Abstract
The most remarkable change of deetiolation in seedling is chlorophyll synthesis and greening. This transition is achieved by photoreduction of dark-accumulated protochlorophyllide (Pchlide) in light. However, overaccumulation of Pchlide results in phototoxicity to plants, so appropriate accumulation and quick reduction of Pchlide are crucial for survival of seedlings during the transition from dark to light. We found that this vital process is tightly regulated by the plant gaseous hormone ethylene. Transgenic analysis using a promoter-GUS reporter system showed that the ethylene signaling was able to activate the expression of PORA (protochlorophyllide oxidoreductase A) gene in seedling cotyledons. We further found that application of ethylene rescued the greening defect of the flu mutant, which over-accumulated Pchlide in the dark. Additionally, genetic studies revealed that Ethylene Insensitive 3 (EIN3) and EIN3-like 1 (EIL1) regulate Pchlide accumulation and cotyledon greening largely independent of Phytochrome-Interacting Factor 1 (PIF1) but partly dependent on PIF3. Therefore, the ethylene signaling via EIN3/EIL1 presents a new pathway to constrain phototoxic Pchlide accumulation in darkness, and simultaneously facilitate Pchlide reduction to synthesize chlorophyll upon light exposure. Our results thus uncover an essential role of ethylene in protecting seedlings from photo-oxidative damage during the process of de-etiolation.
Key words: ethylene, EIN3, FLU, PIF3, chlorophyll synthesis, photooxidation
The gaseous phytohormone ethylene exerts profound and diverse effects on plant development and stress tolerance. Over the past decades, the ethylene signal transduction pathway has been elucidated to a great extent with the adoption of Arabidopsis as a model plant. Ethylene is perceived by a family of membrane-associated receptors and the signal is transduced into the nucleus to cause the accumulation of two master transcriptional activators EIN3 and EIL1, which initiate transcriptional re-programming in various ethylene responses.1–3
In the absence of light, the etiolated seedlings accumulate a suitable amount of chlorophyll precursor, Pchlide, in cotyledons.4 Upon light irradiation when germinating seedlings emerge out of soil, the etiolated cotyledons undergo a dramatic transition to synthesize chlorophyll and become autotrophic (greening), in which Pchlide is photo-reduced in the course of chlorophyll biosynthesis.5,6 On the other hand, the dark-accumulated Pchlide is highly phototoxic and when in excess amount could lead to severe photooxidative damage upon light irradiation.7,8 Therefore, the appropriate amount and rapid reduction of Pchlide to chlorophyllide is crucial for survival via protecting seedlings from photooxidation.
Ethylene Activates the Gene Expression of Photoprotective PORA in Cotyledons
It has been well documented that the reduction from Pchlide to chlorophyllide is catalyzed by protochlorophyllide oxidoreductase (POR, including PORA/B/C in Arabidopsis). Among three PORs, PORA was demonstrated to be the primary enzyme catalyzing Pchlide photoreduction and preventing the production of phototoxic singlet oxygen from Pchlide.9 Our previous study revealed that ethylene treatment induced the transcript level of PORA in etiolated seedlings. We further demonstrated that ethylene-induced PORA expression primarily occurred in cotyledons using PORA promoter-driven GUS reporter system (Fig. 1). When blocking ethylene perception by adding 100 µM AgNO3 (Ag+) in the MS medium, virtually no expression of PORA was detected in the cotyledons (Fig. 1). These results confirm that ethylene is a photoprotective hormone that evidently activates PORA gene expression, particularly in cotyledon tissues.
Figure 1.
