Abstract
Commonly, simple mathematical models can not be used to describe exactly the biological processes due to their higher complexity. In fact, most biological interactions cannot be elucidated by a simple stepwise algorithm or a precise formula, particularly when the data are complex or noisy. ANNs allows an accurate description of those kind of biological processes in plant science, offering new advantages over traditional treatments as the possibility of a model, prediction and optimize results. Different kind of data can be analyzed using a unique and “easy to use” technology. Researchers with a highly specialized mathematical background are not required and ANNs offer the possibility of achieving the whole view of the experimental study with a limited number of experiments and costs. Additionally, it is possible to add new inputs and outputs to the database to reach a new understanding.
Key words: ANNs, artificial intelligence, predicting, optimization, plant model, plant tissue culture
Plant processes are difficult to understand and model because of their higher complexity and their non-linear dynamic behavior. Usually, researchers have had to apply different analytical techniques, such as statistics and mathematical models, to build-up models and an understanding of the factors affecting the processes.1
Data from plant science are usually analyzed through conventional statistical techniques like the analysis of variance (ANOVA). According to Mize and co-workers2 only normal or approximately normal distributed continuous data should be analyze by ANOVA, while discrete data and binary data should to be analyzed by Poisson regression and binary logistic regression, respectively. For this purpose, plant researchers need a high level statistical background therefore, in many cases, have to be assisted by statisticians in spite of it the outputs being difficult to understand.3 Furthermore, techniques to model the whole process or to obtain optimized values taking in account all the factors which influence the study are hard to use or not available with traditional statistics.
Artificial neural networks (ANNs) is a promising technology that mimics the human brain functioning, attempting various cognitive abilities that computers imitate, common in the animal kingdom, such as learning or pattern recognition.4 They entail interlinked nodes that can mathematically abstract neuron function. Each node typically has a number of inputs and one output, which is used as an input for the multiple succeeding nodes. Nodal outputs are usually calculated as some sigmoid function applied to an arithmetic combination of the inputs.5 Plant tissue culture requires optimized processes which can take into account the factors influencing them (genetic, environmental, etc.) and the ANNs together with genetic algorithms can predict the combination of variables (inputs) that yield the optimal result.3
Previously, in other fields, such as in pharmaceutical science, ANNs have been demonstrated as useful tools for modeling complex non linear relationships hidden in data, data mining can offer superior prediction powers compared with the traditional statistical techniques.6,7
In our previous paper we have compared traditional statistics in plant science against the new and promising ANNs technology, selecting a simple in vitro culture proliferation assay with microshoots of kiwifruit Actinidia deliciosa (A. Chev) C.F. Liang et A.R. Ferguson var. deliciosa. Identical conclusions have been obtained by both methods on the effect of two factors or inputs (light conditions and sucrose concentration in the culture medium) on growth parameters (outputs) recorded: response index (binary data), shoots per explant (discrete data) and shoot length (cm, continuous data). However, ANNs offer many advantages and some new features.3
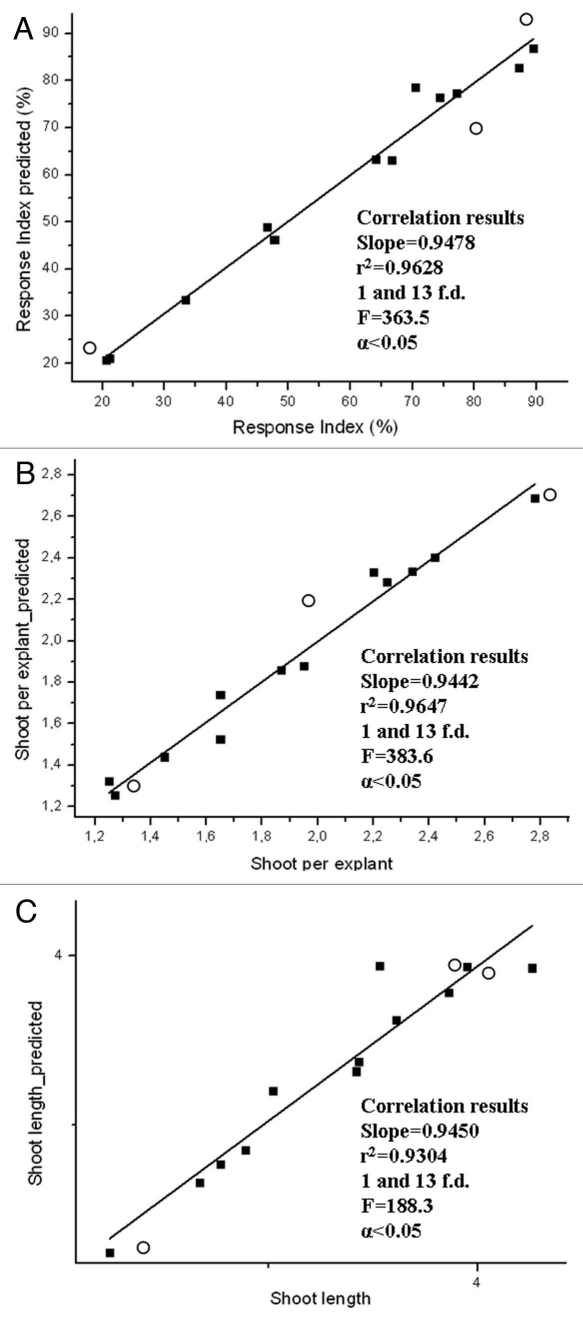
