Abstract
The Gram negative bacterial phytopathogen Pseudomonas syringae employs a molecular syringe termed the type III secretion system (TTSS) to deliver an array of type III secreted effector (TTSE) proteins into plant cells. The major function ascribed to type III effectors of P. syringae is their ability to suppress plant immunity. Because individual pathovars of P. syringae can possess over 30 TTSEs, functional redundancy can provide a hurdle to ascribing functions by TTSE-deletion or -overexpression in such TTSE-rich backgrounds. Approaches to overcome functional redundancy have included the deletion of multiple TTSEs from individual pathovars as well as engineering the plant commensal P. fluorescens strain to express the P. syringae TTSS and deliver P. syringae TTSEs. As we describe here, transgenic Arabidopsis plants expressing individual TTSEs have also been used to overcome problems of functional redundancy and provide invaluable insights into TTSE virulence functions.
Key words: pathogen, virulence, effector, plant immunity, HopF2Pto, RIN4
Functional Insights from TTSE Transgenic Plants
Plant immunity can be triggered by two major classes of pathogen molecules. PAMP-triggered immunity (PTI) is induced by conserved microbial features termed pathogen/microbe associated molecular patterns (PAMPS or MAMPS). Effector-triggered immunity (ETI) induced by pathogen effector proteins is mediated by plant resistance (R) proteins and is often associated with localized cell death termed the hypersensitive response (HR).1,2 TTSE transgenic plants have demonstrated that individual TTSEs can interfere with both branches of plant immunity.
The first Arabidopsis TTSE transgenic plants expressing AvrB or AvrRpt2 demonstrated that TTSE's can function as avirulence and virulence factors inside plant cells.3,4,5 Since then Arabidopsis TTSE transgenic plants have provided invaluable insights into TTSE functions. In a landmark study, Hauck et al. (2003) demonstrated that transgenic expression of AvrPto in Arabidopsis can suppress callose deposition associated with PTI.6 Furthermore, AvrPto can single-handedly promote the growth of non-virulent Pto DC3000 ΔhrcC (lacking a functional TTSS) to levels comparable to wild-type Pto DC3000. Since this study, numerous effectors have been demonstrated to suppress PTI when expressed transgenically, including AvrRpm1, AvrRpt2, AvrB, AvrPtoB, HopAI1, HopF2, HopAO1 and HopG1.7–13 In addition to PTI suppression, TTSE transgenic plants have demonstrated that individual TTSEs can alter plant hormone levels and hormone sensitivity14–16 as well as manipulate miRNA pathways.17 A forward genetic screen was conducted on AvrB transgenic plants to identify potential targets of AvrB operation (TAO genes).18 TAO1 was mapped to a TIR-NB-LRR resistance gene that contributes to AvrB ETI in Arabidopsis.19
We recently investigated the ETI-suppression ability of HopF2Pto using transgenic plants in an attempt to provide clues about its host targets.18 We found that transgenic HopF2Pto differentially inhibited the ETI-associated hypersensitive response induced by various TTSEs in Arabidopsis (ecotype Col-0). HopF2Pto expression compromised AvrRpt2-mediated HR but not the HR induced by AvrRpm1, AvrB or HopZ1a. Interestingly, HopF2Pto also compromised the depletion of RIN4 protein that is normally associated with AvrRpt2-HR suggesting that RIN4 could be a target of HopF2Pto.20,21 In support of this, HopF2Pto interacted with RIN4 both in vitro and in vivo, leading us to investigate whether RIN4 is a virulence target of bacterially delivered HopF2Pto. Pseudomonas syringae growth in Arabidopsis was enhanced by overexpressing HopF2Pto in P. syringae pv. tomato DC3000 (PtoDC3000) lacking endogenous HopF2Pto. This virulence enhancement was not observed in Arabidopsis plants lacking RIN4, confirming that RIN4 is a virulence target of bacterially delivered HopF2Pto.
The crystal structure of HopF1Pph7 displays limited structural similarity to the catalytic domain of the ADP-ribosyltransferase diphtheria toxin.21 Although HopF2Pto is predicted to adopt a similar structure, no HopF2Pto ADP-RT activity could be detected using RIN4 as a substrate in vitro nor from plant extracts of HopF2Pto-expressing plants.19 Nevertheless, a structurally predicted potential catalytic residue (D175) was required for suppression of AvrRpt2-ETI and associated RIN4 depletion, as well as HopF2Pto-enhanced bacterial virulence indicating that HopF2Pto may modify RIN4 in order to promote bacterial virulence. However, the affinity of HopF2PtoD175A for RIN4 has yet to be assessed.
