Abstract
Imaging resource flow in soil-plant systems remains central to understanding plant development and interactions with the environment. Typically, subcellular resolution is required to fully elucidate the compartmentation, behavior, and mode of action of organic compounds and mineral elements within plants. For many situations this has been limited by the poor spatial resolution of imaging techniques and the inability to undertake studies in situ. Here we demonstrate the potential of Nanoscale Secondary Ion Mass Spectrometry (NanoSIMS), which is capable of the quantitative high-resolution spatial imaging of stable isotopes (e.g., 12C, 13C, 14N, 15N, 16O, 18O, 31P, 34S) within intact plant-microbial-soil systems. We present examples showing how the approach can be used to investigate competition for 15N-labelled nitrogen compounds between plant roots and soil microorganisms living in the rhizosphere and the spatial imaging of 31P in roots. We conclude that NanoSIMS has great potential to elucidate the flow of isotopically-labelled compounds in complex media (e.g., soil) and opens up countless new opportunities for studying plant responses to abiotic stress (e.g., 18O3, elevated 13CO2), signal exchange, nutrient flow and plant-microbial interactions.
Key words: mass spectrometry, NanoSIMS, rhizosphere, isotope labelling, soil, nitrogen, carbon, phosphorus, 15N, 13C, 31P
We have used the NanoSIMS technique to investigate the flow of nutrients between microbial and plant cells within the rhizosphere. Secondary Ion Mass Spectrometry (SIMS) involves bombarding a sample with a high-energy ion beam, which sputters atoms, molecules and electrons from the sample surface. Ionized species (secondary ions) are extracted to a mass spectrometer, sorted according to their energy and their mass-to-charge ratio, and counted. NanoSIMS, a recent development in SIMS, combines high sensitivity with high spatial resolution (typically 100 nm) to allow elemental and isotopic imaging of secondary ions, such as 12C-, 16O- and 12C14N-, on a range of biological materials at the sub-cellular scale (Fig. 1A and B). An element map is obtained by scanning the primary ion beam over the sample surface and measuring the secondary ion intensities of any given ion species, at each pixel in the image. The intrinsically high mass resolution allows the separation of different ion species at the same nominal atomic mass (e.g., 12C15N- from 13C14N- at mass 27), while the multi-collection capability allows the simultaneous measurement of up to five ion species. This makes it possible to obtain images of different isotopes from the same area simultaneously, from which quantitative isotope ratios from individual components can then be extracted. As such, NanoSIMS offers a means of elucidating processes involved in the transport of ions and molecules into cells and their distribution within cells, at scales and sensitivities not attainable by other methods.1–5
Figure 1.
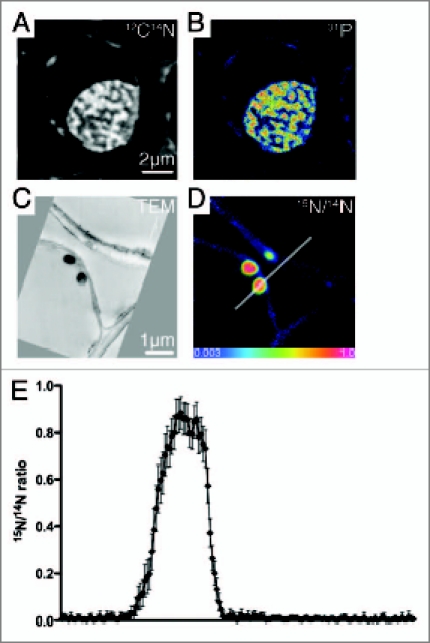
(A) 12C14N- and (B) 31P- images of a wheat root cell nucleus from NanoSIMS illustrating the potential to map different elements at the sub-cellular scale; (C) TEM image of two bacteria attached to a cortical cell wall; (D) corresponding 15N/14N ratio image from NanoSIMS of the same bacteria. The differential uptake of 15N is illustrated by the color scale; ranging from natural abundance (blue) to a 15N/14N ratio = 1.0 (i.e., 50 at% 15N) (pink) for the plant cell and bacteria, respectively; (E) Linescan (3.5 µm) illustrating the variation in 15N/14N across an enriched bacterium and an un-enriched plant cell wall (line in D). Error bars are based on the Poisson counting statistics for each pixel.