In the present addendum, the authors would like to emphasize several features which should contribute, in our opinion, to encourage plant researchers to use and benefit from this technology. Firstly, ANNs treatments have used mean data instead of primary data, even if they were binary, discrete or continuous data. Researchers, who usually apply statistics, may consider this procedure is inappropriate; however from a practical point of view this simple and useful approach allows a model with a high capacity to predict what is going to happen at different conditions not included for the model. The assay can be established, with a limited number of experiments and costs, and this fact is a priority objective of in vitro plant culture. An ANNs model was achieved, by applying the same training parameters simultaneously for the three outputs tested, showing a training set R2, test set R2 and validation set R2 far over 75%, which indicates high predictability and good performance of the model developed.3 Moreover, the accuracy of the ANNs must be high to describe the relevant factors (inputs) studied and to explain complex interrelations and hidden data pattern. In order to illustrate the predictability of the model, correlations between the estimated values from the model and the observed experimental values for the three different parameters (outputs) studied were carried out (Fig. 1A–C).
Figure 1.
Experimental versus predicted values by ANN s for the different parameters studied. (■) Correlation point for data used for model developing, (○) correlation point for data not included for model developing (validation data).
Correlations were statistically significant (F > 100, 1, 10 f.d., α < 0.05) with r2 values close to 1 for three outputs studied. As can be seen, the slopes are also close to 1 indicating that the values estimated by the model are really similar to the experimental values and therefore the excellent predictability of the model is exposed. Moreover, the estimated parameters obtained by the model for validation data lie on the correlation line, even when they are not included for the development of the model.
Secondly, it is interesting to denote the possibility like in INForm® software, of implementing ANNs with genetic algorithms technology to perform process optimization taking in account different kind of data (binary, discrete and continuous) simultaneously. This option is important especially for the in vitro plant research from a practical point of view in order to make better use of resources and to reduce costs when industrial production is carried out.
Our study suggests that ANNs can be a useful tool for plant science offering new advantages over traditional treatments as the possibility of model, predict and optimize results. Different kinds of data can be analyzed using a unique and “easy to use” technology. Researchers do not required a highly specialized mathematical background and ANNs offer the possibility of achieving a whole view of the experimental study with a limited number of experiments and costs. Additionally, it is possible to add new inputs and outputs to the database to reach a new understanding.
Acknowledgements
This work was supported by Xunta de Galicia; exp.2007/097 and PGIDIT02BTF30102PR.
Authors thanks Ms. J. Menis for her help in the correction of the English version of the work.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11702
References
- 1.Jiménez D, Pérez-Uribe A, Satizábal H, Barreto M, Van Damme P, Marco T. A survey of artificial neural network-based modeling in agroecology. In: Prasad B, editor. Soft computing applications in industry, STUDFUZZ 226. Berlin: Springer Verlag; 2008. pp. 247–269. [Google Scholar]
- 2.Mize CW, Koehler KJ, Compton ME. Statistical considerations for in vitro research: II-Data to presentation. In Vitro Cell Dev Biol-Plant. 1999;35:122–126. [Google Scholar]
- 3.Gago J, Martínez-Núñez L, Landín M, Gallego PP. Artificial neural networks as an alternative to the traditional statistical methodology in plant research. J Plant Physiol. 2010;167:23–27. doi: 10.1016/j.jplph.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Kasabov NK. Foundations of neural networks, fuzzy systems and knowledge engineering. Cambridge (MA): MIT Press; 1996. [Google Scholar]
- 5.Welch SM, Roe JL, Dong Z. A genetic neural network model of flowering time control in Arabidopsis thaliana. Agro J. 2003;95:71–81. [Google Scholar]
- 6.Shate PM, Venitz J. Comparison of neural network and multiple linear regression as dissolution predictions. Drug Dev Ind Pharm. 2003;29:349–355. doi: 10.1081/ddc-120018209. [DOI] [PubMed] [Google Scholar]
- 7.Landín M, Rowe RC, York P. Advantages of neuro-fuzzy logic against conventional experimental design and statistical analysis in studying and developing direct compression formulations. Eur J Pharm Sci. 2009;38:325–331. doi: 10.1016/j.ejps.2009.08.004. [DOI] [PubMed] [Google Scholar]