Abstract
NAC (NAM, ATAF and CUC2) is one of the largest families of transcription factors in the plant genome, but the function and regulation of most NAC genes are still largely unknown. We recently isolated a gene encoding a NAC transcription factor designated ANAC078 from Arabidopsis plants and identified 166 genes upregulated in ANAC078-overexpressing plants compared with the wild-type plants under high-light stress. The cyclic amplification and selection of targets (CASTing) technique showed that the ANAC078 recognition sequence contains T[A/T/C] [A/T/G/C] C[T/G] TG[T/G]G as a DNA-binding site. The recognition sequence identified by this technique was detected in the promoter region of 52 upregulated genes, including the gene for a transcription factor, proteasome subunits, peroxidase and a protein kinase. The findings suggest these genes to be directly targeted by the ANAC078 protein.
Key words: Arabidopsis, CASTing, cis-element, environmental stress, NAC, transcription factor
Transcription factors regulate the first step of gene expression. The Arabidopsis genome contains genes for more than 1,500 transcription factors,1 with recent analyses identifying >2,000 transcription factors genes,2–5 more than that in Drosophila melanogaster and Caenorhabditis elegans. Furthermore, the protein of genes encoding transcription factors in Arabidopsis is 5–10% depending on the database, which is higher than that in D. melanogaster (4.7%) and in C. elegans (3.6%),1 although it is comparable with that of human (6.0%).6 These findings might be because plants, unlike animals, are unable to move and therefore experience numerous forms of environmental stress, such as high-light (HL), drought, high temperature, chilling, salinity and air pollution.
Although transcription factors, such as heat stress transcription factor (Hsf), basic helix-loop-helix (bHLH) and MYB, exist not only in the Saccharomyces cerevisiae, D. melanogaster and C. elegans genomes, but also in the Arabidopsis genome, about half of the Arabidopsis transcription factors, such as AP2-ERF, NAC, Dof, YABBY, WRKY, GARP, TCP, SBP, ABI3-VP1 (B3), EIL and LFY, are plant-specific.1,7
NAC (NAM, ATAF and CUC2) is one of the largest families of transcription factors in the plant genome, with 106 and 149 members predicted in Arabidopsis and rice, respectively.8,9 A small number of the Arabidopsis NAC members have been studied,10 but the function and regulation of most NAC genes are still largely unknown.
We previously isolated a gene encoding a NAC transcription factor named ANAC078 according to the nomenclature established for the Arabidopsis NAC family11,12 though it has also been designated NTL11.13 Furthermore, we reported that the transcript levels of 166 genes in ANAC078-overexpressing Arabidopsis plants (Ox-ANAC078) are increased compared with those in the wild-type plants under HL.14
To explore in more detail the system by which putative target genes are regulated via ANAC078, we identified the recognition sequence for the protein using the cycle amplification of targets (CASTing) method.15,16 A His-Trigger factor (TF) tag, as a kind of chaperon, fused with the NAC domain of ANAC078 (TF-NAC domain) was used to select the DNA-binding site from a random pool of double-stranded oligonucleotides. Each round of CASTing consisted of three steps: binding of the double-stranded DNA oligonucleotide pool to the TF-NAC domain protein, removal of unbound DNA, and PCR amplification of DNA that remained bound to the TF-NAC domain protein-Ni sepharose bead complex.