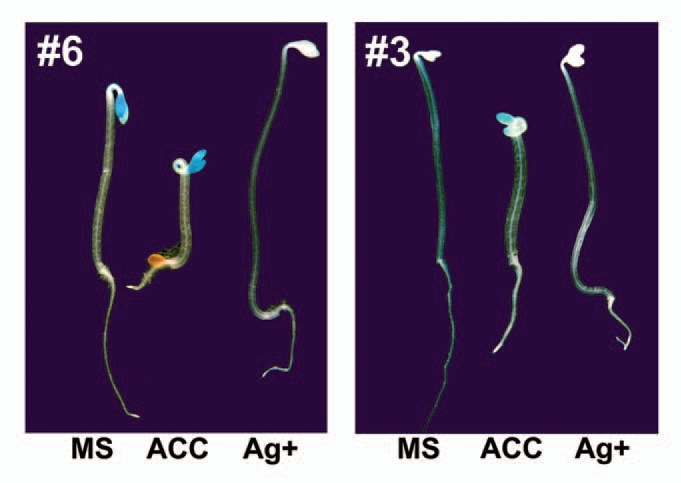
Ethylene activates PORA gene expression in cotyledons of etiolated seedlings. The imaging of GUS staining of 4-day-old dark-grown seedlings (#6, #3 represent two independent transgenic lines containing PORA-promoter-GUS) was shown. The seedlings were grown on MS medium supplemented with or without 10 µM ACC or 100 µM AgNO3 (Ag+).
Ethylene Represses Pchlide Overaccumulation in the flu Mutant
Due to the multifaceted role of Pchlide in chlorophyll biosynthesis and seedling survival, plants have evolved a multitude of strategies to tightly control the amount of Pchlide through the tetrapyrrole biosynthesis pathway. For instance, Heme, a precursor of phytochromobilin, negatively regulates chlorophyll synthesis by inhibiting the conversion of Glutamate to ALA.4 Recent genetic study has identified a negative regulator of chlorophyll biosynthesis, FLU, which directly associates with and inhibits Glu tRNA reductase (HEMA), presenting the second regulatory mechanism controlling chlorophyll biosynthesis.10,11 It has been shown that the excess accumulation of Pchlide in dark-grown FLU mutant lead to the generation of reactive oxygen species (ROS) after dark-to-light shift, which consequently results in severe photooxidative damage.8
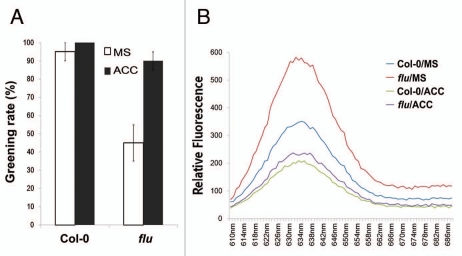
As ethylene application was found to repress the accumulation of Pchlide and ROS,12 we surveyed the impact of ethylene application on the photo-bleaching phenotype observed in the flu mutant. Not surprisingly, ethylene application greatly improved the greening rate of 5 day-old dark grown flu mutant (Fig. 2A). We next detected the effect of ethylene on Pchlide accumulation in flu mutants, and found that ethylene markedly repressed Pchlide overaccumulation in the flu mutant (Fig. 2B). Taken together, our data reveal that the photoprotective effect of ethylene is prominent during de-etiolation by repressing Pchlide accumulation in a FLU-independent mechanism, suggestive of a possible action point downstream of HEMA.
Figure 2.
Ethylene application greatly represses Pchlide overaccumulation in the flu mutant and improves its greening rate. (A) Greening rate quantification of 5-day-old etiolated seedlings transferred to white light (WL) for 1 day. Error bars represent standard error of at least three independent experiments, and more than 50 seedlings were used for calculation each time. (B) Relative fluorescence of Pchlide of 3-day-old etiolated seedlings grown on MS medium supplemented with or without 10 µM ACC.
