]
- 7.Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trindade I, Capitao C, Dalmay T, Fevereiro MP, Santos DM. miR398 and miR408 are upregulated in response to water deficit in Medicago truncatula. Planta. 2010;3:705–716. doi: 10.1007/s00425-009-1078-0. [DOI] [PubMed] [Google Scholar]
- 9.Jagadeeswaran G, Saini A, Sunkar R. Biotic and abiotic stress downregulate miR398 expression in Arabidopsis. Planta. 2009;229:1009–1014. doi: 10.1007/s00425-009-0889-3. [DOI] [PubMed] [Google Scholar]
- 10.Chu CC, Lee WC, Guo WY, Pan SM, Chen LJ, Li HM, et al. A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis. Plant Physiol. 2005;139:425–436. doi: 10.1104/pp.105.065284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdel-Ghany SE, Burkhead JL, Gogolin KA, Andres-Colas N, Bodecker JR, Puig S, et al. AtCCS is a functional homolog of the yeast copper chaperone Ccs1/Lys7. FEBS Letts. 2005;579:2307–2312. doi: 10.1016/j.febslet.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 12.Palatnik JF, Wollmann H, Schommer C, Schwab R, Boisbouvier J, Rodriguez R, et al. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev Cell. 2007;13:115–125. doi: 10.1016/j.devcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 13.German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 14.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 16.Moussian B, Schoof H, Haecker A, Jurgens G, Laux T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mallory AC, Hinze A, Tucker MR, Bouché N, Gasciolli V, Elmayan T, et al. Redundant and specific roles of the ARGONAUTE proteins AGO1 and ZLL in development and small RNA-directed gene silencing. PLoS Genet. 2009;5:1000646. doi: 10.1371/journal.pgen.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 19.Morel JB, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, et al. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dugas DV, Bartel B. Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Mol Biol. 2008;67:403–417. doi: 10.1007/s11103-008-9329-1. [DOI] [PubMed] [Google Scholar]



