Abstract
Rice blast, caused by Magnaporthe oryzae, is a devastating disease of rice (Oryza sativa). The mechanisms involved in resistance of rice to blast have been studied extensively and the rice—M. oryzae pathosystem has become a model for plant—microbe interaction studies. However, the mechanisms involved in nonhost resistance (NHR) of other plants to rice blast are still poorly understood. We have recently demonstrated that AGB1 and PMR5 contribute to PEN2-mediated preinvasion resistance to M. oryzae in Arabidopsis thaliana, suggesting a complex genetic network regulating the resistance. To determine whether other defense factors: RAR1, SGT1 and NHO1, affected the A. thaliana-M. oryzae interactions, double mutants were generated between pen2 and these defense-related mutants. All these double mutants exhibited a level of penetration resistance similar to that of the pen2 mutant, suggesting that none of these mutants significantly compromised resistance to M. oryzae in a pen2 background.
Key words: nonhost resistance, PEN2, RAR1, SGT1, NHO1
Plants face microbial attacks and have evolved innate immunity systems to defend against these threats. The initial step of the immunity signaling pathway is recognition of intra- or extracellular pathogen-derived molecules. Externally oriented transmembrane-type proteins containing leucine-rich repeat (LRR) domains detect extracellular molecules, whereas cytoplasmic sensors possess nucleotide-binding (NB) and LRR domains (NLR).1,2 The LRR domain serves as a pattern-recognition receptor to detect pathogen-derived molecules or host proteins that are targeted by pathogen peptides that have entered the cell, effectors.3 NLR-type sensors are the substrates of a structurally and functionally conserved chaperone complex that consists of HEAT SHOCK PROTEIN 90 (HSP90) and its cochaperone SUPPRESSOR OF THE G2 ALLELE OF SKP1 (SGT1). REQUIRED FOR MLA12 RESISTANCE 1 (RAR1) regulated the HSP90-SGT1 complex, resulting in the stabilization of NLR proteins. Thus, SGT1 and RAR1 are required for the function of multiple and distinct R genes that encode NLR immune sensors in plants.4 Experiments in RAR1-silenced transgenic rice lines showed that RAR1 is not essential for Pib, which encodes an NLR against rice blast fungus.5 In contrast, basal resistance to normally virulent races of rice blast fungus or bacterial blight is significantly reduced in RAR1-silenced lines. This result is consistent with earlier reports that RAR1 is involved in basal resistance to virulent Pseudomonas bacteria in Arabidopsis or blast fungus in barley.6,7 The requirement of SGT1 for immunity in plants is shown mostly by transient silencing of a number of NLR proteins.8,9 In addition, SGT1 is also required for immune responses triggered by non-NLR-type sensors.10 This requirement indicates that either SGT1 function is not limited to the NLR sensors, or some unknown SGT1-dependent NLR proteins also operate downstream of non NLR-type sensors. Furthermore, SGT1 is involved in nonhost resistance, indicating that SGT1 may be a general factor of disease resistance.10 An Arabidopsis mutant, nho1 (nonhost resistance 1), has been isolated on which Pseudomonas syringae pv. phaseolicola grows and causes disease symptoms.11,12 It is significant that this mutant is also compromised in R-gene-mediated resistance to P. syringae.11 Although NHO1 is the flagellin-induced glycerol kinase, whose exact function in NHR remains elusive.12,13 A possible explanation might be that altered plant glycerol pools either directly or indirectly affect nutrient availability for P. syringae. NHO1 is also required for resistance to the fungal pathogen Botrytis cinerea, indicating that NHO1 is not limited to bacterial resistance.12 However, these contributions to NHR to M. oryzae in A. thaliana have not been understood.
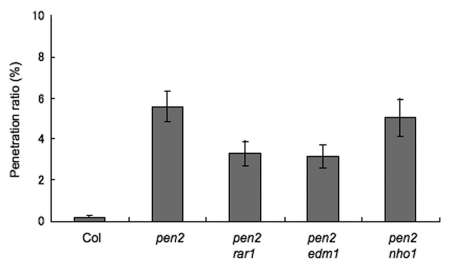
To determine whether these factors were necessary for the resistance to M. oryzae in A. thaliana, the following A. thaliana mutants were inoculated with M. oryzae and monitored by microscopy: rar1-21;14 edm1-1;15 nho1-1,11 (all Col-0 background). All these mutants exhibited a level of penetration resistance similar to that of the wild-type plants (data not shown), suggesting that none of these mutants significantly compromised resistance to M. oryzae. We have recently shown that among the penetration (pen) mutants, only the pen2,16 mutant allowed increased penetration into epidermal cells by M. oryzae.17 Thus, double mutants were generated between pen2 and these mutants to determine whether these factors were necessary for the resistance to M. oryzae in a pen2 background: pen2 rar1-21; pen2 edm1-1; pen2 nho1-1. All these double mutants exhibited a level of penetration resistance similar to that of the pen2 mutant (Fig. 1), suggesting that none of these mutants significantly compromised resistance to M. oryzae in a pen2 background. This might indicate that NHR against M. oryzae may not be conferred by RAR1- and SGT1-dependent NLR immune sensors. Alternatively, since there has been no report that RAR1 is required for any known transmembrane sensors, such as FLS2, EFR or Xa21, RAR1- and SGT1-independent transmembrane-type immune sensors may be required for NHR against M. oryzae. Future studies will be required to reveal the genetic and mechanistic requirements for NHR in A. thaliana-M. oryzae interactions.
