Abstract
In the cell nucleus, the packaging of the DNA into chromatin represses transcription by restricting the access of transcriptional regulators to their binding sites and inhibiting the progression of RNA polymerases during transcript elongation. To efficiently transcribe genes in the context of chromatin, eukaryotes have a variety of transcript elongation factors promoting transcription in vivo. The facilitates chromatin transcription (FACT) complex consisting of the SSRP1 and SPT16 proteins, is a histone chaperone that assists transcription by destabilizing nucleosomes in the path of RNA polymerases. In a recent study, we report that Arabidopsis FACT is critically involved in different aspects of development including leaf growth and the transition to flowering. Moreover, FACT was found to interact genetically with HUB1 that mono-ubiquitinates histone H2B. Depending on the underlying process that is regulated by the two complexes, there appear to be different levels of interaction.
Key words: SPT16, SSRP1, HUB1, transcript elongation, chromatin
The FACT heterodimer purified from human cells was shown to assist transcription by RNA polymerase II (RNAPII) from chromatin templates.1 It promotes transcript elongation by facilitating the removal of histone H2A/H2B dimers from nucleosomes of transcribed genes.2 Yeast FACT (consisting of SPT16 and the SSRP1-like protein POB3) can reorganize nucleosomes without requirement of H2A/H2B removal into a form, in which the nucleosomal DNA is more accessible.3 Thus, FACT is a chromatin factor that in concert with other transcript elongation factors assists the passage of RNA polymerase through chromatin by destabilizing nucleosomes without ATP consumption.4,5
In plants, both subunits of the FACT complex are conserved and SSRP1 has been studied in quite some detail. Mediated by its HMG-box DNA-binding domain, maize SSRP1 binds with high affinity to DNA structures and it interacts preferentially with structurally flexible DNA sites.6 The DNA binding is modulated by phosphorylation catalyzed by the protein kinase CK2.7 SSRP1 interacts with nucleosome particles and associates with nuclease sensitive maize chromatin.8 Consistently, using immunofluorescence Arabidopsis FACT is detected in euchromatin, but not in heterochromatic chromocenters. Typical of transcript elongation factors, the FACT subunits associate with the entire transcribed region of active genes in Arabidopsis, but not with non-transcribed genes or intergenic regions.9
Both genes encoding FACT subunits are essential in yeast and the mouse SSRP1 is critical for cell viability.4,10,11 Since plants are excellent models to study the role of transcript elongation factors in the development of multicellular organisms,12,13 in a recent study we have examined Arabidopsis mutant lines harboring DNA insertions in the SSRP1 and SPT16 genes.14 For one of the analyzed plant lines (ssrp1-1) carrying an insertion in exon 1 of SSRP1, we were unable to obtain plants homozygous for the insertion, suggesting that the gene is also essential in plants. The other tested lines, ssrp1-2, spt16-1 and spt16-2 are viable and have reduced levels of SSRP1 and SPT16. However, the mutant plants are severely affected in various aspects of growth and development including leaf growth and architecture, bolting time, flower architecture and seed production. Moreover, FACT was found to interact genetically with the histone H2B mono-ubiquitinase HUB1.14 Here, we focus on the role of FACT and HUB1 in Arabidopsis leaf development and in the transition to flowering.
Leaf size and shape are species specific and genetically determined by a large number of genes that upon mutation alter the leaf phenotype.15 Local manipulation of cell division and cell expansion showed that these are important parameters for leaf form.16,17 Besides cell cycle regulators and transcription factors, enzymes and complexes that control chromatin status and hence the regulation of RNAPII accessibility to genes, impact leaf size and shape through these cellular parameters.18,19 Indeed, the histone H3 acetyl transferase complex, Elongator and HUB1, affect both cell proliferation during leaf growth, but leaf size and shape are distinct in both mutants. The FACT mutants, ssrp1-2, spt16-1 and spt16-2 also have leaves with reduced lamina area due to reduced cell proliferation, but are asymmetrically serrated with severely reduced venation complexity and different from those of elo or hub1 mutants (Fig. 1). It suggests that the different complexes might regulate different pathways contributing to leaf size and shape. Double mutants were constructed between hub1 and the ssrp1/spt16 mutants and their leaf phenotype was studied to determine genetic interaction.15 The leaf lamina areas of double mutants were strongly reduced as compared to the one of the single mutants; the synergistic effect on leaf growth indicated that FACT and HUB1 act independently on the same growth pathways.14 Double mutants had asymmetrically serrated leaf shapes with reduced venation complexity comparable to the FACT mutants indicating that FACT is epistatic to HUB1 and that both share these developmental pathways. The conclusion is that FACT and HUB1 both affect leaf growth and development through the regulation of common pathways either in a redundant way (growth) or in a sequential way (development). Leaf size and shape depend on environmental conditions such as day length and water deficit.20 This so-called leaf plasticity shows the impact of the environment on the genetic regulation of leaf development. Upon external stimuli, the post-translational modifications of nucleosomal histones change. For example, light regulates histone acetylation levels and consequently expression of specific genes.21,22 Hence, chromatin-modifying complexes that alter histones might act as an interface between environment and RNAPII-mediated gene expression to modulate leaf plasticity. It will be interesting to study whether there is crosstalk between the HUB1 or FACT complexes and signaling components related to the perception of environmental or stress conditions.
