Abstract
As much as there is known about the function of the sodium/proton antiporter SOS1 in plants, recent studies point towards a more general role for this protein. The crucial involvement in salt stress protection is clearly one of its functions—confined to the N-terminus, but the modular structure of the protein includes a segment with several domains that are functionally not studied but comprise more than half of the protein's length. Additional functions of the protein appear to be an influence on vesicle trafficking, vacuolar pH and general ion homeostasis during salt stress. Eliminating SOS1 leads to the expression of genes that are not strictly salinity stress related. Functions that are regulated in sos1 mutants included pathogen responses, and effects on circadian rhythm.
Key words: Arabidopsis thaliana, SOS1 deficiency, crosstalk, calcium-channels, proton gradients, biotic stress
SOS1 (Salt Overly Sensitive 1) is known as a crucial component in the defense of plants against sodium ions that have entered the cytoplasm. The gene encodes a proton/sodium-antiporter whose absence drastically increases salt sensitivity of the salt-sensitive species, Arabidopsis thaliana. Even more interesting is the fact that RNAi-interference with SOS1 in a naturally salt tolerant species, Thellungiella salsuginea converts this halophyte into a plant with pronounced glycophytic characters.1,2 Recently, we have pointed out that SOS1 appears to have additional function.3 We back this conclusion by outlining the function of genes and pathways that are affected when SOS1 is either mutated by T-DNA insertion or by the reduction of SOS1 transcripts through RNAi-interference.
On the level of intracellular physiology, upregulated in wild type are SOS1 and several calcium channels, while moderate downregulation is observed for AQPs, and the TPK1 and HKT1 potassium channels (reviewed in ref. 3). In part, such changes have been observed before.4 This behavior in wild type plants depends on the severity of the salt stress; only at very high concentrations of sodium in wild type can we observe stronger responses in gene expression changes that have been interpreted as chaotic before.5
On the level of cell organization and growth, the consequences of SOS1 deficiency manifested themselves in the disturbance of specific functions. Over time during salt stress the vacuoles disintegrated into small spherical bodies, while the plasma membrane barrier showed gradual degradation. Another consequence of salt stress in the mutant was cessation of membrane trafficking. The pH of vacuoles and cytosol in the atsos1 lines are approximately 0.5 pH units more acidic than in wild type and vacuoles contained higher concentrations of calcium ions in mutant plants. At this stage, the disturbance could well be interpreted as a general breakdown of metabolism and gene expression, but a closer inspection revealed otherwise.
The absence of SOS1 in the atsos1 mutant line of Arabidopsis affected a large number of other ion channels, including calcium transporters and several NHX-type channels. These functions are strongly upregulated (reviewed in ref. 3). Much more strongly downregulated in the mutant than in the wild type were HKT1, and several other K+-transporters, and AQPs. Also, we observed a significant downward adjustment in the expression of a number of plasma-membrane and tonoplast proton ATPases that were not affected in wild type (reviewed in ref. 3). The effect of salt stress in the sos1 mutant, leading to downregulation of the vacuolar H+-PPase AVP1,3 underscored an earlier observation, which had indicated that overexpression of AVP1 improved salt tolerance in Arabidopsis.6 The apparent compensation of the stress-dependent disturbances experienced by the Arabidopsis mutant lines pointed towards a more general function for SOS1.
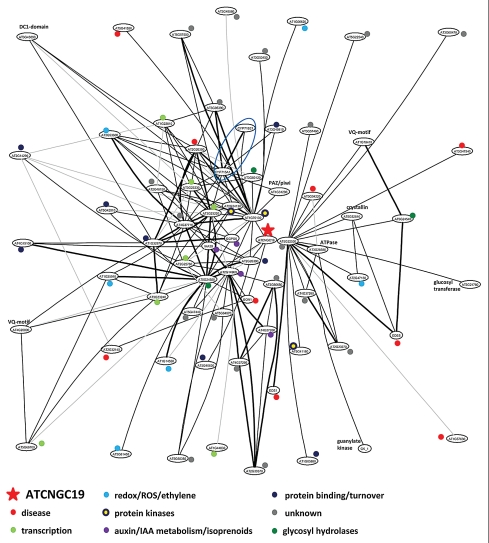
To highlight such functions, we used analysis of transcript co-expression partial-correlation network of the Arabidopsis transcriptome. The program used some 2,000 microarray datasets collected for plants during development, recorded expression in different tissues and included changes in gene expression under a variety of environmental challenges.7,8 Using strongly regulated genes in atsos1 mutants in comparison to wild type as input,3,9 we highlighted the context in which these genes are expressed in plants. In several cases, the analysis highlights structures in the gene networks that appear to point towards a relationship between SOS1 integrity and circadian rhythmicity and flowering/terminal growth (not shown). Here, we present as one example the gene network that surrounds the strongly upregulated CNGC19 gene, encoding a calmodulin-binding cyclic nucleotide gated channel. Recently, CNGC19 has been associated with the classical salt stress response in Arabidopsis.10 However, at high statistical significance, CNGC19 expression is correlated with genes that in their majority are not associated with abiotic stresses (Fig. 1). Rather, the structure and complexity of the network graph suggests that CNGC19 is transcriptionally coupled, in addition to a large number of functionally unknown genes, to functions in disease sensing and resistance, redox/ROS reactions and protein metabolism (Fig. 1). Preliminary data indeed suggest that SOS1 deficiency alters pathogen responses in sos1 mutants (Ali Z, Kim W-Y and Yun D-J, unpublished). In generalizing, integrating transcript behavior in networks (which in the future should be expanded to include other ‘omics’ approaches) can provide clues about crosstalk between different pathways that can lead to new hypotheses. Specifically, that SOS1 includes functions that exceed its primary role as an antiporter in salt stress tolerance reactions has been indicated before.11,12 It is highly likely that the complex structure of the large SOS1 protein is involved in more than one function.