Figure 1.
Double mutant analysis to evaluate the role of the defense related genes on resistance to Magnaporthe oryzae in Arabidopsis thaliana. The frequency of M. oryzae penetration on double mutants at 3 days post-inoculation was expressed as a percentage of total appressoria. Data were collected from six independent plants per line. A minimum of 100 infection sites was inspected per leaf. Results represent mean ± standard error of three independent experiments.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11770
References
- 1.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Shen QH, Schulze-Lefert P. Rumble in the nuclear jungle: compartmentalization, trafficking and nuclear action of plant immune receptors. EMBO J. 2007;26:4293–4301. doi: 10.1038/sj.emboj.7601854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shirasu K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol. 2009;60:139–164. doi: 10.1146/annurev.arplant.59.032607.092906. [DOI] [PubMed] [Google Scholar]
- 5.Thao NP, Chen L, Nakashima A, Hara S, Umemura K, Takahashi A, et al. RAR1 and HSP90 form a complex with Rac/Rop GTPase and function in innate-immune responses in rice. Plant Cell. 2007;19:4035–4045. doi: 10.1105/tpc.107.055517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holt BF, Belkhadir Y, Dangl JL. Antagonistic control of disease resistance protein stability in the plant immune system. Science. 2005;309:929–932. doi: 10.1126/science.1109977. [DOI] [PubMed] [Google Scholar]
- 7.Jarosch B, Collins NC, Zellerhoff N, Schaffrath U. RAR1, ROR1, and the actin cytoskeleton contribute to basal resistance to Magnaporthe grisea in barley. Mol Plant Microbe Interact. 2005;18:397–404. doi: 10.1094/MPMI-18-0397. [DOI] [PubMed] [Google Scholar]
- 8.Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. 2002;295:2073–2076. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- 9.Hein I, Barciszewska-Pacak M, Hrubikova K, Williamson S, Dinesen M, Soenderby IE, et al. Virus-induced gene silencing-based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol. 2005;138:2155–2164. doi: 10.1104/pp.105.062810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peart JR, Lu R, Sadanandom A, Malcuit I, Moffett P, Brice DC, et al. Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc Natl Acad Sci USA. 2002;99:10865–10869. doi: 10.1073/pnas.152330599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu M, Tang X, Zhou JM. Arabidopsis NHO1 is required for general resistance against Pseudomonas bacteria. Plant Cell. 2001;13:437–447. doi: 10.1105/tpc.13.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang L, Li J, Zhao T, Xiao F, Tang X, Thilmony R, et al. Interplay of the Arabidopsis nonhost resistance gene NHO1 with bacterial virulence. Proc Natl Acad Sci USA. 2003;100:3519–3524. doi: 10.1073/pnas.0637377100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Lin H, Zhang W, Zou Y, Zhang J, Tang X, et al. Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc Natl Acad Sci USA. 2005;102:12990–12995. doi: 10.1073/pnas.0502425102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tornero P, Merritt P, Sadanandom A, Shirasu K, Innes RW, Dangl JL. RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell. 2002;14:1005–1015. doi: 10.1105/tpc.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tor M, Gordon P, Cuzick A, Eulgem T, Sinapidou E, Mert-Turk F, et al. Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell. 2002;14:993–1003. doi: 10.1105/tpc.001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310:1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- 17.Maeda K, Houjyou Y, Komatsu T, Hori H, Kodaira T, Ishikawa A. AGB1 and PMR5 contribute to PEN2-mediated preinvasion resistance to Magnaporthe oryzae in Arabidopsis thaliana. Mol Plant Microbe Interact. 2009;22:1331–1340. doi: 10.1094/MPMI-22-11-1331. [DOI] [PubMed] [Google Scholar]