Figure 1.
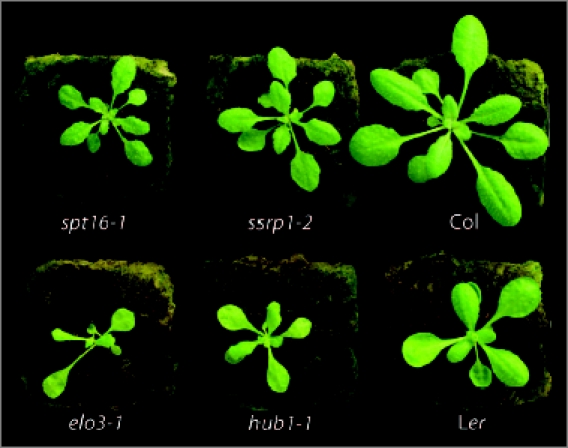
Rosette leaf size and shape of mutants in FACT, Elongator and HUB1 chromatin-related factors. The spt16-1 and ssrp1-2 mutants are in Col background and the elo3-1 and hub1-1 mutants are in Ler background. Twenty-four-day-old plants were grown on rockwool watered with Hyponex solution in growth chamber using conditions of 100 µmol/m−2sec−1 cool white light intensity, 16 hrs light, 21°c and 60% relative humidity.
In Arabidopsis, the transition to flowering is controlled by a network of positive and negative regulators including a variety of chromatin factors. Several protein complexes involved in chromatin remodeling and histone modifications contribute to the regulation of the expression levels of the central floral repressor FLC controlling the transition to flowering.23,24 Among the positive regulators of FLC is HUB1, as FLC expression is downregulated in hub1 mutants and, consistently, the mutant plants show accelerated flowering.25–27 Similarly, mutant plants with reduced expression of the FACT subunits, SSRP1 and SPT16, are early flowering and display reduced FLC transcript levels. Analysis of double mutant plants affected in the expression of both HUB1 and FACT subunits revealed that their flowering time is in between the parental lines,14 suggesting that HUB1 and FACT promote FLC expression and the transition to flowering, but that they act independently on the induction of flowering. Interestingly, mutations of both SSRP1/SPT16 and HUB1 cause reduced transcript levels of FLC, while the expression of reference genes is not decreased by reduced amounts of these chromatin modifiers.14,25 Similarly, an effect on FLC expression by alterations in chromatin structure (while the expression of other genes is not changed) is also observed for defects in other Arabidopsis chromatin modifiers that promote chromatin-regulated transcription, including a chromatin-remodeling complex of the SWR1-type and the histone H3 methyltransferase SDG25.28,29 Consistently, transcript profiling experiments revealed that defects in various chromatin modifiers, including HUB1 and SDG25, cause differential expression of only a subset of genes.18,28 Experiments with various chromatin factors have demonstrated that proper expression of FLC appears to be particularly sensitive to changes in chromatin structure. Therefore, the FLC locus represents a promising model for further studies addressing the role of chromatin-mediated activation and repression of gene transcription.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11646
References
- 1.Orphanides G, LeRoy G, Chang C-H, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 2.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 3.Xin H, Takahata S, Blanksma M, McCullough L, Stillman DJ, Formosa T. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol Cell. 2009;35:365–376. doi: 10.1016/j.molcel.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Formosa T. FACT and the reorganized nucleosome. Mol Bio Syst. 2008;4:1085–1093. doi: 10.1039/b812136b. [DOI] [PubMed] [Google Scholar]
- 5.Reinberg D, Sims RJ. de FACTo nucleosome dynamics. J Biol Chem. 2006;281:23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- 6.Röttgers K, Krohn NM, Lichota J, Stemmer C, Merkle T, Grasser KD. DNA-interactions and nuclear localisation of the chromosomal HMG domain protein SSRP1 from maize. Plant J. 2000;23:395–405. doi: 10.1046/j.1365-313x.2000.00801.x. [DOI] [PubMed] [Google Scholar]
- 7.Krohn NM, Stemmer C, Fojan P, Grimm R, Grasser KD. Protein kinase CK2 phosphorylates the high mobility group domain protein SSRP1, inducing the recognition of UV-damaged DNA. J Biol Chem. 2003;278:12710–12715. doi: 10.1074/jbc.M300250200. [DOI] [PubMed] [Google Scholar]
- 8.Lichota J, Grasser KD. Differential chromatin association and nucleosome binding of the maize HMGA, HMGB and SSRP1 proteins. Biochemistry. 2001;40:7860–7867. doi: 10.1021/bi010548y. [DOI] [PubMed] [Google Scholar]
- 9.Duroux M, Houben A, Ruzicka K, Friml J, Grasser KD. The chromatin remodelling complex FACT associates with actively transcribed regions of the Arabidopsis genome. Plant J. 2004;40:660–671. doi: 10.1111/j.1365-313X.2004.02242.x. [DOI] [PubMed] [Google Scholar]