Figure 1.
Transcriptional network associated with ATCN GC19. The transcriptional network records correlation of expression using an algorithm that establishes partial correlation between the expression characteristics of any pair of genes in relation to the correlation of these genes with all other genes in the dataset. The figure shows the network at the highest level of stringency that sorted some 9,000 genes into a network. When probing for correlations of genes with ATCNGC19, the resulting network graph showed the presence of genes in functional categories that are related to disease resistance, protein turnover, auxin metabolism, and redox/ethylene signaling, next to a surprisingly large number of functionally unknown genes. Information about the network and lists of genes can be retrieved by using the command ‘sub(“ATCN GC19”,2)’ in the R-based GGM workspace included in Ma et al.7
Acknowledgements
The work has been supported by the World Class University Program (grant no. R32-10148), and the Biogreen 21 project of the Rural Development Administration (grant no. 20070301034030) of Korea.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11777
References
- 1.Oh DH, Leidi E, Zhang Q, Hwang SM, Li Y, Quintero FJ, et al. Loss of halophytism by interference with SOS1 expression. Plant Physiol. 2009;151:210–222. doi: 10.1104/pp.109.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oh DH, Ali Z, Yun DJ, Bressan RA, Bohnert HJ. SOS1 and halophytism. Plant Signal Behav. 2009;4:1081–1083. doi: 10.4161/psb.4.11.9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oh DH, Lee S-Y, Bressan RA, Yun DJ, Bohnert HJ. Intracellular consequences of SOS1 deficiency during salt stress. J Exp Bot. 2010;61:1201–1213. doi: 10.1093/jxb/erp391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong Z, Koiwa H, Cushman MA, et al. Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol. 2001;126:363–375. doi: 10.1104/pp.126.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong Q, Li P, Ma S, Rupassara SI, Bohnert HJ. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J. 2005;44:826–839. doi: 10.1111/j.1365-313X.2005.02587.x. [DOI] [PubMed] [Google Scholar]
- 6.Gaxiola RA, Li J, Undurraga S, Dang LM, Allen GJ, Alper SL, et al. Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc Natl Acad Sciences USA. 2001;98:11444–11449. doi: 10.1073/pnas.191389398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma S, Bohnert HJ. Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol. 2007;8:49. doi: 10.1186/gb-2007-8-4-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma S, Gong Q, Bohnert HJ. An Arabidopsis gene network based on the graphical Gaussian model. Genome Res. 2007;17:1614–1625. doi: 10.1101/gr.6911207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh DH, Gong Q, Ulanov A, Zhang Q, Li Y, Bressan RA, et al. Sodium stress in the halophyte Thellungiella halophila and transcriptional changes in a thsos1-RNAi line. J Integr Plant Biol. 2007;49:1484–1496. [Google Scholar]
- 10.Kugler A, Köhler B, Palme K, Wolff P, Dietrich P. Salt-dependent regulation of a CNG channel subfamily in Arabidopsis. BMC Plant Biol. 2009;9:140. doi: 10.1186/1471-2229-9-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu JK. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003;6:441–445. doi: 10.1016/s1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- 12.Katiyar-Agarwal S, Zhu J, Kim K, Agarwal M, Fu X, Huang A, et al. The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:18816–18821. doi: 10.1073/pnas.0604711103. [DOI] [PMC free article] [PubMed] [Google Scholar]