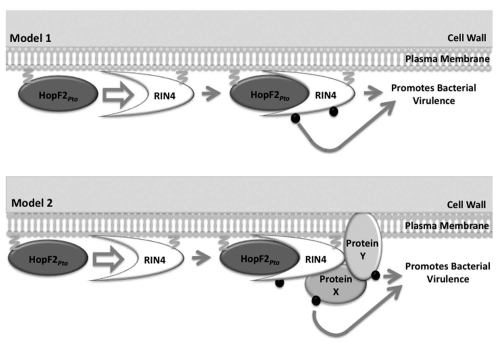
Based on the aforementioned data, we propose two possible models of RIN4-dependent HopF2Pto action (Fig. 1). In the first model HopF2Pto binds and modifies RIN4, and this RIN4-modification directly promotes bacterial virulence. In the second model, HopF2Pto utilizes RIN4 as a scaffold to modify RIN4-associated proteins thereby promoting bacterial virulence. It is important to note that HopF2Pto may have RIN4-independent virulence targets. In support of this, HopF2Pto-mediated PTI suppression is maintained in HopF2Pto transgenic plants lacking RIN4 (Wilton M and Desveaux D, unpublished).11 However, since HopF2Pto-enhanced PtoDC3000 virulence was RIN4-dependent, these targets must be functionally redundant to those of endogenous PtoDC3000 TTSEs.
Figure 1.
Two models of RIN 4-dependent HopF2Pto action. In Model 1 HopF2Pto directly binds and modifies RIN 4 and this RIN 4-modification directly promotes bacterial virulence. In Model 2, HopF2Pto uses RIN 4 as a scaffold to modify RIN 4-associated proteins (hypothetical proteins X and Y) to promote bacterial virulence. Both RIN 4 and HopF2Pto are membrane localized by prenylation and myristoylation, respectively.24,26
ETI-Suppression in TTSE Transgenic Plants—Learning from Specificity
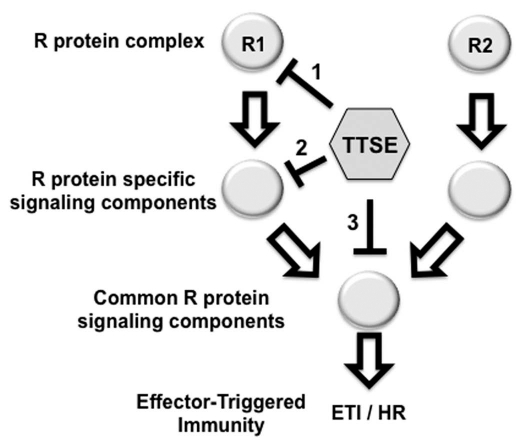
Our results with HopF2Pto emphasize the potential advantage of using TTSE-transgenics to investigate TTSE functions as well as ETI-signaling pathways. We hypothesize that ETI-suppression by TTSEs can occur by targeting three broad categories of ETI-signaling proteins: (1) R proteins or R protein monitored TTSE targets, (2) R protein signaling components that are differentially required by various R protein classes, or (3) R protein signaling components that are required by most or all R proteins (Fig. 2). In the first two cases, ETI-suppression will be specific to certain R protein classes and is exemplified by the AvrRpt2-ETI suppression by HopF2Pto and also by AvrB- and AvrRpm1-ETI suppression by AvrRpt2.19,23,24 In the third case, ETI-suppression will be effective against a broad range of R protein classes. This may be the case for TTSEs that can suppress both ETI and Bax-induced programmed cell death.25 Therefore, important insights into TTSE function can be gained by investigating their specificity of ETI-suppression in transgenic plants. This specificity can also potentially be used to dissect R protein signaling pathways. A continual challenge of TTSE-transgenic plant work will be to confirm that what a TTSE can do when expressed in transgenic plants is actually relevant to the function of that TTSE when delivered from the bacteria.
Figure 2.
