Abstract
OsPHR2, the homolog of AtPHR1, is a central Pi-signaling regulator. The Pi-signaling pathway downstream of AtPHR1, similarly of OsPHR2,1,2 involves a noncoding RNA which targets mimicry of miR399. miRNA399 mediates cleavage of PHO2.3,4 The regulating pathway downstream of OsPHR2 is negatively regulated by the Pi-signaling responsive gene OsSPX1.5,6 Overexpression of AtPHR1 and OsPHR2 leads to an increased concentration of Pi in the shoot tissues with leaf toxic symptom and growth retardation similar as the phenotype of pho2 mutant, especially under Pi abundant conditions.2,6,7 It has been known that the low affinity Pi transporter OsPT2 mainly contributes to the shoot Pi accumulation mediated by OsPHR2, and overexpression of OsPT2 results in shoot Pi accumulation and leaf toxic symptom and growth retardation under Pi abundant conditions.6 Two curious questions are emerging from the reported results: How Os SPX1 functions on the negative regulation of the pathway and what mechanism of the growth retardation mediated by OsPHR2. For the second question, our favored hypothesis is that the growth inhibition mediated by overexpression of OsPHR2 is caused by toxic physiological effects due to excessive Pi accumulation in shoots (Pi toxicity). In fact, the toxic symptoms become diminished with decreased Pi levels in growth medium. However, the plant growth retardation mediated by overexpression of OsPHR2 may be caused by some unknown genetic factor(s) regulated by OsPHR2.
Key words: Oryza Sativa L, OsPHR2, OsSPX1, pi-signaling, plant growth
Plant Growth Inhibition Mediated by OsPHR2 is not Restricted to Shoot Pi Concentration
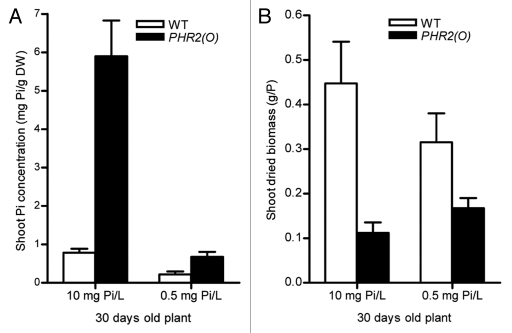
To determine whether the plant growth inhibition mediated by OsPHR2 is restricted to shoot Pi concentration, we performed a experiment with several Pi levels in a solution culture to found a condition under which the shoot Pi concentration in PHR2(O) plants is similar as in shoots of the wild type plants at 10 mg Pi/L level. At the supplied Pi level of 0.5 mg Pi/L, the shoot Pi concentration in OsPHR2(O) plants is similar as that in the shoots of wild type plants at the supplied Pi level of 10 mg Pi/L (Fig. 1A), that the shoot Pi concentration in PHR2(O) is not toxic level in term of physiology, while the biomass of PHR2(O) plants is still much lower compared with that of the wild type (Fig. 1B).
Figure 1.
Shoot Pi concentration (A) and shoot biomass of 30 d-old plants of wild type (WT) and the transgenic plants (B) with overexpression of OsPHR2 under solution culture with abundant supplied Pi (10 mg Pi/L) and low Pi level (0.5 mg Pi/L). The solution pH is 5.5 and each plant occupied one liter of culture solution. The culture solution was replaced every two days.
Isolation of Mutant with Rescue of Plant Growth Inhibition Under Overexpression of OsPHR2
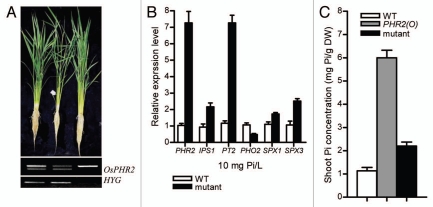
Because the plant growth inhibition mediated by OsPHR2 is not restricted to shoot Pi concentration, we reasoned that there should be some genetic factor(s) under the control of OsPHR2. The upregulation or repression of the factor(s) by OsPHR2 may limit the plant growth. In fact, some mutants with rescued growth under Pi-supplied condition were isolated from an EMS-generated mutant library of seeds under background of overexpression of OsPHR2 (Fig. 2A). To confirm the background of the isolated mutants, the expression patterns of OsPHR2 and the PSI (Pi-starvation induced) genes downstream of OsPHR2 were investigated. The results indicate that the recovery of mutant plant growth is under background of overexpression of OsPHR2 (Fig. 2B). The Pi concentration analysis also showed that the shoot Pi accumulation mediated by OsPHR2 is remarkably reduced in the mutant (Fig. 2C). The present results provide the evidence that OsPHR2 plays multiple functions on Pi-signaling, Pi uptake and translocation and the factor(s) which may negatively regulate plant growth.