We previously demonstrated the use of NanoSIMS to image and map the location of 15N-labelled bacterial communities artificially introduced into soil microhabitats.6,7 We extended this approach to a natural ecosystem, by examining the differential partitioning of 15N-labelled ammonium (15NH4+) between plant roots and soil microbial communities at the nanometer scale (Fig. 1C and D).8 It was shown that introduced 15N could be detected, and more importantly, mapped, in individual bacterial cells found in the soil matrix, within the rhizosphere, within root hairs, and intra-cellular within the root. The 15N/14N ratio data (determined as the ratio between the 12C15N- and the 12C14N- signals) could then be extracted from specific regions of interest—groups of pixels bounding a particular feature, such as a bacterium or a root cell wall, or linescans (Fig. 1E). This unique approach allows the visualization of nutrient flows and metabolic pathways through complex, multi-component ecosystems. Here we consider further the application of the technique to study nutrient availability in plant cell research.
Uptake of Amino Acids by Plants and Microorganisms
Amino acids represent a large input of organic N to rhizosphere soil, constituting an important source of N to both plants and microorganisms, and they have been implicated as a major factor regulating ecosystem productivity.9 Recently, we, and others, have confirmed that higher plants can capture amino acids from soil.10,11 This challenges the paradigm that N must first be microbially processed to inorganic N before becoming plant-available. This direct uptake of organic N by some plants provides an effective shortcut in the N cycle.11,12 Most experiments to date have used either 15N- and/or 13C-labelled amino acids to determine the relative degree of plant-microbe competition for dissolved organic N in the rhizosphere using bulk mass spectrometry methods.11 This approach has been criticized, however, as it still remains difficult to distinguish between direct uptake and indirect uptake, where the amino acids are first mineralized to NH4+ before being taken up by the roots.9 We propose that by using dual-labelled 15N and 13C amino acids it will be possible to image the location of both isotopes within plant cells, associated microbial communities, and fungal associations at the sub-cellular scale. Where both isotopes are co-located within the same pixels of the image we can infer uptake as an organic molecule, while spatial separation of the isotopes in the image map would indicate a split of the molecule. Simultaneous labelling with 13C and 15N has proved effective on other biological systems analyzed by NanoSIMS,13–15 however, these experiments did not attach the two isotopes to the same starting molecule. Combining this with the use of halogenated DNA probes that are detectable by the NanoSIMS (e.g., Iodine),16 it should be possible to study such rhizosphere functions along with molecular identification of target organisms.
Spatial Mapping of Phosphorus (P) within the Soil Matrix
The spatial imaging of phosphorus (P) around roots also represents a real challenge within intact plant-soil systems. This is due to the poor solubility and slow diffusion of P in soil, which consequently produces steep and very narrow depletion zones around roots. Previous research in rhizosphere systems has tended to focus on the use of micro-autoradiography, using 32P and 33P radioisotopes.17,18 This technique is typically limited to a spatial resolution of 0.25 to 1 mm, due to (1) the inability to get close contact between the soil particles and the screen and (2) the hemispherical spread of β-particles, which leads to image blurring. Although only one stable isotope of P exists (31P), NanoSIMS has great potential to help spatially resolve P depletion and transfer in the plant-soil system. For example, in model systems, using sand grains coated with goethite [Fe(OH)3\ to which 31P is uniformly adsorbed, it would be possible to determine the exact spatial extent of the depletion zone in soil with high precision. In addition, it would enable examination of P depletion with root hairs of different ages, or whether the depletion zone extends past the root hair tips, which would imply mobilization by root exudates.19 NanoSIMS would also allow the spatial heterogeneity of P in the rhizosphere to be determined, as well as the spatial distribution of P within mycorrhizas associated with roots.20
Existing work clearly proves that NanoSIMS has tremendous potential to allow the study of assimilatory processes at the sub-micron level in a wide range of biochemical and molecular applications. The ability to acquire quantitative data at high spatial resolution opens up countless new opportunities for ecosystems involving plant cells, microorganisms and animals.