- 10.Cao S, Bendall H, Hicks GG, Nashabi A, Sakano H, Shinkai Y, et al. The high-mobility-group box protein SSRP1/T160 is essential for cell viability in day 3.5 mouse embryos. Mol Cell Biol. 2003;23:5301–5307. doi: 10.1128/MCB.23.15.5301-5307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singer RA, Johnston GC. The FACT chromatin modulator: genetic and structure/function relationships. Biochem. Cell Biol. 2004;82:419–427. doi: 10.1139/o04-050. [DOI] [PubMed] [Google Scholar]
- 12.Grasser KD. Emerging role for transcript elongation in plant development. Trends Plant Sci. 2005;10:484–490. doi: 10.1016/j.tplants.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Nelissen H, Boccardi TM, Himanen K, Van Lijsebettens M. Impact of core histone modifications on transcriptional regulation and plant growth. Crit Rev Plant Sci. 2007;26:243–263. [Google Scholar]
- 14.Lolas IB, Himanen K, Grønlund JT, Lynggaard C, Houben A, Melzer M, et al. The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J. 2010;61:686–697. doi: 10.1111/j.1365-313X.2009.04096.x. [DOI] [PubMed] [Google Scholar]
- 15.Pérez-Pérez JM, Candela H, Micol JL. Understanding synergy in genetic interactions. Trends Genet. 2009;25:368–376. doi: 10.1016/j.tig.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Pien S, Wyrzykowska J, McQuee-Mason S, Smart C, Fleming A. Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc Natl Acad Sci USA. 2001;98:11812–11817. doi: 10.1073/pnas.191380498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wyrzykowska J, Pien S, Shen W-H, Fleming A. Manipulation of leaf shape by modulation of cell division. Development. 2002;129:957–964. doi: 10.1242/dev.129.4.957. [DOI] [PubMed] [Google Scholar]
- 18.Fleury D, Himanen K, Cnops G, Nelissen H, Boccardi TM, Maere S, et al. The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell. 2007;19:417–432. doi: 10.1105/tpc.106.041319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelissen H, De Groeve S, Fleury D, Neyt P, Bruno L, Bitonti MB, et al. Plant Elongator regulates auxinrelated genes during RNA polymerase II-mediated transcription elongation. Proc Natl Acad Sci USA. 2010;107:1678–1683. doi: 10.1073/pnas.0913559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granier C, Aguirrezabal L, Chenu K, Cookson SJ, Dauzat M, Hamard P, et al. PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol. 2006;169:623–635. doi: 10.1111/j.1469-8137.2005.01609.x. [DOI] [PubMed] [Google Scholar]
- 21.Chua YL, Brown APC, Gray JC. Targeted histone acetylation and altered nuclease accessibility over short regions of the pea plastocyanin gene. Plant Cell. 2001;13:599–612. doi: 10.1105/tpc.13.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Offermann S, Danker T, Dreymüller D, Kalamajka R, Töpsch S, Weyand K, Peterhänsel C. Illumination is necessary and sufficient to induce histone acetylation independent of transcriptional activity at the C4-specific phospoenolpyruvate carboxylase promoter in maize. Plant Physiol. 2006;141:1078–1088. doi: 10.1104/pp.106.080457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farrona S, Coupland G, Turck F. The impact of chromatin regulation on the floral transition. Sem Cell Dev Biol. 2008;19:560–573. doi: 10.1016/j.semcdb.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 24.He Y. Control of the transition to flowering by chromatin modifications. Mol Plant. 2009;2:554–564. doi: 10.1093/mp/ssp005. [DOI] [PubMed] [Google Scholar]
- 25.Cao Y, Dai Y, Cui S, Ma L. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell. 2008;20:2586–2602. doi: 10.1105/tpc.108.062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu X, Jiang D, Wang Y, Bachmair A, He Y. Repression of the floral transition via histone H2B monoubiquitination. Plant J. 2009;57:522–533. doi: 10.1111/j.1365-313X.2008.03709.x. [DOI] [PubMed] [Google Scholar]
- 27.Xu L, Ménard R, Berr A, Fuchs J, Cognat V, Meyer D, Shen WH. The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J. 2009;57:279–288. doi: 10.1111/j.1365-313X.2008.03684.x. [DOI] [PubMed] [Google Scholar]
- 28.Berr A, Xu L, Gao J, Cognat V, Steinmetz A, Dong A, Shen WH. SET DOMAIN GROUP25 encodes a histone methyltransferase and is involved in FLOWERING LOCUS C activation and repression of flowering. Plant Physiol. 2009;151:1476–1485. doi: 10.1104/pp.109.143941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lázaro A, Gómez-Zambrano A, López-González L, Piñeiro M, Jarillo JA. Mutations in the Arabidopsis SWC6 gene, encoding a component of the SWR1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J Exp Bot. 2008;59:653–666. doi: 10.1093/jxb/erm332. [DOI] [PubMed] [Google Scholar]