Figure 2.
Mutants showed rescue of plant growth under background of overexpression of OsPHR2. (A) Ninety days old plants under solution culture with Pi-supplied condition (10 mg Pi/L). PCR analysis using the primers flanking 3, 4, 5 introns of OsPHR2 and HYG (hygromycin gene) (below). (B) Relative expression levels of OsPHR2 and the PSI (Pi-starvation induced) genes downstream of OsPHR2. (C) Shoot Pi concentration in the wild type, transgenic plants with overexpression of OsPHR2 and the mutant.
Genetic Factor(s) Involved in the OsPHR2-Mediated Plant Growth Inhibition May be Unknown Factor(s)
Several SPX domain (SYG1/PHO81/XPR1) genes in plants were found to be involved in responses to environmental cues or internal regulation of nutrition homeostasis in plants.8–12 It has been demonstrated that OsSPX1 is a negative regulator of OsPHR2 and involved in the feedback Pi-signaling network in roots defined by OsPHR2 and OsPHO2.6 At least six genes with an exclusive SPX domain in rice have been based on the present rice genome database. Among them, overexpression of OsSPX3 inhibits plant growth and OsSPX1 positively regulates OsSPX3 in shoots.13 To determine whether OsSPX3 is involved in the rescue of plant growth in the mutant under background of OsPHR2(O), the expression pattern of OsSPX3 in the mutant was compared with that in the wild type and OsPHR2(O) plants. qRT-PCR analysis showed upregulation of OsSPX3 in the mutant as in PHR2(O) plants (Fig. 2B). The result indicates that the rescue of the mutant plant growth under background of PHR2(O) may be independent of OsSPX3.
Addendum to: Liu F, Wang ZY, Ren HY, et al. OsSPX1 suppresses function of OsPHR2 on expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J. 2010 doi: 10.1111/j.1365-313X.2010.04170.x. In press.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11645
References
- 1.Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, et al. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15:2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou J, Jiao F, Wu Z, Wang X, He X, Zhong W, et al. OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 2008;146:1673–1686. doi: 10.1104/pp.107.111443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 4.Schachtman DP, Shin R. Nutrient sensing and signaling: NPKS. Annu Rev Plant Biol. 2007;58:47–69. doi: 10.1146/annurev.arplant.58.032806.103750. [DOI] [PubMed] [Google Scholar]
- 5.Wang C, Ying S, Huang H, Li K, Wu P, Shou H. Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J. 2009;57:895–904. doi: 10.1111/j.1365-313X.2008.03734.x. [DOI] [PubMed] [Google Scholar]
- 6.Liu F, Wang ZY, Ren HY, Shen CJ, Li Y, Ling HQ, et al. OsSPX1 suppresses function of OsPHR2 on expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J. 2010 doi: 10.1111/j.1365-313X.2010.04170.x. In press. [DOI] [PubMed] [Google Scholar]
- 7.Nilsson L, Muller R, Nielsen TH. Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environm. 2007;30:1499–1512. doi: 10.1111/j.1365-3040.2007.01734.x. [DOI] [PubMed] [Google Scholar]
- 8.Duan K, Yi KK, Dang L, Huang HJ, Wu W, Wu P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 2008;54:965–975. doi: 10.1111/j.1365-313X.2008.03460.x. [DOI] [PubMed] [Google Scholar]
- 9.Hamburger D, Rezzonico E, MacDonald-Comber Petetot J, Somerville C, Poirier Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell. 2002;14:889–902. doi: 10.1105/tpc.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakanishi H, Okumura N, Umehara Y, Nishizawa N, Chino M, Mori S. Expression of a gene specific for iron deficiency (Ids3) in the roots of Hordeum vulgare. Plant Cell Physiol. 1993;34:401–410. [PubMed] [Google Scholar]
- 11.Poirier Y, Thoma S, Somerville C, Schiefelbein J. A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 1991;97:1087–1093. doi: 10.1104/pp.97.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Ribot C, Rezzonico E, Poirier Y. Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 1994;135:400–411. doi: 10.1104/pp.103.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Hu H, Huang H, Duan K, Wu Z, Wu P. Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. J Integr Plant Biol. 2009;51:663–674. doi: 10.1111/j.1744-7909.2009.00834.x. [DOI] [PubMed] [Google Scholar]