The Co-Action of EIN3/EIL1 with PIF3 on Photo-Protection may be Different from that with PIF1
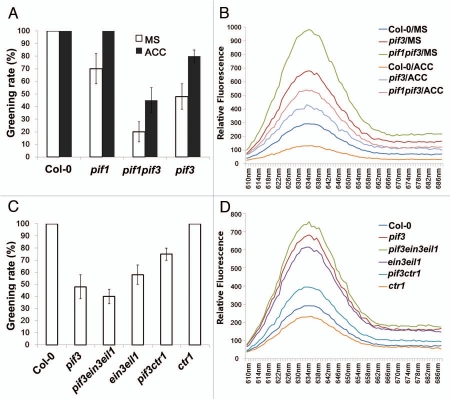
Previous studies led to the identification of a family of bHLH (helix-loop-helix) transcription factors, phytochrome-interacting factors (PIFs, including PIF1 and PIF3-7), which play central roles in phytochrome-mediated light responses, including cotyledon greening.13–15 PIF1 was shown to induce PORC expression by directly binding to its promoter, and also modulate several other genes involved in the regulation of chlorophyll biosynthesis.16 PIF3 was also found to be required for early chloroplast development and regulate expression of a discrete set of key tetrapyrrole metabolic genes.14,17 In etiolated seedling stage, both PIF1 and PIF3 loss of function mutants accumulate higher amounts of Pchlide compared to wild type, which lead to photooxidative damage upon light illumination. As we found that EIN3/EIL1 cooperate with PIF1 to prevent photooxidation and promote greening,12 we speculated that EIN3/EIL1 might also be able to rescue the greening defect of the pif3 mutant, as for the pif1 mutant. To assess this possibility, we transferred 4-day-old etiolated pif3 mutant seedlings grown on MS medium with or without 10 µM ACC to white light for 1 day exposure. As expected, ACC treatment notably improved the greening rate of pif3 mutants (Fig. 3A). Next we detected the accumulation of Pchlide in 3-day-old etiolated seedlings of pif3 mutants grown on MS supplemented with or without 10 µM ACC. In the presence of ACC, the high level of Pchlide in the pif3 mutant was dramatically reduced (Fig. 3B). To further understand how ethylene enhances the greening of etiolated seedlings, we crossed pif3 with ethylene-insensitive mutant ein3eil1 and constitutive triple-response mutant ctr1. After transferring 4-day-old etiolated seedlings to white light irradiation, the pif3ein3eil1 triple mutant exhibited a severer greening defect than pif3 mutant, whereas most of the pif3ctr1 double mutants turned green normally (Fig. 3C). Consistently, we found that pif3ein3eil1 and pif3ctr1 seedlings accumulated higher and lower levels of Pchlide, respectively, compared with the pif3 mutant (Fig. 3D).
Figure 3.
Ethylene application represses Pchlide overaccumulation and improves greening rates of the pif3 mutant via the action of EIN3/EIL1. (A and C) Greening rate quantification of 4-day-old etiolated seedlings transferred to white light (WL) for 1 day. Error bars represent standard error of at least three independent experiments, and more than 50 seedlings were used for calculation each time. (B and D) Relative fluorescence of Pchlide of 3-day-old etiolated seedlings grown on MS medium supplemented with or without 10 µM ACC.
However, ethylene application or loss of CTR1 function failed to rescue the greening defect of pif3 to the same degree as for pif1. In previous research, we showed that 4-day-old etiolated ctr1pif1 or ACC-treated pif1 fully turned green upon light illumination.12 By comparison, ctr1pif3 or ACC-treated pif3 still displayed a portion of greening impairment (Fig. 3A and C). Furthermore, pif3ein3eil1 triple mutant displayed a much milder additive effect than pif1ein3eil1 when compared with single or double mutants. For instance, 4-day-old pif1ein3eil1 etiolated seedlings accumulated almost the doubled amount of Pchlide compared with pif1 and ein3eil1 mutants, and more than 90% of etiolated seedlings were photo-bleached and died upon light exposure.12 In contrast, Pchlide accumulation and the greening defect phenotype of pif3ein3eil1 were just slightly severer than pif3 or ein3eil1 mutants (Fig. 3C and D). Moreover, while PIF1 and EIN3/EIL1 seemed to regulate chlorophyll synthesis quite independently,12 we found that EIN3/EIL1 was able to upregulate the expression of PIF3 (data not shown). Collectively, these results suggest that EIN3/EIL1 mediate the effect of ethylene to repress Pchlide overaccumulation and to enhance cotyledon greening largely independently of PIF1 but partly dependent on PIF3. We are currently in the process of investigating how EIN3/EIL1 work in cooperation with distinctive PIFs in the regulation of seedling de-etiolation.