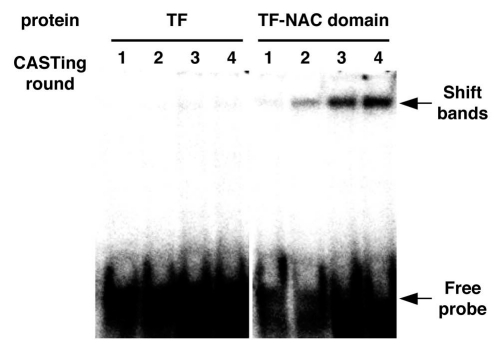
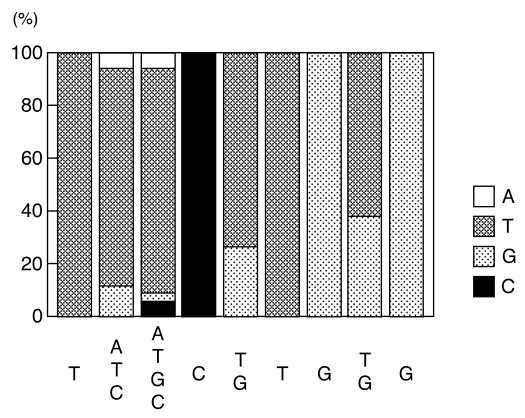
To confirm enrichment of the population with high-affinity binding sites, an electrophoretic mobility shift analysis (EMSA) was performed after each round of CASTing to assess the efficiency of selection. Figure 1 shows the progressive enrichment of the recognition sequence of ANAC078 obtained by this procedure. During the first cycle, the fraction of specific targets within the population was so small that it was not associated with the TF-NAC domain protein. However, a small signal was detected during the two cycles, and thus became a major fraction after four cycles. Figure 2 indicates the sequences obtained after cloning the four-cycle-selected material. Thirty four clones, sequenced independent of each other, contained the T[A/T/C] [A/T/G/C] C[T/G] TG[T/G]G sequence.
Figure 1.
EMSA analysis of CASTing-enriched probes. The material obtained from each cycle of CASTing was radioactively labeled by including [α-32P]ATP and subjected to EMSA in the presence of 2 µg of bacterially expressed TF-fused ANAC078 NAC domain protein.
Figure 2.
ANAC078-binding sequence. The numbers represent the frequency with which a nucleotide was found at each position (expressed as a percentage of the total).
It has been reported that ANAC019, ANAC055 and ANAC072 bound to the 63-bp promoter region of the ERD1 gene which contained the CATGTG MYC-like motif.17 The recognition sequence of ANAC019, ANAC055 and ANAC072 was determined as ANN NNN TCN NNN NNN ACA CGC ATGT, containing CATGT and harboring CACG as the core DNA-binding site.17 Although C[T/G] TG[T/G] similar to CATGT motif was contained in the 3′ region of ANAC078 recognition sequence, no CACG motif was contained in the recognition sequence. Previously, it has been reported that the Arabidopsis NAC1 (ANAC022) protein binds to a 21-bp segment (CTGA CGT AAG GGA TGA CGC AC) within the cauliflower mosaic virus (CaMV) 35S-90 promoter.18 Duval et al. (2002)19 reported that purified AtNAM (ANAC018) protein bound to a region of the CaMV 35S promoter between -70 and -76, which was located in the 21-bp segment. No CACG motif was contained in the NAC1 or AtNAM recognition sequence either. Further analysis of the function of the CACG motif in gene regulation via the NAC family is needed.
A search was made for the recognition sequence of ANAC078 in the promoter regions (within 1,000 bp upstream of the coding region) of 166 putative target of ANAC078. The sequence was detected in the putative promoter region of 52 target genes (Suppl. Table 1). These genes encoded transcription factors, proteasome subunits, peroxidase, a protein kinase and so on, suggesting their expression of 52-genes to be directly regulated by ANAC078.
In fact, the transcript levels of transcription factor genes related to flavonoid biosynthesis and the levels of anthocyanins were significantly increased in Ox-ANAC078 plants and reduced in knockout ANAC078 plants (KO-ANAC078) compared with wild-type plants under HL stress.14 Proteasomes are an essential component of the quality-control system that limits the accumulation of non-functional proteins in the cell.20–23 Recently, it has been reported that the 20S proteasome plays an important role in responses to several types of stress, such as salinity, and oxidative stress.24,25 As shown in Supplementary Table 1, there are several genes (AT1G56650, AT1G62085, AT1G73870, AT2G47460, AT3G01600 and AT5G58610) encoding transcription factors. It seems likely that the expression of other ANAC078-target genes which do not have the ANAC078-recognition sequence in their promoter region is regulated by these transcription factor under HL stress.
The present findings and results reported previously suggest that ANAC078 is a key regulator in the induction of not only the defence system, but also the growth and developmental processes of vegetative organs in response to external signals, including environmental stress.