Acknowledgements
This research was funded through an ARC Discovery Project (DP0985832) and the Grains Research and Development Corporation (GRDC). The authors acknowledge the facilities, scientific and technical assistance of the Australian Microscopy & Microanalysis Research Facility at the Centre for Microscopy, Characterisation & Analysis, The University of Western Australia, a facility funded by The University, State and Commonwealth Governments.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11775
References
- 1.Lechene C, Hillion F, McMahon G, Benson D, Kleinfeld AM, Kampf JP, et al. High-resolution quantitative imaging of mammalian and bacterial cells using stable isotope mass spectrometry. J Biol. 2006;5:20. doi: 10.1186/jbiol42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lechene C, Luyten Y, McMahon G, Distel D. Quantitative imaging of Nitrogen fixation by individual bacteria in animal cells. Science. 2007;317:1563–1566. doi: 10.1126/science.1145557. [DOI] [PubMed] [Google Scholar]
- 3.Guerquin-Kern J-L, Wu T-D, Qunitana C, Croisy A. Progress in analytical imaging of the cell by dynamic secondary ion mass spectrometry (SIMS microscopy) Biochim Biophys Acta. 2005;1724:228–238. doi: 10.1016/j.bbagen.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Eybe T, Audinot JN, Bohn T, Guignard C, Migeon HN, Hoffmann L. NanoSIMS 50 elucidation of the natural element composition in structures of cyanobacteria and their exposure to halogen compounds. J App Microbiol. 2008;105:1502–1510. doi: 10.1111/j.1365-2672.2008.03870.x. [DOI] [PubMed] [Google Scholar]
- 5.Smart KE, Kilburn MR, Salter CJ, Smith JAC, Grovenor CRM. NanoSIMS and EPMA analysis of nickel localisation in leaves of the hyperaccumulator plant Alyssum lesbiacum. Int J Mass Spec. 2007;260:107–114. [Google Scholar]
- 6.Herrmann AM, Clode PL, Fletcher IR, Nunan N, Stockdale EA, O'Donnell AG, et al. A novel method for the study of the biophysical interface in soils using nano-scale secondary ion mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:29–34. doi: 10.1002/rcm.2811. [DOI] [PubMed] [Google Scholar]
- 7.Herrmann AM, Ritz K, Nunan N, Clode PL, Pett-Ridge J, Kilburn MR, et al. Nano-scale secondary ion mass spectroscopy—a new analytical tool in biogeochemistry and soil ecology: a review article. Soil Biol Biochem. 2007;39:1835–1850. [Google Scholar]
- 8.Clode PL, Kilburn MR, Jones DL, Stockdale EA, Cliff JB, Herrmann AM, et al. In situ mapping of nutrient uptake in the rhizosphere using nanoscale secondary ion mass spectrometry. Plant Physiol. 2009;151:1751–1757. doi: 10.1104/pp.109.141499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones DL, Healey JR, Willett VB, Farrar JF, Hodge A. Dissolved organic nitrogen uptake by plants—an important N uptake pathway? Soil Biol Biochem. 2005;37:413–423. [Google Scholar]
- 10.Jones DL, Shannon D, Junvee-Fortune T, Farrar JF. Plant capture of free amino acids is maximized under high soil amino acid concentrations. Soil Biol Biochem. 2005;37:179–181. [Google Scholar]
- 11.Kielland K. Amino acid absorption by arctic plants: implications for plant nutrition and nitrogen cycling. Ecology. 1994;75:2373–2383. [Google Scholar]
- 12.Schimel JP, Bennett J. Nitrogen mineralization: Challenges of a changing paradigm. Ecology. 2004;85:591–602. [Google Scholar]
- 13.Musat N, Halm H, Winterholler B, Hoppe P, Peduzzi S, Hillion F, et al. A single-cell view on the ecophysiology of anaerobic photrophic bacteria. Proc Natl Acad Sci USA. 2008;105:17861–17866. doi: 10.1073/pnas.0809329105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finzi-Hart JA, Pett-Ridge J, Weber PK, Popa R, Fallon SJ, Gunderson T, et al. Fixation and fate of C and N in the cyanobacterium Trichodesmium using nanometer-scale secondary ion mass spectrometry. Proc Natl Acad Sci USA. 2009;106:6345–6350. doi: 10.1073/pnas.0810547106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behrens S, Lösekann T, Pett-Ridge J, Weber PK, Ng W-O, Stevenson BS, et al. Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition flourescence in situ hybridization and nanometer-scale secondary ion mass spectometry. App Environ Microbiol. 2008;74:10. doi: 10.1128/AEM.00191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li T, Wu TD, Mazéas L, Toffin L, Guerquin-Kern J-L, Leblon G, et al. Simultaneous analysis of microbial identity and function using NanoSIMS. Environ Microbiol. 2008;10:580–588. doi: 10.1111/j.1462-2920.2007.01478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhat KKS, Nye PH. Diffusion of phosphate to plant roots in soil I. Quantitative autoradiography of the depletion zone. Plant Soil. 1973;38:161–175. [Google Scholar]
- 18.Misra RK, Alston AM, Dexter AR. Role of root hairs in phosphorus depletion from a macrostructured soil. Plant Soil. 1988;107:11–18. [Google Scholar]
- 19.Jones DL. Organic acids in the rhizosphere—a critical review. Plant Soil. 1998;205:25–44. [Google Scholar]
- 20.Smits MM, Herrmann AM, Duane M, Duckworth OW, Bonneville S, Benning LG, et al. The fungal-mineral interface: Challenges and considerations of micro-analytical developments. Fungal Biol Rev. 2010 doi: 10.1016/j.fbr.2009.11.00. In press. [DOI] [Google Scholar]