Conclusions and Perspectives
Taken together, we conclude that plant gaseous hormone ethylene is essential for the survival of seedlings during deetiolation by promoting the production of photoprotective PORA and repressing over-accumulation of phototoxic Pchlide in etiolated seedlings. The finding that ethylene rescued the greening defect of flu that over-accumulates Pchlide suggests that ethylene acts in parallel with FLU or downstream of HEMA, the target enzyme of FLU. In addition to PIFs that have been demonstrated to play central roles in seedling de-etiolation,18 we have discovered that EIN3/EIL1 are a new class of key regulators of chlorophyll biosynthesis, which work largely independent of PIF1 but partly dependent on PIF3. Therefore, elucidation of the co-actions between the two classes of regulators will largely improve our understanding on how etiolated seedlings become autotrophically greening and adapt to their ambient environment. Moreover, delineation of the transcriptional network regulated by these transcription factors would no doubt provide further insights and depth into such fundamental process of plant development and adaptation.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11698
References
- 1.Solano R, Ecker JR. Ethylene gas: perception, signaling and response. Curr Opin Plant Biol. 1998;1:393–398. doi: 10.1016/s1369-5266(98)80262-8. [DOI] [PubMed] [Google Scholar]
- 2.Guo H, Ecker JR. The ethylene signaling pathway: new insights. Curr Opin Plant Biol. 2004;7:40–49. doi: 10.1016/j.pbi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Yoo SD, Cho YH, Sheen J. Emerging connections in the ethylene signaling network. Trends Plant Sci. 2009;14:270–279. doi: 10.1016/j.tplants.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grimm B. Novel insights in the control of tetrapyrrole metabolism of higher plants. Curr Opin Plant Biol. 1998;1:245–250. doi: 10.1016/s1369-5266(98)80112-x. [DOI] [PubMed] [Google Scholar]
- 5.Fujita Y. Protochlorophyllide reduction: A key step in the greening of plants. Plant Cell Physiol. 1996;37:411–421. doi: 10.1093/oxfordjournals.pcp.a028962. [DOI] [PubMed] [Google Scholar]
- 6.Lebedev N, Timko MP. Protochlorophyllide photoreduction. Phot Res. 1998;58:5–23. [Google Scholar]
- 7.Reinbothe S, Reinbothe C, Apel K, Lebedev N. Evolution of chlorophyll biosynthesis—the challenge to survive photooxidation. Cell. 1996;86:703–705. doi: 10.1016/s0092-8674(00)80144-0. [DOI] [PubMed] [Google Scholar]
- 8.den Camp RO, Przybyla D, Ochsenbein C, Laloi C, Kim CH, Danon A, et al. Rapid induction of distinct stress responses after the release of singlet oxygen in arabidopsis. Plant Cell. 2003;15:2320–2332. doi: 10.1105/tpc.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buhr F, El Bakkouri M, Valdez O, Pollmann S, Lebedev N, Reinbothe S, et al. Photoprotective role of NADPH: protochlorophyllide oxidoreductase A. Proc Natl Acad Sci USA. 2008;105:12629–12634. doi: 10.1073/pnas.0803950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meskauskiene R, Nater M, Goslings D, Kessler F, den Camp RO, Apel K. FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2001;98:12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goslings D, Meskauskiene R, Kim CH, Lee KP, Nater M, Apel K. Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J. 2004;40:957–967. doi: 10.1111/j.1365-313X.2004.02262.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhong SW, Zhao MT, Shi TY, Shi H, An FY, Zhao Q, et al. EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci USA. 2009;106:21431–21436. doi: 10.1073/pnas.0907670106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, et al. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, et al. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA. 2009;106:7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duek PD, Fankhauser C. bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 2005;10:51–54. doi: 10.1016/j.tplants.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Moon J, Zhu L, Shen H, Huq E. PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci USA. 2008;105:9433–9438. doi: 10.1073/pnas.0803611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephenson PG, Fankhauser C, Terry MJ. PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci USA. 2009;106:7654–7659. doi: 10.1073/pnas.0811684106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castillon A, Shen H, Huq E. Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007;12:514–521. doi: 10.1016/j.tplants.2007.10.001. [DOI] [PubMed] [Google Scholar]