Supplementary Material
Acknowledgements
This work was supported by a Grant-in-aid for Scientific Research (A) (S.S: 19208031) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and by a Grant-in-aid for Young Scientists (B) (Y.Y.: 21780310) from MEXT of Japan. It was also supported by CREST, JST (S.S.: 2005–2010).
Abbreviations
- CASTing
cyclic amplification and selection of targets
- EMSA
electrophoretic mobility shift analysis NAC, NAM, ATAF and CUC2
- TF
trigger factor
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11577
References
- 1.Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 2.Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, et al. AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics. 2003;4:25. doi: 10.1186/1471-2105-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo A, He K, Liu D, Bai S, Gu X, Wei L, et al. DATF: a database of Arabidopsis transcription factors. Bioinformatics. 2005;21:2568–2569. doi: 10.1093/bioinformatics/bti334. [DOI] [PubMed] [Google Scholar]
- 4.Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, et al. RARTF: database and tools for complete sets of Arabidopsis transcription factors. DNA Res. 2005;12:247–256. doi: 10.1093/dnares/dsi011. [DOI] [PubMed] [Google Scholar]
- 5.Riano-Pachon DM, Ruzicic S, Dreyer I, Mueller-Roeber B. PlnTFDB: an integrative plant transcription factor database. BMC Bioinformatics. 2007;8:42. doi: 10.1186/1471-2105-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 7.Mitsuda N, Ohme-Takagi M. Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 2009;50:1232–1248. doi: 10.1093/pcp/pcp075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong W, Shen YP, Ma G, Pan Y, Du YL, Wang DH, et al. Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 2004;135:773–782. doi: 10.1104/pp.104.042176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong Y, Liu T, Tian C, Sun S, Li J, Chen M. Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Mol Biol. 2005;59:191–203. doi: 10.1007/s11103-005-6503-6. [DOI] [PubMed] [Google Scholar]
- 10.Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 11.Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2006;48:535–547. doi: 10.1111/j.1365-313X.2006.02889.x. [DOI] [PubMed] [Google Scholar]
- 12.Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 13.Kim SY, Kim SG, Kim YS, Seo PJ, Bae M, Yoon HK, et al. Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucl Acids Res. 2007;35:203–213. doi: 10.1093/nar/gkl1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morishita T, Kojima Y, Maruta T, Nishizawa-Yokoi A, Yabuta Y, Shigeoka S. Arabidopsis NAC transcription factor, ANAC078, regulates flavonoids biosynthesis under high-light. Plant Cell Physiol. 2009;50:2210–2222. doi: 10.1093/pcp/pcp159. [DOI] [PubMed] [Google Scholar]
- 15.Wright WE, Binder M, Funk W. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol Cell Biol. 1991;11:4104–4110. doi: 10.1128/mcb.11.8.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newhouse CD, Silverstein SJ. Orientation of a novel DNA binding site affects human papillomavirus-mediated transcription and replication. J Virol 200. 75:1722–1735. doi: 10.1128/JVI.75.4.1722-1735.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Q, Frugis G, Colgan D, Chua NH. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14:3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duval M, Hsieh TF, Kim SY, Thomas TL. Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol Biol. 2002;50:237–248. doi: 10.1023/a:1016028530943. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 21.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 22.Ron D. Stressed cells cope with protein overload. Science. 2006;313:52–53. doi: 10.1126/science.1130469. [DOI] [PubMed] [Google Scholar]
- 23.Kurepa J, Smalle JA. To misfold or to lose structure? Detection and degradation of oxidized proteins by the 20S proteasome. Plant Signal Behav. 2008;3:386–388. doi: 10.4161/psb.3.6.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurepa J, Toh-E A, Smalle JA. 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J. 2008;53:102–114. doi: 10.1111/j.1365-313X.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Kurepa J, Smalle JA. The Arabidopsis 26S proteasome subunit RPN1a is required for optimal plant growth and stress responses. Plant Cell Physiol. 2009;50:1721–1725. doi: 10.1093/pcp/pcp105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.