ETI-suppression in TTSE transgenic plants. Individual TTSE can potentially suppress ETI by targeting various components of R protein signaling pathways including: (1) R proteins or R protein-associated proteins, (2) ETI signaling components that are differentially required by R proteins, or (3) ETI signaling components required by most or all R proteins. These possible scenarios can be investigated using TTSE transgenic plants in order gain insight into TTSE functions as well as R protein signaling pathways.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11703
References
- 1.Dangl JL, Jones JDG. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 2.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Gopalan S, Bauer DW, Alfano JR, Loniello AO, He SY, Collmer A. Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McNellis TW, Mudgett MB, Li K, Aoyama T, Horvath D, Chua NH, et al. Glucocortoid-inducible expression of a bacterial avirulence gene in transgenic Arabidopsis induces hypersensitive cell death. Plant J. 1998;14:247–257. doi: 10.1046/j.1365-313x.1998.00106.x. [DOI] [PubMed] [Google Scholar]
- 5.Chen ZY, Kloek AP, Boch J, Katagiri F, Kunkel BN. The Pseudomonas syringae AvrRpt2 gene product promotes pathogen virulence from inside plant cells. Mol Plant Microbe Interact. 2000;13:1312–1321. doi: 10.1094/MPMI.2000.13.12.1312. [DOI] [PubMed] [Google Scholar]
- 6.Hauck P, Thilmony R, He SY. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci USA. 2003;100:8577–8582. doi: 10.1073/pnas.1431173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim MG, da Cunha L, McFall AJ, Belkhadir Y, DebRoy S, Dangl JL, et al. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 8.Shang YL, Li XY, Cui HT, He P, Thilmony R, Chintamanani S, et al. RAR1, a central player in plant immunity, is targeted by Pseudomonas syringae effector AvrB. Proc Natl Acad Sci USA. 2006;103:19200–19205. doi: 10.1073/pnas.0607279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Torres M, Mansfield JW, Grabov N, Brown IR, Ammouneh H, Tsiamis G, et al. Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis. Plant J. 2006;47:368–382. doi: 10.1111/j.1365-313X.2006.02798.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Shao F, Cui H, Chen LJ, Li HT, Zuo Y, et al. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-Induced immunity in plants. Cell Host Microbe. 2007;1:175–185. doi: 10.1016/j.chom.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Guo M, Tian F, Wamboldt Y, Alfano JR. The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol Plant Microbe Interact. 2009;22:1069–1080. doi: 10.1094/MPMI-22-9-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Underwood W, Zhang S, He SY. The Pseudomonas syringae type III effector tyrosine phosphatase HopAO1 suppresses innate immunity in Arabidopsis thaliana. Plant J. 2007;52:658–672. doi: 10.1111/j.1365-313X.2007.03262.x. [DOI] [PubMed] [Google Scholar]
- 13.Block A, Guo M, Li G, Elowsky C, Clemente TE, Alfano JR. The Pseudomonas syringae type III effector HopG1 targets mitochondria, alters plant development and suppresses plant innate immunity. Cell Microbiol. 2010;12:318–330. doi: 10.1111/j.1462-5822.2009.01396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Egea PR, et al. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway. EMBO J. 2007;26:1434–1443. doi: 10.1038/sj.emboj.7601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen ZY, Agnew JL, Cohen JD, He P, Shan LB, Sheen J, et al. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci USA. 2007;104:20131–20136. doi: 10.1073/pnas.0704901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goel AK, Lundberg D, Torres MA, Matthews R, Akimoto-Tomiyama C, Farmer L, et al. The Pseudomonas syringae type III effector HopAM1 enhances virulence on water-stressed plants. Mol Plant Microbe Interact. 2008;21:361–370. doi: 10.1094/MPMI-21-3-0361. [DOI] [PubMed] [Google Scholar]
- 17.Navarro L, Jay F, Nomura K, He SY, Voinnet O. Suppression of the microRNA pathway by bacterial effector proteins. Science. 2008;321:964–967. doi: 10.1126/science.1159505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eitas TK, Nimchuk ZL, Dangl JL. Arabidopsis TAO1 is a TIR-NB-LRR protein that contributes to disease resistance induced by the Pseudomonas syringae effector AvrB. Proc Natl Acad Sci USA. 2008;105:6475–6480. doi: 10.1073/pnas.0802157105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilton M, Subramaniam R, Elmore J, Felsensteiner C, Coaker G, Desveaux D. The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc Natl Acad Sci USA. 2010;107:2349–2354. doi: 10.1073/pnas.0904739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 21.Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 22.Singer AU, Desveaux D, Betts L, Chang JH, Nimchuk Z, Grant SR, et al. Cyrstal structures of the type III effector protein AvrPphF and its chaperone reveal residues required for plant pathogenesis. Structure. 2004;12:1669–1681. doi: 10.1016/j.str.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 23.Ritter C, Dangl JL. Interference between two specific pathogen recognition events mediated by distinct plant disease resistance genes. Plant Cell. 1996;8:251–257. doi: 10.1105/tpc.8.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL. The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc Natl Acad Sci USA. 2005;102:6496–6501. doi: 10.1073/pnas.0500792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jamir Y, Guo M, Oh H-S, Petnicki-Ocwieja T, Chen S, Tang X, et al. Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J. 2004;37:554–565. doi: 10.1046/j.1365-313x.2003.01982.x. [DOI] [PubMed] [Google Scholar]
- 26.Robert-Seilaniantz A, Shan L, Zhou J-M, Tang X. The Pseudomonas syringae pv. tomato DC3000 type III effector HopF2 has a putative myristoylation site required for its avirulence and virulence functions. Mol Plant Microbe Interact. 2006;19:130–138. doi: 10.1094/MPMI-19-0130. [DOI] [PubMed] [Google Scholar]